Abstract
Problem
The fetal inflammatory response syndrome (FIRS) is considered the counterpart of the systemic inflammatory response syndrome (SIRS), but similarities in their regulatory mechanisms are unclear. This study characterizes the fetal mRNA transcriptome of peripheral leukocytes to identify key biological processes and pathways involved in FIRS.
Method of Study
Umbilical cord blood from preterm neonates with FIRS (funisitis, plasma IL-6>11 pg/ml; n=10) and neonates with no evidence of inflammation (n=10) was collected at birth.
Results
Microarray analysis of leukocyte RNA revealed differential expression of 541 unique genes, changes confirmed by qRT-PCR for 41 or of 44 genes tested. Similar to SIRS and sepsis, ontological and pathway analyses yielded significant enrichment of biological processes including antigen processing and presentation, immune response, and processes critical to cellular metabolism. Results are comparable with microarray studies of endotoxin challenge models and pediatric sepsis, identifying 25 genes across all studies.
Conclusions
This study is the first to profile genome-wide expression in FIRS, which demonstrates a substantial degree of similarity with SIRS despite differences in fetal and adult immune systems.
Keywords: prematurity, preterm birth, chorioamnionitis, FIRS, microarray, transcriptomics
INTRODUCTION
The fetal inflammatory response syndrome (FIRS), considered to be the fetal counterpart of systemic inflammatory response syndrome (SIRS), is frequently present in neonates delivered as a result of spontaneous preterm labor.1;2 Intrauterine infection is one of the most important mechanisms of disease in preterm birth for which a causal association with prematurity has been recognized.3–8 Intra-amniotic infection and/or inflammation is present in approximately one third of the patients with spontaneous preterm labor9–11 and it is associated with FIRS and fetal injury.12–21 Indeed, FIRS is an independent risk factor for perinatal morbidity/mortality2;22 and has also been associated with infection-related neonatal complications,23 bronchopulmonary dysplasia,2;12;24–26 and impaired neurological outcomes27 including cerebral palsy.13;28–35 Moreover, fetal microbial invasion or other insults may result in a systemic fetal inflammatory response36 that can progress toward multiple organ dysfunction, including the hematopoietic system,37–39 the adrenals,40 heart,41;42 kidneys,43 thymus44;45 and skin.46
Currently, FIRS is defined by an elevated umbilical cord plasma interleukin (IL)-6 concentration2 and/or the presence of funisitis.23;47;48 Another approach to diagnose FIRS is to measure C-reactive protein concentration in umbilical cord blood, which has been shown to be elevated in patients with funisitis and congenital neonatal sepsis.49 Alterations in the abundance of IL-6 as well as additional serum cytokines have better characterized SIRS and sepsis50–52 but leukocyte biology in both FIRS and SIRS is not completely understood.
The fetal immune system develops and matures over the course of gestation. Fetal white blood cell counts change with gestational age with lymphocytes being the most prevalent leukocyte through approximately 37 weeks.53 Whereas lymphocytes and monocytes increase linearly with gestational age, neutrophils increase exponentially after 31 weeks of gestation to become the predominant leukocyte at term. In addition, the majority of cord blood lymphocytes are CD45RA+ (naive T lymphocytes) whereas adult lymphocytes are primarily CD45RO+.54;55 These variations may be just one factor contributing to the altered immune status of neonates versus adults.
Differences beyond white blood cell counts and cellular subsets have also been described for fetal, neonatal, and adult immune systems. Functional differences in migration, phagocytosis, oxidative burst, and cytokine production have been identified using ex vivo assays of leukocyte function. For example, migration of un-stimulated neonatal granulocytes is higher than that of adults but leukotriene B4 stimulation leads to greater migration by adult neutrophils.56 Phagocytosis of opsonized Escherichia coli was lower in fetal granulocytes and monocytes relative to neonatal granulocytes; cells from both populations demonstrated less activity than those from adults.57 This is in contrast to the increased intracellular oxidative burst observed in unstimulated as well as N-formyl methionyl-leucyl-phenylalanine (fMLF) or Escherichia coli stimulated fetal granulocytes. Cytokine responses to ex vivo stimuli are also altered in neonatal leukocytes with some studies finding similarities in the abundance of cytokine mRNA (IL-1β, IL-6, and IL-8) in stimulated umbilical cord blood mononuclear cells (CBMC) and adult peripheral blood mononuclear cells (PBMC).58 Decreased concentrations of other cytokines (IL-12, IL-15, GM-CSF, and M-CSF) have also been observed in CBMC and found to result from decreased mRNA stability.55 A direct comparison of unstimulated adult and cord blood monocyte gene expression by microarray has demonstrated that at least 20 genes are increased in abundance in adult monocytes whereas 3 are decreased relative to cord blood monocytes.59 Additional differences were observed upon stimulation with LPS. Thus, fetal and neonatal leukocytes are responsive to their environment but that response is sometimes altered, and perhaps immature, relative to what is found in an adult immune system.
With the wealth of knowledge available regarding a “normal” immune response in both neonatal and adult leukocytes, there is a paucity of information regarding the understanding of leukocyte biology in both SIRS and FIRS. Delineation of stereotypic changes in leukocyte gene expression may provide more objective tools for the identification and treatment of these inflammatory response syndromes. This has been the focus of genome-wide expression profiling studies in an effort to characterize leukocyte changes both in patients with SIRS/sepsis51;60–62 and by studying the effects of in vivo endotoxin challenge in adult human volunteers.52;63 However, the degree of similarity of molecular mechanisms contributing to FIRS and SIRS is unknown. Thus, we pursued mRNA profiling in umbilical cord blood from neonates with documented FIRS to begin addressing this question.
MATERIALS AND METHODS
Sample Collection and Study Subjects
This retrospective study included preterm neonates with (n=10) and without (n=10) FIRS. Blood collection for RNA, plasma, and serum isolation was obtained from the umbilical vein prior to placenta detachment. Mothers of neonates provided written informed consent for the collection of biological materials and clinical data under protocols approved by the institutional review boards of Wayne State University (Detroit, MI), Sotero del Rio Hospital (Santiago, Chile), and the National Institute of Child Health and Human Development of the National Institutes of Health (NIH/DHHS; Bethesda, MD). All neonates were born to mothers experiencing spontaneous preterm labor with intact membranes at the time of enrollment. The presence or absence of FIRS was determined by pathologic analysis of the umbilical cord for the presence of neutrophils in the wall of the umbilical cord vessels and/or Wharton’s jelly48 and measurement of IL-6 in cord plasma by high sensitivity ELISA (R&D Systems; Minneapolis, MN). Neonates were included in the FIRS group if the umbilical cord plasma IL-6 concentrations were ≥11pg/ml2 and funisitis was observed in the umbilical cord.48
Multiplex Analysis of Cord Serum Cytokines
Blood collected into evacuated tubes (BD Vacutainer; BD Diagnostics; Franklin Lakes, NJ) was allowed to clot for 30 minutes at room temperature. Tubes were centrifuged at 1300×g for 10 minutes and serum transferred to cryovials for storage at −70°C until the time of assay. Cord serum levels of IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12p70, IL-13, interferon (IFN)-γ, granulocyte-macrophage colony stimulating factor (GM-CSF), and TNF-α were assessed with a high sensitivity human cytokine kit (LINCO Research; St. Charles, MO). The assay was performed in duplicate with 50μl of serum. Additional cytokines were measured using the Beadlyte Human Multi-Cytokine Detection System 7 (CCL22, sFasL, G-CSF, GROα, IFNα2a, IL-16, IL-1Ra, sIL-2Rα, IL-9, MCP-3, MIP-1β, TNFβ) and multiplex beads for eotaxin, IP-10, MCP-1, MIP-1α; all were purchased from Upstate USA (Charlottesville, VA). Prior to assay, serum samples were diluted 1:2 with serum diluent (Upstate USA; Charlottesville, VA). All assays were performed according to manufacturer’s recommendations. Briefly, 50 μl of prepared standards, whole serum, or diluted serum were incubated with antibody-coated beads in a 96-well filter plate at 4°C on a plate shaker overnight. Wells were vacuum-washed with wash buffer followed by incubation with biotinylated detection antibody at room temperature with agitation for 1 hour (Linco kit) or 1.5 hour (Upstate kit). This was followed by 30 minute incubation with phycoerythrin-conjugated streptavidin at room temperature with agitation. Finally, samples were resuspended in sheath fluid (Luminex Corporation, Austin, TX) containing 0.5% phosphate buffered formalin (Fisher Scientific, Pittsburg, PA) prior to data acquisition with a Luminex 100 analyzer (Luminex Corporation, Austin, TX).
Statistical analysis was performed when cytokine levels were above the limit of detection in at least 16 of 20 samples. Serum cytokine data were analyzed using a Wilcoxon test. Spearman correlations were calculated for umbilical cord blood IL-6 concentrations obtained using the conventional plate ELISA and data obtained from microparticle multiplex assays utilizing the statistical program SPSS version 14.0 (SPSS Inc., Chicago, IL). Differences were considered significant at P<0.05.
RNA Preparation
Umbilical vein cord blood was collected prior to placenta detachment into PAXgene blood RNA tubes (PreAnalytiX GmbH, distributed by Becton and Dickinson Company; Franklin Lakes, NJ) from all preterm neonates. Blood tubes were maintained at room temperature for 24 hours and then frozen at −70°C until further processing. RNA was isolated using the PAXgene Blood RNA kit (Qiagen; Valencia, CA) which includes an on-column DNase I treatment for digestion of contaminating genomic DNA. Quantity and quality of isolated RNA was assessed by UV spectrophotometry (NanoDrop Technologies; Wilmington, DE) and Agilent Bioanalyzer RNA Nano-Chip (Agilent Technologies, Inc.; Santa Clara, CA), respectively.
