Abstract
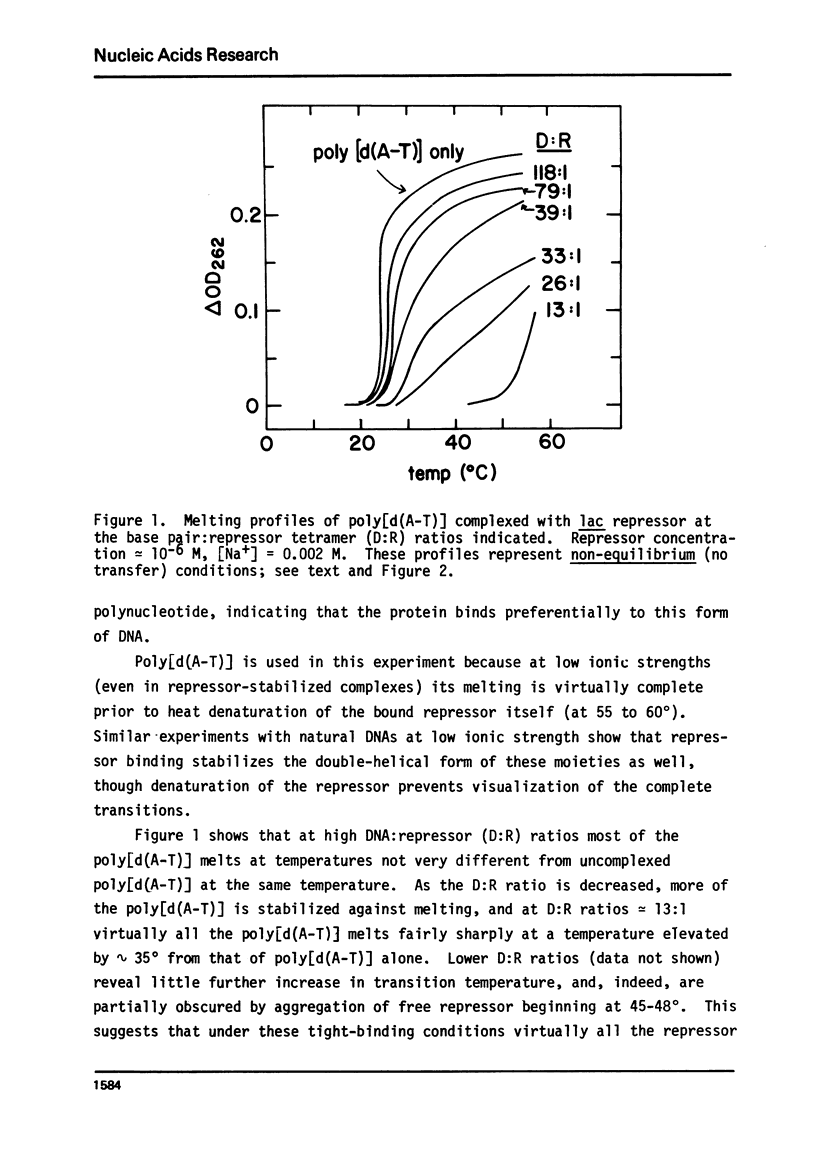
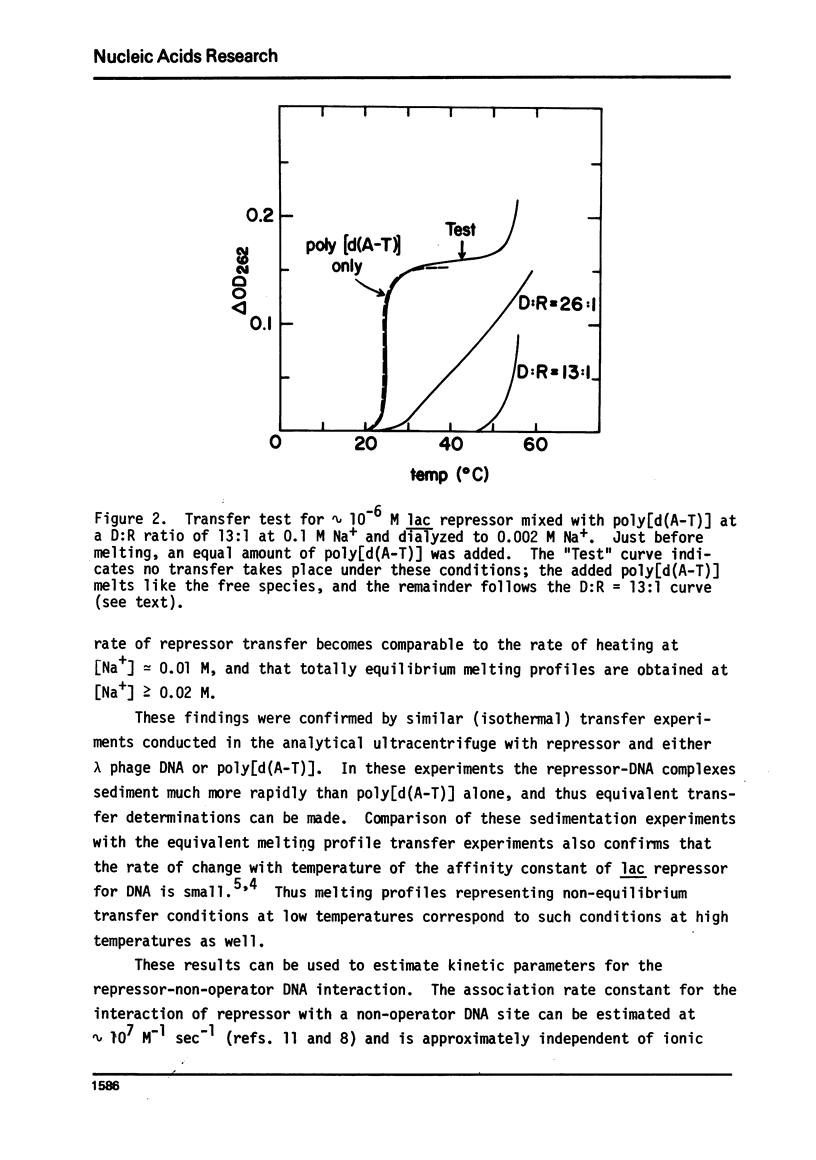
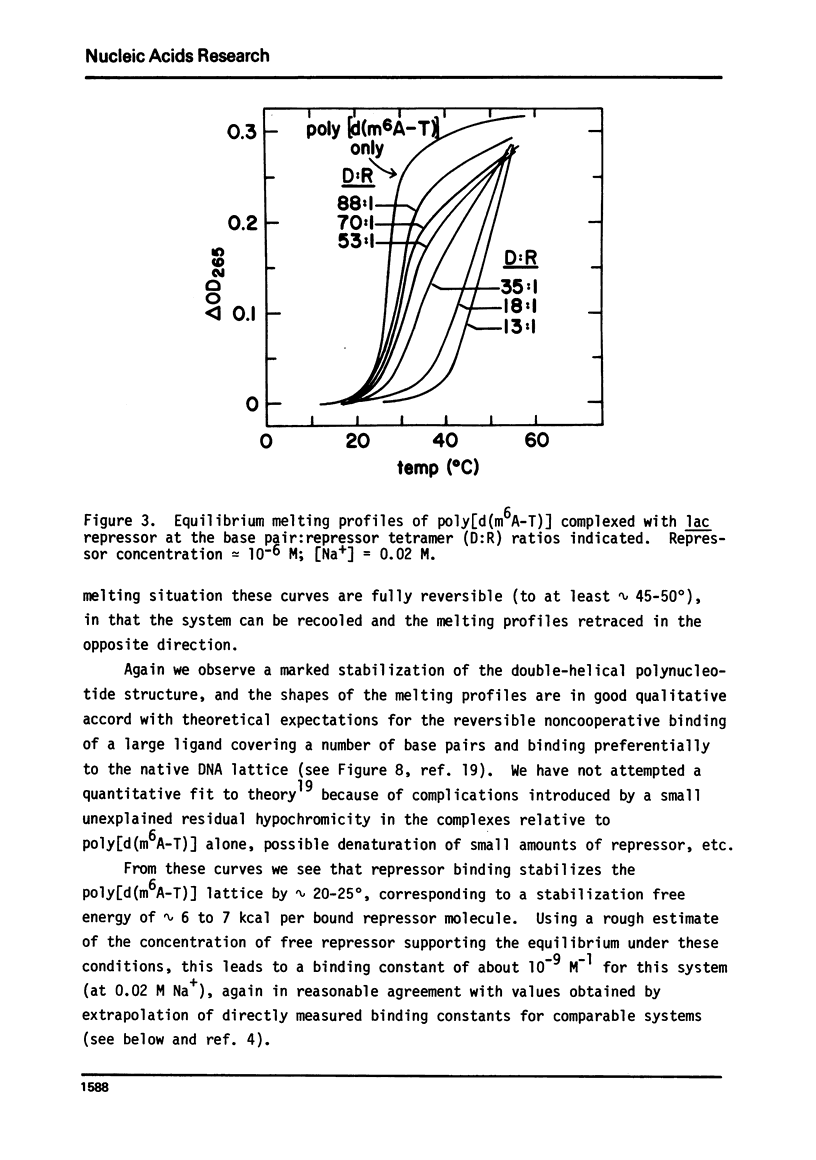
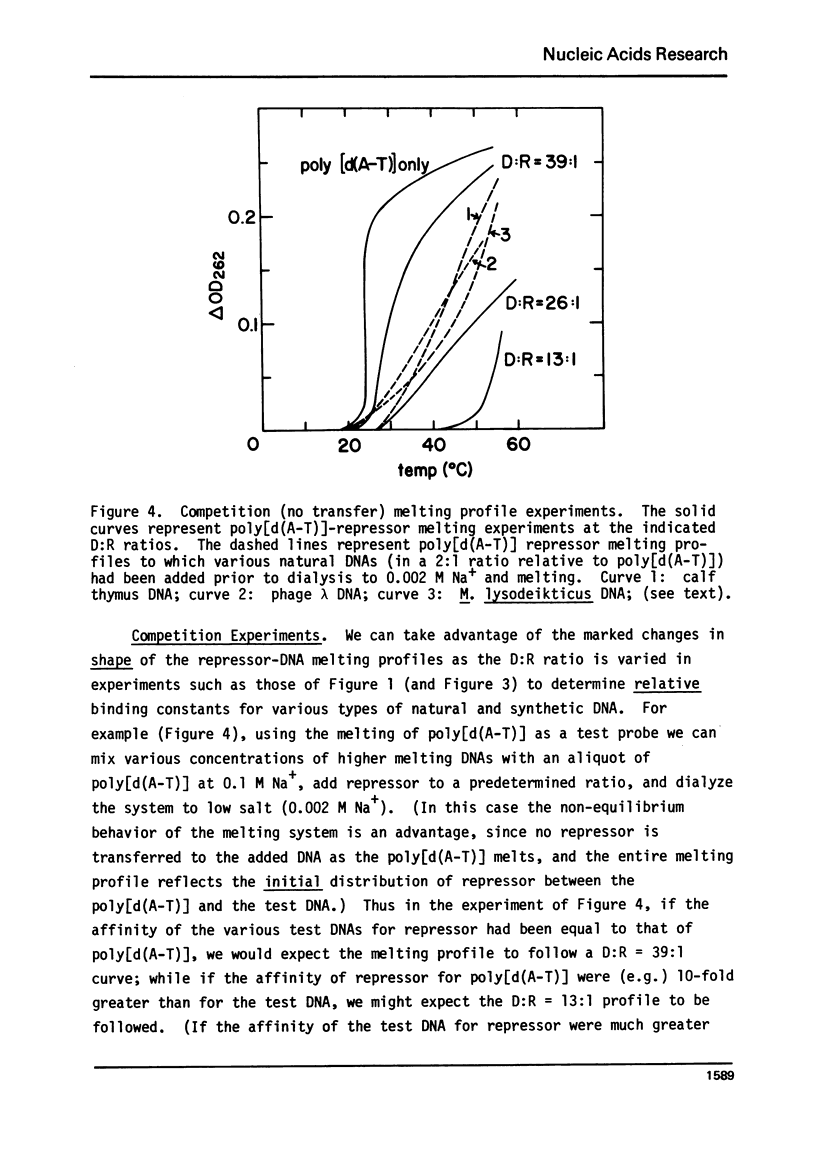
It is shown by melting profile analysis of lac repressor-DNA complexes that repressor binds tightly and preferentially (relative to single-stranded DNA) to double-stranded non-operator DNA. This binding stabilizes the DNA against melting and the repressor against thermal denaturation. Analysis of the extent of stabilization and the rate of dissociation of repressor from non-operator DNA as a function of sodium ion concentration shows, in confirmation of other studies,3,4 that the binding constant (KRD) is very ionic strength dependent; KRD increases from ∼ 106 M−1 at ∼ 0.1 M Na+ to values in excess of 1010 M−1 at 0.002 M Na+. Repressor bound to non-operator DNA is not further stabilized against thermal denaturation by inducer binding, indicating that the inducer and DNA binding sites probably represent separately stabilized local conformations. Transfer melting experiments are used to measure the rate of dissociation of repressor from operator DNA. These experiments show that most of the ionic strength dependence of the binding constant is in the dissociation process; the estimated