Microarray Analysis
Whole genome expression analysis was performed on leukocyte RNA from 10 neonates with FIRS and 7 neonates without systemic inflammation. Limited RNA was available for 3 additional neonates without systemic inflammation. These samples were preserved for inclusion in subsequent confirmation experiments. Prior to microarray analysis, 3μg total RNA was globin reduced using GLOBINclear Whole Blood Globin Reduction Kit (Ambion; Austin, TX) according to manufacturer’s instructions. In addition to capture oligos targeting alpha and beta globin, a gamma (fetal) globin oligo was generated by Ambion (Austin, TX) and included in the capture oligo mix. Confirmation of globin reduction was performed in triplicate on select samples using TaqMan inventoried gene expression assays (Applied Biosystems; Foster City, CA) for alpha, beta, and gamma globin.
Complementary RNA (cRNA) synthesis, microarray hybridization, and chip imaging were performed at the Applied Genomics Technology Center of Wayne State University (www.agtc.wayne.edu; Detroit, MI). Globin reduced RNA (500ng) was amplified and biotin-labeled with the Illumina TotalPrep RNA Amplification kit (Ambion; Austin, TX). Labeled cRNA were hybridized to Sentrix-Human 6 Expression BeadChips (Illumina, Inc.; San Diego, CA). BeadChips were imaged using a BeadArray Reader (Illumina, Inc.; San Diego, CA). Initial array data processing, including background correction and intensity averaging across multiple beads for each gene, was performed using BeadStudio Software (Illumina, Inc.; San Diego, CA). Average intensity values underwent a log-transformation followed by quantile normalization. Differential expression was inferred using a moderated two-sample t-test64 with false discovery rate (FDR) adjustment of P-values.65 Differences were considered significant at P<0.05. All microarray data preprocessing and analysis was performed using the correspondent R (www.r-project.org) and Bioconductor (www.bioconductor.org) software packages.
The list of differentially expressed genes was further analyzed for enrichment of ontological clusters using Onto-Express,66 available within the Onto-Tools software suite.67 Enriched biological processes, molecular functions, and cellular components were identified using an over-representation analysis, which tests if the number of genes directly associated to a particular gene ontology term was significantly higher than the number expected by random chance. Pathway analysis was performed using a similar over-representation analysis on the metabolic and signaling pathway collection from MetaCore database (GeneGo Inc.; St. Joseph, MI). Another, conceptually different, pathway analysis method, the impact analysis, was performed on the signaling pathways from KEGG (http://www.genome.jp/kegg/). In addition to counting how many differentially expressed genes are involved in a particular pathway, this analysis also takes into account the position of the genes in the pathways, their measured fold changes, as well as the interactions among genes as described by the topology of each pathway. This “impact analysis” was performed using Pathway Express68 available within the OntoTools software collection at (http://vortex.cs.wayne.edu/projects.htm).
Quantitative Real Time RT-PCR (qRT-PCR)
Differences in mRNA abundance were verified using total leukocyte RNA from the same neonates studied by microarray analysis with 3 additional neonates in the no inflammation group (n=10 for each group). cDNA was created using TaqMan Reverse Transcription Reagents and mRNA abundance was assayed in triplicate with TaqMan inventoried gene expression assays (both from Applied Biosystems; Foster City, CA) (supplemental Table I). Two different approaches were utilized to test differential expression between groups. In the first approach, the three technical replicates for each individual were averaged and differences were assessed using a t-test. Data (Cttarget–Ctref) were also modeled using Generalized Estimating Equations (GEE)69 taking into account that the repeated observations from each individual are clustered (i.e. correlated) and not independent measurements.
Comparative analysis with published microarray studies
Microarray data from two publications were obtained through supplementary information available online (https://www.gluegrant.org/pubsupport/Nature_1/ and http://physiolgenomics.physiology.org/).52;63 Data from a third study was kindly provided by Dr. H.R. Wong.62 For all three studies, differentially expressed probe sets were converted to gene symbols and intersected with the list of differentially expressed genes from the current study. The directions of gene expression changes provided by the authors52;62;63 were also compared for consistency with the results from the current study.
RESULTS
Characteristics of study population
All neonates included were delivered preterm as a result of spontaneous labor. The median gestational age of neonates with FIRS (funisitis positive and plasma IL-6≥11pg/ml) was 30.3 weeks whereas neonates with no evidence of inflammation (funisitis negative and plasma IL-6<11pg/ml) were delivered at the median gestational age of 31.2 weeks (Table I). In both groups, 80% of the mothers received antibiotics prior to delivery whereas 50% received tocolysis in an effort to suppress preterm labor. All mothers received glucocorticoids prior to delivery to aid in fetal lung maturation. Five of the 10 mothers delivering neonates with FIRS underwent amniocentesis; all were diagnosed with intra-amniotic infection. In contrast, negative amniotic fluid culture results were obtained from amniocentesis performed on 7 of 10 mothers delivering preterm neonates with no evidence of fetal systemic inflammation. Among neonates with FIRS, sepsis was diagnosed in 7 cases within 28 days of delivery; one additional case was diagnosed at 42 days of birth. All but two of the neonates survived. Three of the neonates without FIRS developed sepsis within 28 days of delivery; all survived.
Table I.
Clinical characteristics of the study population
| No FIRSa (n=10)b | FIRS (n=10) | P-value | |
|---|---|---|---|
| Median gestational age at delivery, weeks (range)c | 31.2 (26.7–33.1) | 30.3 (25.1–33) | NS |
| Median cord blood IL-6 concentration, pg/ml (range)c | 1.9 (0.5–9.8) | 59.5 (19.7–2155.9) | <0.001 |
| Median steroid administration-to-delivery interval, days (range)c | 3.5 (0–52) | 0 (0–42) | NS |
| Received antibiotics (%)d | 8 (80%) | 8 (80%) | NS |
| Received tocolytics (%)d | 5 (50%) | 5 (50%) | NS |
| Gender (Male/Female)d | 7/3 | 5/5 | NS |
| Amniocentesis performed prior to deliveryd | 7 (70%) | 5 (50%) | NS |
| Positive Amniotic Fluid Cultured | 0 (0%) | 5 (50%) | 0.03 |
| Neonatal sepsis diagnosis within 28 days of deliveryd | 3 (30%) | 7 (70%) | NS |
Fetal Systemic Inflammatory Response Syndrome
Samples from 7 of 10 neonates used for microarray experiment; all samples used for qRT-PCR
Significance tested using Mann-Whitney U test, NS = Not Significant
Significance tested using Fisher’s exact test, NS = Not Significant
Increased pro- and anti-inflammatory cytokines in neonates with FIRS
To evaluate whether increases in both pro- and anti-inflammatory cytokines occur in the immature fetal immune system, multiplex immunoassays were utilized to measure various cytokines (Table II). Serum concentrations of a number of pro-inflammatory cytokines were significantly (P<0.05) increased in cord blood from FIRS neonates in comparison to those without FIRS, including IL-6, IL-8, TNF-α, IP-10 (CXCL10), and MCP-1 (CCL2). Plasma IL-6 concentrations as measured by high sensitivity ELISA were utilized for the initial categorization of samples into FIRS and no FIRS groups. Slightly higher concentrations were observed in serum using the multiplex assay but a significant positive correlation between the results of the two assays was observed (r2=0.798; P<0.001). Anti-inflammatory cytokines such as IL-10, IL-13, and IL-1Ra were all increased in neonates with FIRS, with IL-10 increases observed as significant. Statistical analysis was not performed on IL-13 and IL-1Ra as detectable concentrations of these cytokines were only present in a subset of samples. Several pro-inflammatory factors, including G-CSF, GM-CSF, and IL-1β were also elevated in FIRS but did not meet the criteria for statistical analysis.
Table II.
Multiplex analysis of serum cytokines in FIRS
| Cytokine | No FIRS
|
FIRS
|
P-Value | ||
|---|---|---|---|---|---|
| # samplesa | Median (Range) | # samples | Median (Range) | ||
| IL-1β | 0 | NDb | 5 | 1.18 (0.7–5.77) | ND |
| IL-2 | 0 | ND | 4 | 1.83 (0.34–4.19) | ND |
| IL-4 | 4 | 35.07 (14.58–44.66) | 3 | 7.44 (3.73–25.45) | ND |
| IL-5 | 7 | 0.49 (0.19–1.28) | 6 | 0.44 (0.13–25.15) | ND |
| IL-6 | 10 | 5.01 (1.42–18.73) | 9c | 157.96 (33.17–1954.93) | <0.001 |
| IL-7 | 9 | 0.45 (0.17–2.71) | 10 | 1.32 (0.24–4.62) | NSd |
| IL-8 | 10 | 9.87 (1.73–20.57) | 10 | 68.9 (17.86–759.26) | <0.001 |
| IL-10 | 10 | 2.65 (1.4–34.09) | 10 | 26.61 (12.34–113.89) | 0.003 |
| IL-12p70 | 0 | ND | 1 | 1.75 | ND |
| IL-13 | 1 | 0.59 | 5 | 1.63 (0.59–2.21) | ND |
| IFN-γ | 1 | 0.43 | 2 | 29.01 (9.75–48.26) | ND |
| GM-CSF | 3 | 0.59 (0.45–1.84) | 7 | 1.22 (0.24–11.48) | ND |
| TNF-α | 10 | 11.02 (6.43–21.09) | 10 | 27.77 (8.34–90.29) | 0.008 |
| IL-1Ra | 6 | 49.5 (31–66) | 9 | 473 (47–4719) | ND |
| IL-16 | 10 | 108 (48–216) | 10 | 125 (65–614) | NS |
| sIL-2Ra | 10 | 457.5 (311–594) | 10 | 1130 (370–2993) | 0.019 |
| IFN- α2a | 0 | ND | 2 | 42.8 (36.9–48.7) | ND |
| IP-10 | 10 | 66.4 (41–107.4) | 10 | 147.15 (114–409) | <0.001 |
| Eotaxin | 8 | 48 (35–59) | 9 | 47 (32–70) | NS |
| G-CSF | 0 | ND | 8 | 16.4 (8.2–672.9) | ND |
| GRO | 10 | 818 (534–5298) | 10 | 1937 (304–2903) | NS |
| MCP-1 | 10 | 76.5 (29–182) | 10 | 169.5 (79–940) | 0.019 |
| MDC(CCL22) | 10 | 787 (332–1189) | 10 | 623.5 (194–1010) | NS |
| MIP-1α | 8 | 71 (27–121) | 10 | 85.5 (34–165) | NS |
| MIP-1β | 5 | 16.3 (14.2–39.3) | 7 | 18.9 (14.9–43.7) | ND |
| sFasL | 0 | ND | 2 | 125.5 (70–181) | ND |
Number of samples with cytokine concentrations within the range of the standard curve
ND = Not Determined
IL-6 concentrations for one of the FIRS cases was greater than the upper limit of the assay (this data was not included)
NS = Not Significant by Wilcoxon test
Differential gene expression in FIRS
Previous studies have demonstrated the importance of globin reduction when using RNA collected with the PAXgene system.70;71 In our samples, alpha, beta, and gamma globin RNAs were reduced by greater than 90% using a modified GLOBINclear procedure (supplemental Figure 1). Others have observed that changes in leukocyte abundance contribute to changes in mRNA abundance.72 Thus, we evaluated total white blood cell counts, percent lymphocytes, percent monocytes, and percent neutrophils in all samples where differentials were available. The overall white blood cell count was higher in neonates with FIRS but the difference was not significant (supplemental Table II); percentages of lymphocytes, monocytes, and neutrophils were similar between the two groups. Thus, leukocyte differential was not utilized as a covariate for analysis of microarray data.
Differential abundance of total leukocyte RNA in preterm neonates with and without FIRS was assessed using the Illumina microarray platform. Of the greater than 26,000 well annotated probes representing genes on the array, 296 demonstrated increases and 252 were decreased for a total of 548 significantly differentially expressed probes (P<0.05, Figure 1). These corresponded to 541 unique leukocyte genes differentially expressed in FIRS relative to no inflammation. Principal component analysis performed using expression data from all annotated probes (~26,000) showed a clear distinction between the cluster of no inflammation (No FIRS) samples and the cluster of FIRS samples (Figure 2). Ontological analysis of the 541 differentially expressed genes with Onto-Express/Onto-Tools demonstrated significant enrichment of 27 biological processes (Table III) and 23 molecular functions (Table IV). Key enriched biological processes included antigen processing and presentation, anti-apoptosis, immune and inflammatory responses as well as processes critical to cellular metabolism. A number of enriched molecular functions were related to signal transduction and transferase activity. Metabolic and signaling pathways significantly associated with gene expression data were also identified. Out of the nearly 50 signaling pathways annotated by KEGG, the “impact analysis” identified 5 pathways as significant. These included pathways for antigen processing and presentation (P≤0.0001), phosphatidylinositol signaling system (P≤0.0001), type I diabetes mellitus (P=0.0014), cell adhesion molecules (P=0.0014), and B cell receptor signaling (P=0.0017). Similar pathways were included in the more than one hundred of the in-house annotated pathways from the MetaCore database that were significantly enriched in the set of differentially expressed FIRS genes. Additional pathways such as transcription regulation of granulocyte development, glycolysis and gluconeogenesis, as well as those important to apoptosis (anti-apoptotic TNFs/NF-κB/IAP, apoptotic TNF-family, p53 independent apoptotic signaling, and anti-apoptotic TNFs/NF-κB/Bcl-2 pathways) were identified using MetaCore.
Figure 1. Genes differentially expressed in FIRS.
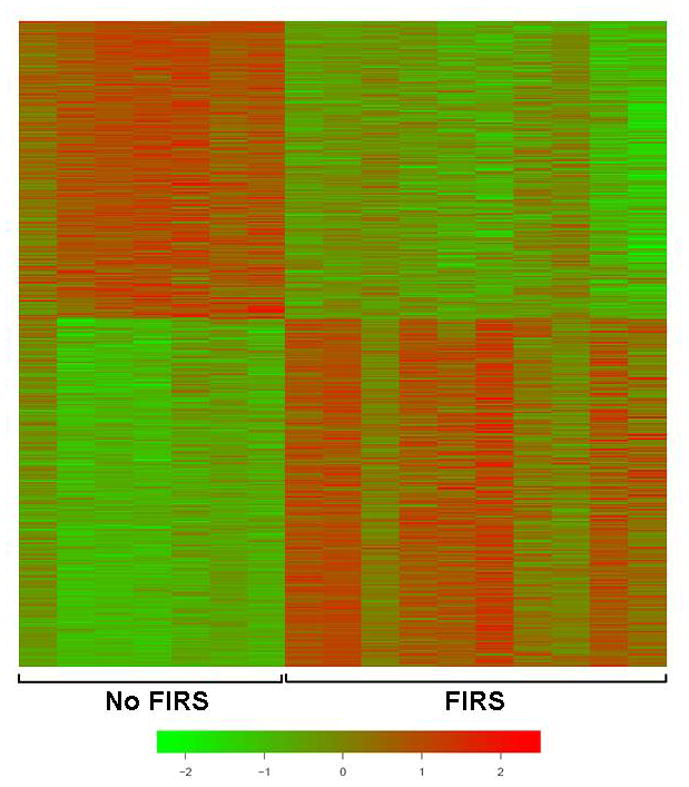
Cord blood RNA from preterm neonates with (n=10) and without (n=7) FIRS was analyzed using the Illumina microarray platform. The color scale represents normalized log2 gene expression levels; data are 0 centered by rows (genes) and sorted as a function of the t-scores between the two groups. Displayed are the 296 genes that were significantly increased in leukocytes from neonates with FIRS and the 252 that were decreased (False discovery rate <0.05).
Figure 2. Principal Component Analysis of gene expression data.

The intensity of 26,000 well annotated probes on the Illumina arrays were used to compute the main directions of variability within the data (principal components). This unsupervised analysis demonstrates that gene expression values can be used to delineate the two groups of samples. More details on this representation can be found elsewhere (Machine Learning and Its Applications to Biology, Tarca AL, Carey VJ, Chen Xw, Romero R, Draghici S, PLoS Comput Biol, 3(6): e116 doi:10.1371/journal.pcbi.0030116.)
Table III.
Twenty-seven biological processes were significantly enriched for within the list of differentially expressed genes
| GO ID | Biological Process | # of Genes on Array | # of Genes Differentially Expressed | P-value |
|---|---|---|---|---|
| GO:0006955 | immune response | 255 | 26 | 0.0000 |
| GO:0019884 | antigen presentation, exogenous antigen | 15 | 7 | 0.0000 |
| GO:0019886 | antigen processing, exogenous antigen via MHC class II | 16 | 7 | 0.0000 |
| GO:0007166 | cell surface receptor linked signal transduction | 129 | 17 | 0.0000 |
| GO:0009596 | detection of pest, pathogen or parasite | 5 | 3 | 0.0000 |
| GO:0006096 | glycolysis | 31 | 6 | 0.0001 |
| GO:0019735 | antimicrobial humoral response (sensu Vertebrata) | 73 | 9 | 0.0001 |
| GO:0006959 | humoral immune response | 26 | 5 | 0.0002 |
| GO:0006916 | anti-apoptosis | 102 | 10 | 0.0002 |
| GO:0005975 | carbohydrate metabolism | 185 | 14 | 0.0002 |
| GO:0042981 | regulation of apoptosis | 72 | 8 | 0.0003 |
| GO:0007243 | protein kinase cascade | 51 | 6 | 0.0011 |
| GO:0006954 | inflammatory response | 170 | 12 | 0.0011 |
| GO:0006952 | defense response | 70 | 7 | 0.0012 |
| GO:0008654 | phospholipid biosynthesis | 18 | 3 | 0.0033 |
| GO:0006633 | fatty acid biosynthesis | 31 | 4 | 0.0033 |
| GO:0016192 | vesicle-mediated transport | 50 | 5 | 0.0047 |
| GO:0015986 | ATP synthesis coupled proton transport | 35 | 4 | 0.0053 |
| GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB cascade | 56 | 5 | 0.0077 |
| GO:0006935 | chemotaxis | 99 | 7 | 0.0086 |
| GO:0006461 | protein complex assembly | 108 | 7 | 0.0141 |
| GO:0006928 | cell motility | 111 | 7 | 0.0159 |
| GO:0007242 | intracellular signaling cascade | 285 | 13 | 0.0252 |
| GO:0006468 | protein amino acid phosphorylation | 413 | 17 | 0.0287 |
| GO:0006886 | intracellular protein transport | 135 | 7 | 0.0432 |
| GO:0008152 | metabolism | 281 | 12 | 0.0447 |
| GO:0006457 | protein folding | 165 | 8 | 0.0447 |
Table IV.
Twenty-three molecular functions were significantly enriched for within the list of differentially expressed genes
| GO ID | Molecular Function | # of Genes on Array | # of Genes Differentially Expressed | P-value |
|---|---|---|---|---|
| GO:0045012 | MHC class II receptor activity | 15 | 7 | 0.0000 |
| GO:0004888 | transmembrane receptor activity | 80 | 11 | 0.0000 |
| GO:0005515 | protein binding | 2206 | 83 | 0.0001 |
| GO:0016798 | hydrolase activity, acting on glycosyl bonds | 43 | 7 | 0.0001 |
| GO:0005031 | tumor necrosis factor receptor activity | 7 | 3 | 0.0002 |
| GO:0016853 | isomerase activity | 81 | 9 | 0.0003 |
| GO:0045028 | purinergic nucleotide receptor activity, G-protein coupled | 18 | 4 | 0.0006 |
| GO:0005057 | receptor signaling protein activity | 34 | 5 | 0.0016 |
| GO:0008415 | acyltransferase activity | 88 | 8 | 0.0029 |
| GO:0004298 | threonine endopeptidase activity | 17 | 3 | 0.0044 |
| GO:0016740 | transferase activity | 838 | 35 | 0.0046 |
| GO:0046872 | metal ion binding | 1438 | 52 | 0.0059 |
| GO:0004713 | protein-tyrosine kinase activity | 107 | 8 | 0.0072 |
| GO:0004871 | signal transducer activity | 228 | 13 | 0.0078 |
| GO:0005529 | sugar binding | 110 | 8 | 0.0085 |
| GO:0008138 | protein tyrosine/serine/threonine phosphatase activity | 21 | 3 | 0.0095 |
| GO:0008270 | zinc ion binding | 1405 | 50 | 0.0096 |
| GO:0016491 | oxidoreductase activity | 344 | 16 | 0.0147 |
| GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | 28 | 3 | 0.0171 |
| GO:0004497 | monooxygenase activity | 51 | 4 | 0.0249 |
| GO:0020037 | heme binding | 78 | 5 | 0.0340 |
| GO:0016787 | hydrolase activity | 622 | 23 | 0.0398 |
| GO:0005506 | iron ion binding | 192 | 9 | 0.0479 |
Validation of differential expression by quantitative real time RT-PCR
Forty-four genes were chosen for validation by qRT-PCR. Gene selection was based on rank within the list of significantly differentially expressed genes (1=most significant) and inclusion in at least one of the significantly enriched ontological clusters. Forty-one of the 44 genes tested by qRT-PCR were significantly different in neonates with FIRS relative to those without systemic inflammation by both statistical analysis methods (GEE and t-test; P<0.05) (Figure 3 and Table V). The correlation between log-fold changes obtained with microarray and qRT-PCR was high (92.5%) while the regression slope was 0.52 indicating the expected compression of the expression range with microarray technology.73 Validation of inflammation and immune response genes in umbilical cord blood leukocytes exhibited changes often seen in the presence of systemic and chronic inflammation (Figure 3A). Calgranulins B and C (S100A9 and S100A12) were both significantly up-regulated in neonates with FIRS. Increases in arachidonate 5-lipoxygenase-activating protein (ALOX5AP) and leukotriene B4 receptor (LTB4R) were also found. Similar to previous observations in endotoxin challenge models of sepsis,51;52 mRNA abundance for IL-1 receptor antagonist (IL1RN, also known as IL-1Ra) was significantly higher in FIRS and coincides with increases in serum IL-1Ra (Table II). The observed decrease in IL-16 was the only gene that demonstrated significant differences by GEE analysis alone (GEE P=0.0448; t-test P=0.0806) and these changes were not confirmed at the serum protein concentration.