dissociation rate constant decreases from greater than 10−1 sec−1 at [Na+] ≥ 0.02 M to less than 10−4 sec−1 at [Na+] ≤ 0.002 M. Competition melting experiments are used to show that at 0.02 to 0.002 M Na+ the affinity of lac repressor for various natural DNAs and synthetic double-stranded polynucleotides (including poly[d(m6A-T)], which carries a methyl group in the large groove) are approximately independent of base composition, except that the affinity of repressor for poly[d(A-T)] is ∼ 2- to 3-fold greater than for the other DNAs tested. The affinity for single-stranded polynucleotides is atleast 50-fold less than for the doublehelical forms.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bourgeois S., Pfahl M. Repressors. Adv Protein Chem. 1976;30:1–99. doi: 10.1016/s0065-3233(08)60478-7. [DOI] [PubMed] [Google Scholar]
- Hinkle D. C., Chamberlin M. J. Studies of the binding of Escherichia coli RNA polymerase to DNA. I. The role of sigma subunit in site selection. J Mol Biol. 1972 Sep 28;70(2):157–185. doi: 10.1016/0022-2836(72)90531-1. [DOI] [PubMed] [Google Scholar]
- Kolchinsky A. M., Mirzabekov A. D., Gilbert W., Li L. Preferential protection of the minor groove of non-operator DNA by lac repressor against methylation by dimethyl sulphate. Nucleic Acids Res. 1976 Jan;3(1):11–18. doi: 10.1093/nar/3.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiken S. L., Gross C. A., Von Hippel P. H. Equilibrium and kinetic studies of Escherichia coli lac repressor-inducer interactions. J Mol Biol. 1972 Apr 28;66(1):143–155. doi: 10.1016/s0022-2836(72)80012-3. [DOI] [PubMed] [Google Scholar]
- Lin S. Y., Riggs A. D. Lac repressor binding to non-operator DNA: detailed studies and a comparison of eequilibrium and rate competition methods. J Mol Biol. 1972 Dec 30;72(3):671–690. doi: 10.1016/0022-2836(72)90184-2. [DOI] [PubMed] [Google Scholar]
- Lin S., Riggs A. D. The general affinity of lac repressor for E. coli DNA: implications for gene regulation in procaryotes and eucaryotes. Cell. 1975 Feb;4(2):107–111. doi: 10.1016/0092-8674(75)90116-6. [DOI] [PubMed] [Google Scholar]
- McGhee J. D. Theoretical calculations of the helix-coil transition of DNA in the presence of large, cooperatively binding ligands. Biopolymers. 1976 Jul;15(7):1345–1375. doi: 10.1002/bip.1976.360150710. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., von Hippel P. H. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974 Jun 25;86(2):469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- Olins D. E., Olins A. L., Von Hippel P. H. Model nucleoprotein complexes: studies on the interaction of cationic homopolypeptides with DNA. J Mol Biol. 1967 Mar 14;24(2):157–176. doi: 10.1016/0022-2836(67)90324-5. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Lohman M. L., De Haseth P. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976 Oct 25;107(2):145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Steitz T. A. Protein-DNA interaction investigated by binding Escherichia coli lac repressor protein to poly(d(A-U-HgX)). J Mol Biol. 1976 May 5;103(1):25–38. doi: 10.1016/0022-2836(76)90050-4. [DOI] [PubMed] [Google Scholar]
- Richter P. H., Eigen M. Diffusion controlled reaction rates in spheroidal geometry. Application to repressor--operator association and membrane bound enzymes. Biophys Chem. 1974 Oct;2(3):255–263. doi: 10.1016/0301-4622(74)80050-5. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970 Nov 14;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Revzin A., Gross C. A., Wang A. C. Non-specific DNA binding of genome regulating proteins as a biological control mechanism: I. The lac operon: equilibrium aspects. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4808–4812. doi: 10.1073/pnas.71.12.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]