Figure 3. Confirmation of differential gene expression by qRT-PCR.
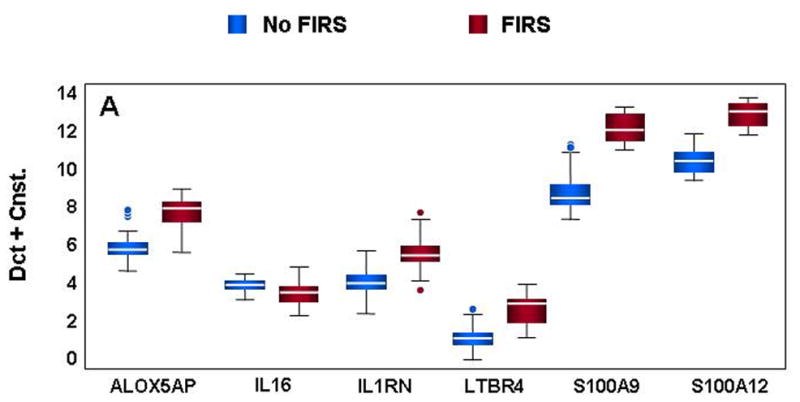
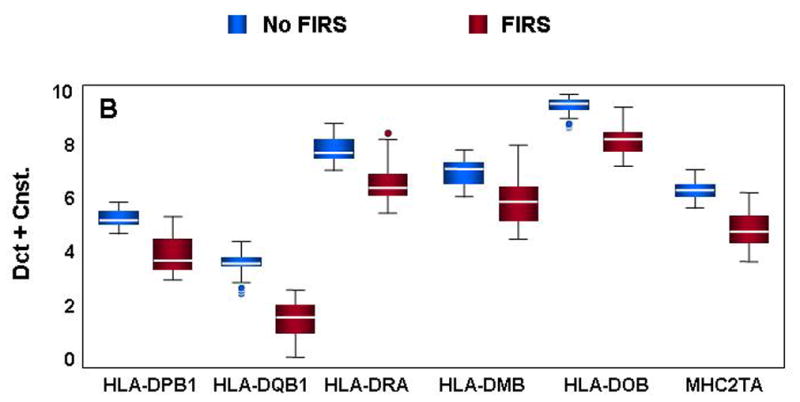
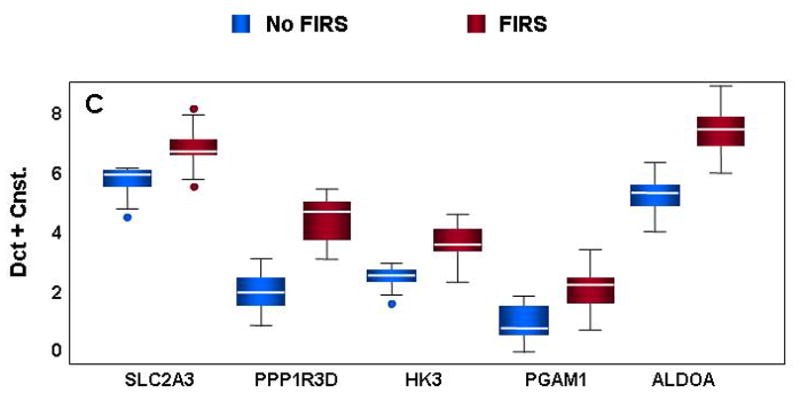
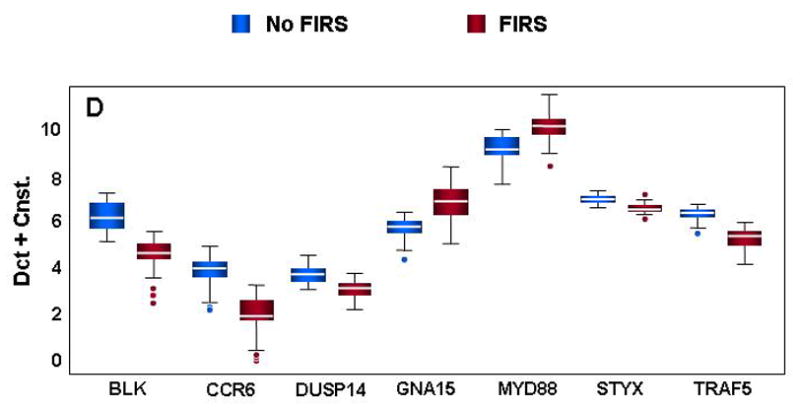
mRNA abundance was assessed in cord blood RNA from neonates with and without FIRS (n=10/group). Altered abundance of genes within ontological categories of immune response and inflammation (A), MHC II receptor activity (B), carbohydrate metabolism (C) and signal transduction (D) was confirmed using qRT-PCR. Box-plots include 50% of the data with the middle line showing the median value; P<0.05 for all genes shown when modeled using GEE. Depending on the statistical method used, 93% to 95% of the genes tested confirmed the change observed by microarray analysis.
Table V.
Additional genes tested by quantitative real time RT-PCR
| Symbol | Gene Name | Microarray Rank | Fold-Difference Microarraya | Fold-Difference qRT-PCR | P-value t-testb | P-value GEE | Direction in FIRS |
|---|---|---|---|---|---|---|---|
| BIRC3 | baculoviral IAP repeat containing 3 | 16 | 1.865 | 2.390 | 0.0000 | 0.0000 | ↓ |
| SERPINB2 | Plasminogen activator inhibitor-2 (serpin peptidase inhibitor, clade B) | 21 | 1.341 | 6.407 | 0.0000 | >0.0001 | ↑ |
| SEMA4D | semaphorin 4D (CD100) | 52 | 1.348 | 1.174 | 0.4779 | 0.4443 | ↓ |
| FAIM3 | Fas apoptosis inhibitory molecule 3 | 81 | 2.120 | 2.682 | 0.0000 | 0.0000 | ↓ |
| BIRC5 | baculoviral IAP repeat containing 5 | 399 | 1.412 | 3.245 | 0.0000 | 0.0000 | ↑ |
| PRKCZ | protein kinase c, zeta | 518 | 1.150 | 1.576 | 0.0031 | 0.0003 | ↓ |
| MS4A1 | CD20 | 65 | 1.769 | 3.455 | 0.0003 | 0.0000 | ↓ |
| CD79b | B-cell specific Ig beta | 9 | 2.012 | 3.177 | 0.0001 | 0.0000 | ↓ |
| CST7 | Cystatin F (leukostatin) | 36 | 3.615 | 6.004 | 0.0000 | 0.0000 | ↑ |
| PLA2G4B | phospholipase A2, group IVB | 70 | 1.541 | 1.574 | 0.0021 | 0.0001 | ↓ |
| ALDH3B1 | aldehyde dehydrogenase 3 family member B1 | 493 | 1.398 | 2.151 | 0.0042 | 0.0004 | ↑ |
| ACSL5 | acyl-CoA synthetase long-chain family member 5 | 527 | 1.216 | 1.055 | 0.6481 | 0.6241 | ↓ |
| ST3GAL5 | ST3 beta galactoside alpha-2,3-sialyltransferase 5 | 486 | 1.237 | 1.397 | 0.0121 | 0.0031 | ↓ |
| TLR10 | toll-like receptor 10 | 39 | 1.258 | 2.903 | 0.0005 | 0.0000 | ↓ |
| LDLR | low density lipoprotein receptor | 68 | 1.629 | 2.198 | 0.0041 | 0.0001 | ↑ |
| CD72 | CD72 | 87 | 1.458 | 2.933 | 0.0000 | 0.0000 | ↓ |
| CRLS1 | cardiolipin synthase 1 | 529 | 1.439 | 1.716 | 0.0006 | 0.0000 | ↑ |
| CD19 | CD19 | 5 | 2.153 | 3.721 | 0.0000 | 0.0000 | ↓ |
| PLCG1 | phospholipase C, gamma 1 | 520 | 1.801 | 2.151 | 0.0020 | 0.0001 | ↓ |
| PASK | PAS domain containing ser/thr kinase | 478 | 1.552 | 1.667 | 0.0016 | 0.0001 | ↓ |
All genes were significantly different at P ≤0.05 by microarray analysis
qRT-PCR confirmation of gene expression changes were statistically analyzed by both GEE and t-test
A number of genes important to the MHC type II processing pathway were also significantly decreased in preterm neonates with FIRS (Figure 3B). Critical components of MHC antigen presentation were found at lower levels in neonates with FIRS (HLA-DPB1, HLA-DQB1, and HLA-DRA). In addition, antigen processing was putatively altered as indicated by a decrease in HLA-DMB, an important component of MHC peptide loading. Interestingly, an inhibitor of HLA-DM activity (HLA-DOB) was also lower in neonates with FIRS. The down-regulation of various HLA genes could directly result from decreases in MHC2TA (CIITA), a positive regulator of MHC II gene transcription.
Genes important to carbohydrate metabolism including glucose transporter 3 (SLC2A3), protein phosphatase 1 regulatory subunit 3D (PPP1R3D), hexokinase (HK3), phosphoglycerate mutase 1 (PGAM1), and fructose bisphosphate-aldolase (ALDOA) were also significantly up-regulated in umbilical cord blood leukocytes from neonates with FIRS (Figure 3C). In contrast, genes mediating signal transduction and intracellular transferase activity were decreased in FIRS (Figure 3D). Decreases in CCR6, TRAF5, and B lymphoid tyrosine kinase (BLK) may play a role in suppressing a lymphocyte response in conjunction with the observed decreases in antigen processing and presentation as well as IL-16 (see Figure 3A). Two additional genes with “protein tyr/ser/thr phosphatase activity” (DUSP14 and STYX) were also down-regulated during FIRS suggesting an overall decrease in de-phosphorylation activity in cord blood leukocytes. Decreased intracellular phosphatase activity may be one mechanism by which cells regulate potential increases in signal transduction elicited by greater MYD88 and g-protein receptor signaling molecule (GNA15) levels observed in this study.
Similarities and differences between FIRS and sepsis or endotoxin challenge
Several studies have focused on genome-wide expression profiling in an effort to characterize leukocyte changes both in patients with SIRS/sepsis62 and by studying the effects of in vivo endotoxin challenge in human volunteers52;63 To evaluate similarities and differences between the data of the current study and that of recently published works, a comparative analysis was performed. The 3776 differentially expressed probe sets detected by Calvano and colleagues63 using the Affymetrix HG-U133 PLUS 2.0 array were converted to gene symbols. An overall direction of change was inferred by the direction of change observed across most of the time point post endotoxin challenge. One hundred ninety-three differentially expressed genes were in common between this endotoxin challenge study and the FIRS results of the current study; 74.1% of the genes were differentially expressed in the same direction (e.g. increased due to endotoxin challenge or FIRS). A second endotoxin challenge model of sepsis identified 201 differentially expressed probe sets observed using the Affymetrix HG-U 95Av2 array at the 6 hour time point post challenge.52 Only 35 genes were in common between the two studies but all gene expression changes were in the same direction. The third comparison examined the similarities and differences between the current FIRS study and that of pediatric septic shock.62 All patients included in the study were less than 10 years of age with a median age of 2.4 in the control group, 2.7 in the survivor group, and 1.6 in the non-survivor group.62 A total of 1710 differentially expressed probe sets were observed using the Affymetrix HG-U133 PLUS 2.0 array. Similar to the study by Calvano et al.,63 approximately 35% of the genes changed in FIRS were also changed in pediatric septic shock. All 195 genes were observed to be changed in the same direction in both studies. In total, 25 common genes were observed across all studies (Table VI) with a high degree of consistency in the direction of change observed.
Table VI.
Genes differentially expressed* in FIRS, pediatric sepsis, and endotoxin challenge models of sepsis.
| Gene Symbol | FIRS | Calvano et al., 2005 | Talwar et al., 2006 | Wong et al., 2007 |
|---|---|---|---|---|
| ADM | ↑ | ↑ | ↑ | ↑ |
| ALOX5AP | ↑ | ↑ | ↑ | ↑ |
| ALPL | ↑ | ↓ | ↑ | ↑ |
| ANXA3 | ↑ | ↑ | ↑ | ↑ |
| BAZ1A | ↑ | ↑ | ↑ | ↑ |
| CEACAM1 | ↑ | ↑ | ↑ | ↑ |
| CKAP4 | ↑ | ↑ | ↑ | ↑ |
| EMR1 | ↑ | ↓ | ↑ | ↑ |
| FGR | ↑ | ↓ | ↑ | ↑ |
| FLOT1 | ↑ | ↑ | ↑ | ↑ |
| HLA-DPA1 | ↓ | ↓ | ↓ | ↓ |
| HLA-DPB1 | ↓ | ↓ | ↓ | ↓ |
| HLA-DQB1 | ↓ | ↓ | ↓ | ↓ |
| HP | ↑ | ↑ | ↑ | ↑ |
| IL1RN | ↑ | ↑ | ↑ | ↑ |
| LIMK2 | ↑ | ↑ | ↑ | ↑ |
| MAPK14 | ↑ | ↑ | ↑ | ↑ |
| PGD | ↑ | ↑ | ↑ | ↑ |
| PGLYRP1 | ↑ | ↑ | ↑ | ↑ |
| S100A12 | ↑ | ↑ | ↑ | ↑ |
| SERPINB1 | ↑ | ↑ | ↑ | ↑ |
| SLC2A3 | ↑ | ↑ | ↑ | ↑ |
| SPOCK2 | ↓ | ↓ | ↓ | ↓ |
| UGCG | ↑ | ↑ | ↑ | ↑ |
| UPP1 | ↑ | ↑ | ↑ | ↑ |
Arrows indicate direction of change relative to healthy controls
DISCUSSION
Complexity of systemic inflammatory response syndromes – either in the fetus, neonate, child, or adult – has challenged researchers and clinicians alike. The results of this study are the first to describe global gene expression differences in preterm neonates with FIRS relative to those without a systemic inflammatory response at the time of birth. In addition, a number of the differentially expressed genes in FIRS coincide with those previously described in studies conducted in adults using endotoxin challenge models52;63 as well as in pediatric sepsis.62 Observed increases in umbilical cord serum concentrations of pro- and anti-inflammatory cytokines also echoed changes observed in adult sepsis/SIRS.50–52 Inflammation-associated increases in pro-inflammatory cytokines are a hallmark of adult SIRS and sepsis.74 This pro-inflammatory aspect of sepsis has been the target of a number of potential treatments with little success.75 Potent inflammatory mediators such as TNF-α, IL-6,28 IL-8, and MCP-1 were all increased in neonates with FIRS. These cytokines and others are also increased in amniotic fluid76–92 as well as in adults with SIRS/sepsis50;74 or endotoxin challenge models52 but to a much greater extent. In vitro LPS stimulation studies of monocytes have demonstrated that less TNF-α and IL-6 are produced from umbilical cord blood leukocytes versus adult blood cells.57;93 Therefore, it is not surprising that serum cytokine concentrations in FIRS, although increased, are not as high as found in adults with sepsis/SIRS.
Blood leukocytes both respond and contribute to inflammatory response syndromes. Changes were readily detectable at the gene expression level in neonates with FIRS with a number of genes indicative of innate immune activation. Several factors important to leukocyte adhesion and chemotaxis (LTB4R, ALOX5AP, CD11b, S100A9, S100A12) were up-regulated during FIRS. The leukotriene B4 receptor (LTB4R, also called BLT1) is a high affinity receptor found on neutrophils, monocytes, and some lymphocyte subsets.94 Binding of the receptor with leukotriene B4 or other agonistic ligands results in increased leukocyte chemotaxis, calcium flux, and adhesion.95 The increase observed in FIRS is interesting in light of evidence demonstrating decreased expression of LTB4R in neutrophils and monocytes stimulated with inflammatory mediators such as LPS and TNF-α.94;96 Pettersson and colleagues hypothesized this down regulation to be a mechanism of limiting excessive inflammatory cell accumulation at sites of infection.96 Leukotrienes are further implicated in FIRS, SIRS, and sepsis via modulation of ALOX5AP (also called FLAP), a molecule intimately involved in leukotriene synthesis.97 Not only was ALOX5AP increased in leukocytes from the neonates with FIRS but increases have also been observed in endotoxin challenged adults52;63 and in pediatric sepsis.62 Chemical inhibitors of ALOX5AP result in improved survival in mice subjected to a cecal ligation and puncture model of peritonitis and severe sepsis.98 Thus, aberrant control of leukotriene mediated adhesion and chemotaxis may play a critical role in exacerbating the inflammation of FIRS and SIRS.
Leukocyte contributions and responses are not limited to leukotriene-induced cell adhesion and migration during systemic inflammatory response. Neutrophils contain several antimicrobial peptides, which are small molecular weight proteins with broad spectrum antimicrobial activity against bacteria, viruses, and fungi.99–101 Calgranulins are phagocyte-specific proteins with a variety of functions including antimicrobial activity, enhancement of migration and chemotaxis, as well as up-regulation of adhesion molecules.102 Up-regulation of calgranulin C (S100A12) is consistent across studies of endotoxin challenged adults,52;63 pediatric sepsis,62 and the current study (Figure 3 and Table VI). Protein concentrations of S100A12 are also increased in the amniotic fluid of neonates with funisitis,103 indicating that the fetal inflammatory response is limited to the fetal blood compartment. Increases in calprotectin, a heterodimer of calgranulins A (S100A8, also called MRP8) and B (S100A9, also called MRP14), is also associated with intra-amniotic infection in preterm labor and preterm premature rupture of membranes.99 Thus, the FIRS-associated increases in cord blood leukocyte mRNA for S100A9 and S100A12 are consistent both with adult SIRS/sepsis and inflammation in the amniotic cavity. As a pro-inflammatory mediator, S100A9 has been implicated in induction of CD11b expression and has a role in increased neutrophil adherence.104 Surface CD11b is increased on neutrophils from patients with sepsis and associates with decreased chemotaxis relative to healthy adults.105 In preterm infants, neutrophil surface CD11b is lower than in their term counterparts as well as adults and it does not increase with in vitro stimulation.106 Leukocytes from the preterm neonates of the current study indicate that the level of CD11b is altered in response to environmental changes, possibly including increased S100A9, which is in contrast to the study of leukocytes from preterm versus term infants and adults. These conflicting results may be due to evaluating cells exposed to multiple stimuli in vivo versus individual in vitro treatment. Despite aspects of immaturity and limited in vitro responsiveness of fetal leukocytes, the fetal systemic inflammatory response has many characteristics of that found in adult inflammation.
Fetal systemic inflammation is associated with phenotypic and metabolic changes consistent with activation in fetal immune cells. In a recent study,107 funisitis was associated with a significant increase in the median mean channel brightness (MCB) of CD14, CD64, and CD66b on granulocytes and the MCB of CD64 on monocytes collected from umbilical cord blood. The basal intracellular reactive oxygen species production and oxidative burst were higher in the umbilical cord monocytes of neonates with funisitis than in those without funisitis.107
Systemic inflammatory response syndromes are also characterized by the dysregulation of pro- and anti-inflammatory responses.108 As gauged by the anti-inflammatory associated changes in FIRS, the fetal inflammatory response syndrome exhibits gene expression changes similar to those found in SIRS. One predominant theme identified through ontological and pathway analyses was that antigen processing and presentation by the MHC II pathway was clearly affected in FIRS. Changes in gene expression were observed at all major steps of the pathway. Gamma interferon-inducible lysosomal thiol reductase (GILT/IFI30) was increased in FIRS. This enzyme plays a vital role in reducing disulfide bonds to unfold and degrade proteins in antigen presenting cells (APC).109 While breakdown of endocytosed antigens is potentially increased in APC’s during FIRS, the subsequent steps of antigen processing and presentation appear to be down-regulated. Antigen presentation molecules HLA-DPB1, HLA-DQB1, and HLA-DRA, as well as molecules important to MHC peptide loading such as HLA-DR and CD74 (also called the invariant chain)110 were lower at the mRNA level in cord blood leukocytes when systemic inflammation was present. Observed decreases in the transcriptional regulator of MHC II gene expression MHC2TA may play a key role in HLA mRNA abundance changes. Similar changes have been observed previously at the mRNA and surface protein levels in adult SIRS/sepsis. Changes in HLA also appear to associate with patient survival with substantially lower levels found in non-surviving patients versus those that survive.111;112 A third study demonstrated that differences in HLA were not observed upon initial diagnosis, but levels three to four days post diagnosis demonstrated a correlation with survivors versus non-survivors.113 HLA levels may remain low for several weeks past the initial sepsis diagnosis.114 Interestingly, total HLA was not significantly different in patients with sepsis versus healthy adults but the cellular distribution was drastically altered. A punctate intracellular distribution was observed during sepsis compared to the normal periphery location. IL-10, a cytokine that was increased in the current study, was identified as a contributing factor in HLA-DR redistribution.114 Thus, patients who survive the initial hyper-inflammatory response of SIRS may continue to experience problems due to the compensatory anti-inflammatory response that follows. If it is indeed the case that the compensatory anti-inflammatory response is already present in preterm neonates with FIRS, the vulnerability of newborns to infection or insults may contribute to increased incidence of sepsis within the first 28 days of life such as was observed in neonates of the current study.
The anti-inflammatory limb of the immune response is crucial for dampening intra-amniotic inflammation.115–119 Additional alterations indicate the presence of a compensatory anti-inflammatory response in neonates with FIRS. Fundamental B cell receptor signaling pathway genes were also decreased in umbilical cord blood leukocytes from FIRS neonates. Initiation of this pathway by antigen is potentially altered due to decreases in CD79A and CD79B (Igα and Igβ). These two molecules have a role in B cell activation and are at higher levels in cord blood B cells from healthy term neonates relative to the levels found on adult B cells.120 CD19, a positive B cell receptor regulator involved in signaling for proliferation, was also decreased in leukocytes during FIRS. This is in contrast to increased CD19 surface levels observed by Weinschenk and colleagues in preterm neonates with confirmed sepsis.121 Time and type of sampling (cord blood at delivery versus neonatal blood) may contribute to the appeared difference of the current and previous observations. To further support the hypothesis that B cell receptor signaling is diminished in FIRS neonates, important NF-κB activation molecules were also decreased. These included CARD11 (CARMA1), a mediator of antigen receptor-induced NF-κB activation,122 and IκB kinase (IKBKB) which is also involved in NF-κB activation.123 Decreases in CD22 and CD72 in leukocytes from FIRS neonates may be one mechanism to combat this putative decrease in B cell receptor signaling. CD22 is an inhibitor of B cell receptor signaling124 while CD72 prevents differentiation of naive B cells into plasma cells.125 Thus, although receptor signaling is lowered, there is also less inhibition of any signals that may be initiated. Finally, the anti-inflammatory aspect of FIRS was not limited to changes in mRNA abundance. Increases in serum IL-10 as well as serum and mRNA IL-1Ra observed in this study were also found within the first 10 hours post endotoxin challenge of healthy adults.51 In addition, higher concentrations of IL-10 and IL-1Ra associate with adverse outcomes for adults with sepsis.50;74 High concentrations of cord plasma IL-1Ra in preterm neonates is a risk factor for adverse outcomes and neonatal morbidity.126 Thus, an anti-inflammatory response has a profound impact on neonatal health and appears to be an important component of FIRS.
Genome-wide expression profiling in patients with SIRS/sepsis51;60–62 or in endotoxin challenge models of sepsis52;63 have begun to provide a better understanding of the molecular mechanisms associated with disease. With the many similarities in alterations of both pro- and anti-inflammatory leukocyte genes found in FIRS and studies with adult leukocytes during inflammation, the list of genes found across several reports and those reported herein is 25 genes. A key factor contributing to this disparity may be differences between the fetal and adult immune systems. Other important factors could include the RNA isolation methods and/or the microarray platforms and analysis methods used. For this reason, it would be difficult to say that the 275 genes found specific to this study, when compared to those of Calvano et al.,63 Talwar et al.,52 and Wong et al.,62 are truly specific to fetal inflammatory response syndrome. The high success rate of confirmation lends confidence in the differentially expressed genes of the current study. These results also indicate that the modified globin reduction procedure did not have any deleterious effects on the sample preparation as globin reduced samples were utilized for the microarray experiment only. With a list of targets in hand, confirmation of mRNA changes in independent groups of neonates with FIRS by qRT-PCR will only further substantiate the current observations. In closing, the similarities in the transcriptomes of FIRS and SIRS/sepsis may provide new targets for strategies designed to modulate inflammation despite differences in the maturity and cellular composition of fetal and adult immune systems.
Conclusions
This is the first study to profile genome-wide expression in FIRS, which demonstrates a substantial degree of similarity with SIRS despite differences in fetal and adult immune systems.
Supplementary Material
Globin RNA transcript abundance was assayed in triplicate with TaqMan inventoried gene expression assays (Applied Biosystems) for alpha, beta, and gamma globin. The currently available GlobinClear-Human kit (Ambion) results in >90% depletion of alpha (HBA) and beta (HBB) globin but minimal reduction in gamma globin (HBG, fetal globin) as is demonstrated with the solid bars (representative n=4). With the addition of gamma globin capture oligo, this transcript is also reduced by >90% (hatched bars; representative n=10).
Acknowledgments
The authors wish to thank the nursing staffs of the Perinatology Research Branch, the Detroit Medical Center, and Sotero del Rio Hospital for their contributions to this work. This research was supported (in part) by the Perinatology Research Branch, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH, DHHS, by NSF DBI-0234806, NIH(NCRR) 1S10 RR017857-01, MLSC MEDC-538 and MEDC GR-352, NIH 1R21 CA10074001, 1R21 EB00990-01 and 1R01 NS045207-01.
Reference List
- 1.Romero R, Gomez R, Ghezzi F, Yoon BH, Mazor M, Edwin SS, Berry SM. A fetal systemic inflammatory response is followed by the spontaneous onset of preterm parturition. Am J Obstet Gynecol. 1998;179:186–193. doi: 10.1016/s0002-9378(98)70271-6. [DOI] [PubMed] [Google Scholar]
- 2.Gomez R, Romero R, Ghezzi F, Yoon BH, Mazor M, Berry SM. The fetal inflammatory response syndrome. Am J Obstet Gynecol. 1998;179:194–202. doi: 10.1016/s0002-9378(98)70272-8. [DOI] [PubMed] [Google Scholar]
- 3.Naeye RL, Ross SM. Amniotic fluid infection syndrome. Clin Obstet Gynaecol. 1982;9:593–607. [PubMed] [Google Scholar]
- 4.Minkoff H. Prematurity: infection as an etiologic factor. Obstet Gynecol. 1983;62:137–144. [PubMed] [Google Scholar]
- 5.Romero R, Mazor M. Infection and preterm labor. Clin Obstet Gynecol. 1988;31:553–584. doi: 10.1097/00003081-198809000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Goldenberg RL, Hauth JC, Andrews WW. Intrauterine infection and preterm delivery. N Engl J Med. 2000;342:1500–1507. doi: 10.1056/NEJM200005183422007. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch E, Wang H. The molecular pathophysiology of bacterially induced preterm labor: insights from the murine model. J Soc Gynecol Investig. 2005;12:145–155. doi: 10.1016/j.jsgi.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Koga K, Cardenas I, Aldo P, Abrahams VM, Peng B, Fill S, Romero R, Mor G. Activation of TLR3 in the trophoblast is associated with preterm delivery. Am J Reprod Immunol. 2009;61:196–212. doi: 10.1111/j.1600-0897.2008.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romero R, Salafia CM, Athanassiadis AP, Hanaoka S, Mazor M, Sepulveda W, Bracken MB. The relationship between acute inflammatory lesions of the preterm placenta and amniotic fluid microbiology. Am J Obstet Gynecol. 1992;166:1382–1388. doi: 10.1016/0002-9378(92)91609-e. [DOI] [PubMed] [Google Scholar]
- 10.Goncalves LF, Chaiworapongsa T, Romero R. Intrauterine infection and prematurity. Ment Retard Dev Disabil Res Rev. 2002;8:3–13. doi: 10.1002/mrdd.10008. [DOI] [PubMed] [Google Scholar]
- 11.Yoon BH, Romero R, Moon JB, Shim SS, Kim M, Kim G, Jun JK. Clinical significance of intra-amniotic inflammation in patients with preterm labor and intact membranes. Am J Obstet Gynecol. 2001;185:1130–1136. doi: 10.1067/mob.2001.117680. [DOI] [PubMed] [Google Scholar]
- 12.Yoon BH, Romero R, Jun JK, Park KH, Park JD, Ghezzi F, Kim BI. Amniotic fluid cytokines (interleukin-6, tumor necrosis factor-alpha, interleukin-1 beta, and interleukin-8) and the risk for the development of bronchopulmonary dysplasia. Am J Obstet Gynecol. 1997;177:825–830. doi: 10.1016/s0002-9378(97)70276-x. [DOI] [PubMed] [Google Scholar]
- 13.Yoon BH, Jun JK, Romero R, Park KH, Gomez R, Choi JH, Kim IO. Amniotic fluid inflammatory cytokines (interleukin-6, interleukin-1beta, and tumor necrosis factor-alpha), neonatal brain white matter lesions, and cerebral palsy. Am J Obstet Gynecol. 1997;177:19–26. doi: 10.1016/s0002-9378(97)70432-0. [DOI] [PubMed] [Google Scholar]
- 14.Yoon BH, Romero R, Kim CJ, Koo JN, Choe G, Syn HC, Chi JG. High expression of tumor necrosis factor-alpha and interleukin-6 in periventricular leukomalacia. Am J Obstet Gynecol. 1997;177:406–411. doi: 10.1016/s0002-9378(97)70206-0. [DOI] [PubMed] [Google Scholar]
- 15.Ghezzi F, Gomez R, Romero R, Yoon BH, Edwin SS, David C, Janisse J, Mazor M. Elevated interleukin-8 concentrations in amniotic fluid of mothers whose neonates subsequently develop bronchopulmonary dysplasia. Eur J Obstet Gynecol Reprod Biol. 1998;78:5–10. doi: 10.1016/s0301-2115(97)00236-4. [DOI] [PubMed] [Google Scholar]
- 16.Yoon BH, Romero R, Park JS, Chang JW, Kim YA, Kim JC, Kim KS. Microbial invasion of the amniotic cavity with Ureaplasma urealyticum is associated with a robust host response in fetal, amniotic, and maternal compartments. Am J Obstet Gynecol. 1998;179:1254–1260. doi: 10.1016/s0002-9378(98)70142-5. [DOI] [PubMed] [Google Scholar]
- 17.Park JS, Romero R, Yoon BH, Moon JB, Oh SY, Han SY, Ko EM. The relationship between amniotic fluid matrix metalloproteinase-8 and funisitis. Am J Obstet Gynecol. 2001;185:1156–1161. doi: 10.1067/mob.2001.117679. [DOI] [PubMed] [Google Scholar]
- 18.Gibbs RS. The relationship between infections and adverse pregnancy outcomes: an overview. Ann Periodontol. 2001;6:153–163. doi: 10.1902/annals.2001.6.1.153. [DOI] [PubMed] [Google Scholar]
- 19.Chaiworapongsa T, Romero R, Kim JC, Kim YM, Blackwell SC, Yoon BH, Gomez R. Evidence for fetal involvement in the pathologic process of clinical chorioamnionitis. Am J Obstet Gynecol. 2002;186:1178–1182. doi: 10.1067/mob.2002.124042. [DOI] [PubMed] [Google Scholar]
- 20.Moon JB, Kim JC, Yoon BH, Romero R, Kim G, Oh SY, Kim M, Shim SS. Amniotic fluid matrix metalloproteinase-8 and the development of cerebral palsy. J Perinat Med. 2002;30:301–306. doi: 10.1515/JPM.2002.044. [DOI] [PubMed] [Google Scholar]
- 21.Lee SE, Romero R, Jung H, Park CW, Park JS, Yoon BH. The intensity of the fetal inflammatory response in intraamniotic inflammation with and without microbial invasion of the amniotic cavity. Am J Obstet Gynecol. 2007;197:294–296. doi: 10.1016/j.ajog.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Gotsch F, Romero R, Kusanovic JP, Mazaki-Tovi S, Pineles BL, Erez O, Espinoza J, Hassan SS. The fetal inflammatory response syndrome. Clin Obstet Gynecol. 2007;50:652–683. doi: 10.1097/GRF.0b013e31811ebef6. [DOI] [PubMed] [Google Scholar]
- 23.Yoon BH, Romero R, Park JS, Kim M, Oh SY, Kim CJ, Jun JK. The relationship among inflammatory lesions of the umbilical cord (funisitis), umbilical cord plasma interleukin 6 concentration, amniotic fluid infection, and neonatal sepsis. Am J Obstet Gynecol. 2000;183:1124–1129. doi: 10.1067/mob.2000.109035. [DOI] [PubMed] [Google Scholar]
- 24.Yoon BH, Romero R, Kim CJ, Jun JK, Gomez R, Choi JH, Syn HC. Amniotic fluid interleukin-6: a sensitive test for antenatal diagnosis of acute inflammatory lesions of preterm placenta and prediction of perinatal morbidity. Am J Obstet Gynecol. 1995;172:960–970. doi: 10.1016/0002-9378(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 25.Yoon BH, Romero R, Kim KS, Park JS, Ki SH, Kim BI, Jun JK. A systemic fetal inflammatory response and the development of bronchopulmonary dysplasia. Am J Obstet Gynecol. 1999;181:773–779. doi: 10.1016/s0002-9378(99)70299-1. [DOI] [PubMed] [Google Scholar]
- 26.Mittendorf R, Covert R, Montag AG, elMasri W, Muraskas J, Lee KS, Pryde PG. Special relationships between fetal inflammatory response syndrome and bronchopulmonary dysplasia in neonates. J Perinat Med. 2005;33:428–434. doi: 10.1515/JPM.2005.076. [DOI] [PubMed] [Google Scholar]
- 27.Mittendorf R, Montag AG, MacMillan W, Janeczek S, Pryde PG, Besinger RE, Gianopoulos JG, Roizen N. Components of the systemic fetal inflammatory response syndrome as predictors of impaired neurologic outcomes in children. Am J Obstet Gynecol. 2003;188:1438–4. doi: 10.1067/mob.2003.380. [DOI] [PubMed] [Google Scholar]
- 28.Yoon BH, Romero R, Yang SH, Jun JK, Kim IO, Choi JH, Syn HC. Interleukin-6 concentrations in umbilical cord plasma are elevated in neonates with white matter lesions associated with periventricular leukomalacia. Am J Obstet Gynecol. 1996;174:1433–1440. doi: 10.1016/s0002-9378(96)70585-9. [DOI] [PubMed] [Google Scholar]
- 29.Dammann O, Leviton A. Maternal intrauterine infection, cytokines, and brain damage in the preterm newborn. Pediatr Res. 1997;42:1–8. doi: 10.1203/00006450-199707000-00001. [DOI] [PubMed] [Google Scholar]
- 30.Leviton A, Paneth N, Reuss ML, Susser M, Allred EN, Dammann O, Kuban K, Van Marter LJ, Pagano M, Hegyi T, Hiatt M, Sanocka U, Shahrivar F, Abiri M, Disalvo D, Doubilet P, Kairam R, Kazam E, Kirpekar M, Rosenfeld D, Schonfeld S, Share J, Collins M, Genest D, Shen-Schwarz S. Maternal infection, fetal inflammatory response, and brain damage in very low birth weight infants. Developmental Epidemiology Network Investigators. Pediatr Res. 1999;46:566–575. doi: 10.1203/00006450-199911000-00013. [DOI] [PubMed] [Google Scholar]
- 31.Dammann O, Leviton A. Role of the fetus in perinatal infection and neonatal brain damage. Curr Opin Pediatr. 2000;12:99–104. doi: 10.1097/00008480-200004000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Yoon BH, Romero R, Park JS, Kim CJ, Kim SH, Choi JH, Han TR. Fetal exposure to an intra-amniotic inflammation and the development of cerebral palsy at the age of three years. Am J Obstet Gynecol. 2000;182:675–681. doi: 10.1067/mob.2000.104207. [DOI] [PubMed] [Google Scholar]
- 33.Patrick LA, Smith GN. Proinflammatory cytokines: a link between chorioamnionitis and fetal brain injury. J Obstet Gynaecol Can. 2002;24:705–709. doi: 10.1016/s1701-2163(16)30325-5. [DOI] [PubMed] [Google Scholar]
- 34.Hagberg H, Mallard C, Jacobsson B. Role of cytokines in preterm labour and brain injury. BJOG. 2005;112 (Suppl 1):16–18. doi: 10.1111/j.1471-0528.2005.00578.x. [DOI] [PubMed] [Google Scholar]
- 35.Bashiri A, Burstein E, Mazor M. Cerebral palsy and fetal inflammatory response syndrome: a review. J Perinat Med. 2006;34:5–12. doi: 10.1515/JPM.2006.001. [DOI] [PubMed] [Google Scholar]
- 36.Romero R, Athayde N, Gomez R, Mazor M, Yoon BH, Edwin SS, Ghezzi F, Berry SM. The fetal inflammatory response syndrome is characterized by the outpouring of a potent extracellular matrix degrading enzyme into the fetal circulation. Am J Obstet Gynecol. 1998;178:S3. [Google Scholar]
- 37.Berry SM, Romero R, Gomez R, Puder KS, Ghezzi F, Cotton DB, Bianchi DW. Premature parturition is characterized by in utero activation of the fetal immune system. Am J Obstet Gynecol. 1995;173:1315–1320. doi: 10.1016/0002-9378(95)91378-5. [DOI] [PubMed] [Google Scholar]
- 38.Berry SM, Gomez R, Athayde N, Ghezzi F, Mazor M, Yoon BH, Edwin SS, Romero R. The role of granulocyte colony stimulating factor in the neutrophilia observed in the fetal inflammatory response syndrome. Am J Obstet Gynecol. 1998;178:S202. Ref Type: Abstract. [Google Scholar]
- 39.Gomez R, Berry S, Yoon BH, Mazor M, Athayde N, Ghezzi F, Romero R. The hematologic profile of the fetus with systemic inflammatory response syndrome. Am J Obstet Gynecol. 1998;178:S202. [Google Scholar]
- 40.Yoon BH, Romero R, Jun JK, Maymon E, Gomez R, Mazor M, Park JS. An increase in fetal plasma cortisol but not dehydroepiandrosterone sulfate is followed by the onset of preterm labor in patients with preterm premature rupture of the membranes. Am J Obstet Gynecol. 1998;179:1107–1114. doi: 10.1016/s0002-9378(98)70114-0. [DOI] [PubMed] [Google Scholar]
- 41.Yanowitz TD, Jordan JA, Gilmour CH, Towbin R, Bowen A, Roberts JM, Brozanski BS. Hemodynamic disturbances in premature infants born after chorioamnionitis: association with cord blood cytokine concentrations. Pediatr Res. 2002;51:310–316. doi: 10.1203/00006450-200203000-00008. [DOI] [PubMed] [Google Scholar]
- 42.Romero R, Espinoza J, Goncalves LF, Gomez R, Medina L, Silva M, Chaiworapongsa T, Yoon BH, Ghezzi F, Lee W, Treadwell M, Berry SM, Maymon E, Mazor M, DeVore G. Fetal cardiac dysfunction in preterm premature rupture of membranes. J Matern Fetal Neonatal Med. 2004;16:146–157. doi: 10.1080/14767050400009279. [DOI] [PubMed] [Google Scholar]
- 43.Yoon BH, Kim YA, Romero R, Kim JC, Park KH, Kim MH, Park JS. Association of oligohydramnios in women with preterm premature rupture of membranes with an inflammatory response in fetal, amniotic, and maternal compartments. Am J Obstet Gynecol. 1999;181:784–788. doi: 10.1016/s0002-9378(99)70301-7. [DOI] [PubMed] [Google Scholar]
- 44.Di Naro E, Cromi A, Ghezzi F, Raio L, Uccella S, D’Addario V, Loverro G. Fetal thymic involution: a sonographic marker of the fetal inflammatory response syndrome. Am J Obstet Gynecol. 2006;194:153–159. doi: 10.1016/j.ajog.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 45.Yinon Y, Zalel Y, Weisz B, Mazaki-Tovi S, Sivan E, Schiff E, Achiron R. Fetal thymus size as a predictor of chorioamnionitis in women with preterm premature rupture of membranes. Ultrasound Obstet Gynecol. 2007;29:639–643. doi: 10.1002/uog.4022. [DOI] [PubMed] [Google Scholar]
- 46.Kim YM, Romero R, Chaiworapongsa T, Espinoza J, Mor G, Kim CJ. Dermatitis as a component of the fetal inflammatory response syndrome is associated with activation of Toll-like receptors in epidermal keratinocytes. Histopathology. 2006;49:506–514. doi: 10.1111/j.1365-2559.2006.02542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim CJ, Yoon BH, Romero R, Moon JB, Kim M, Park SS, Chi JG. Umbilical arteritis and phlebitis mark different stages of the fetal inflammatory response. Am J Obstet Gynecol. 2001;185:496–500. doi: 10.1067/mob.2001.116689. [DOI] [PubMed] [Google Scholar]
- 48.Pacora P, Chaiworapongsa T, Maymon E, Kim YM, Gomez R, Yoon BH, Ghezzi F, Berry SM, Qureshi F, Jacques SM, Kim JC, Kadar N, Romero R. Funisitis and chorionic vasculitis: the histological counterpart of the fetal inflammatory response syndrome. J Matern Fetal Neonatal Med. 2002;11:18–25. doi: 10.1080/jmf.11.1.18.25. [DOI] [PubMed] [Google Scholar]
- 49.Yoon BH, Romero R, Shim JY, Shim SS, Kim CJ, Jun JK. C-reactive protein in umbilical cord blood: a simple and widely available clinical method to assess the risk of amniotic fluid infection and funisitis. J Matern Fetal Neonatal Med. 2003;14:85–90. doi: 10.1080/jmf.14.2.85.90. [DOI] [PubMed] [Google Scholar]
- 50.Gogos CA, Drosou E, Bassaris HP, Skoutelis A. Pro-versus anti-inflammatory cytokine profile in patients with severe sepsis: a marker for prognosis and future therapeutic options. J Infect Dis. 2000;181:176–180. doi: 10.1086/315214. [DOI] [PubMed] [Google Scholar]
- 51.Prabhakar U, Conway TM, Murdock P, Mooney JL, Clark S, Hedge P, Bond BC, Jazwinska EC, Barnes MR, Tobin F, mian-Iordachi V, Greller L, Hurle M, Stubbs AP, Li Z, Valoret EI, Erickson-Miller C, Cass L, Levitt B, Davis HM, Jorkasky DK, Williams WV. Correlation of protein and gene expression profiles of inflammatory proteins after endotoxin challenge in human subjects. DNA Cell Biol. 2005;24:410–431. doi: 10.1089/dna.2005.24.410. [DOI] [PubMed] [Google Scholar]
- 52.Talwar S, Munson PJ, Barb J, Fiuza C, Cintron AP, Logun C, Tropea M, Khan S, Reda D, Shelhamer JH, Danner RL, Suffredini AF. Gene expression profiles of peripheral blood leukocytes after endotoxin challenge in humans. Physiol Genomics. 2006;25:203–215. doi: 10.1152/physiolgenomics.00192.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davies NP, Buggins AG, Snijders RJ, Jenkins E, Layton DM, Nicolaides KH. Blood leucocyte count in the human fetus. Arch Dis Child. 1992;67:399–403. doi: 10.1136/adc.67.4_spec_no.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen SB, Perez-Cruz I, Fallen P, Gluckman E, Madrigal JA. Analysis of the cytokine production by cord and adult blood. Hum Immunol. 1999;60:331–336. doi: 10.1016/s0198-8859(98)00126-8. [DOI] [PubMed] [Google Scholar]
- 55.Satwani P, Morris E, van d V, Cairo MS. Dysregulation of expression of immunoregulatory and cytokine genes and its association with the immaturity in neonatal phagocytic and cellular immunity. Biol Neonate. 2005;88:214–227. doi: 10.1159/000087585. [DOI] [PubMed] [Google Scholar]
- 56.Dos Santos C, Davidson D. Neutrophil chemotaxis to leukotriene B4 in vitro is decreased for the human neonate. Pediatr Res. 1993;33:242–246. doi: 10.1203/00006450-199303000-00006. [DOI] [PubMed] [Google Scholar]
- 57.Strunk T, Temming P, Gembruch U, Reiss I, Bucsky P, Schultz C. Differential maturation of the innate immune response in human fetuses. Pediatr Res. 2004;56:219–226. doi: 10.1203/01.PDR.0000132664.66975.79. [DOI] [PubMed] [Google Scholar]
- 58.Berner R, Welter P, Brandis M. Cytokine expression of cord and adult blood mononuclear cells in response to Streptococcus agalactiae. Pediatr Res. 2002;51:304–309. doi: 10.1203/00006450-200203000-00007. [DOI] [PubMed] [Google Scholar]
- 59.Jiang H, van d V, Satwani P, Baxi LV, Cairo MS. Differential gene expression patterns by oligonucleotide microarray of basal versus lipopolysaccharide-activated monocytes from cord blood versus adult peripheral blood. J Immunol. 2004;172:5870–5879. doi: 10.4049/jimmunol.172.10.5870. [DOI] [PubMed] [Google Scholar]
- 60.Prucha M, Ruryk A, Boriss H, Moller E, Zazula R, Herold I, Claus RA, Reinhart KA, Deigner P, Russwurm S. Expression profiling: toward an application in sepsis diagnostics. Shock. 2004;22:29–33. doi: 10.1097/01.shk.0000129199.30965.02. [DOI] [PubMed] [Google Scholar]
- 61.Pachot A, Lepape A, Vey S, Bienvenu J, Mougin B, Monneret G. Systemic transcriptional analysis in survivor and non-survivor septic shock patients: a preliminary study. Immunol Lett. 2006;106:63–71. doi: 10.1016/j.imlet.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 62.Wong HR, Shanley TP, Sakthivel B, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, Penfil S, Monaco M, Tagavilla MA, Odoms K, Dunsmore K, Barnes M, Aronow BJ. Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007;30:146–155. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 64.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 65.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 66.Draghici S, Khatri P, Martins RP, Ostermeier GC, Krawetz SA. Global functional profiling of gene expression. Genomics. 2003;81:98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
- 67.Khatri P, Bhavsar P, Bawa G, Draghici S. Onto-Tools: an ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments. Nucleic Acids Res. 2004;32:W449–W456. doi: 10.1093/nar/gkh409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fu WJ, Hu J, Spencer T, Carroll R, Wu G. Statistical models in assessing fold change of gene expression in real-time RT-PCR experiments. Comput Biol Chem. 2006;30:21–26. doi: 10.1016/j.compbiolchem.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 70.Liu J, Walter E, Stenger D, Thach D. Effects of globin mRNA reduction methods on gene expression profiles from whole blood. J Mol Diagn. 2006;8:551–558. doi: 10.2353/jmoldx.2006.060021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Field LA, Jordan RM, Hadix JA, Dunn MA, Shriver CD, Ellsworth RE, Ellsworth DL. Functional identity of genes detectable in expression profiling assays following globin mRNA reduction of peripheral blood samples. Clin Biochem. 2007;40:499–502. doi: 10.1016/j.clinbiochem.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 72.Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Draghici S, Khatri P, Eklund AC, Szallasi Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006;22:101–109. doi: 10.1016/j.tig.2005.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cavaillon JM, dib-Conquy M, Fitting C, Adrie C, Payen D. Cytokine cascade in sepsis. Scand J Infect Dis. 2003;35:535–544. doi: 10.1080/00365540310015935. [DOI] [PubMed] [Google Scholar]
- 75.Russell JA. Management of sepsis. N Engl J Med. 2006;355:1699–1713. doi: 10.1056/NEJMra043632. [DOI] [PubMed] [Google Scholar]
- 76.Romero R, Durum S, Dinarello C, Hobbins JC, Mitchell M. Interleukin-1. A signal for the initiation of labor in chorioamnionitis. Society for Gynecologic Investigation (33rd Annual Meeting); 1986. Ref Type: Abstract. [Google Scholar]
- 77.Romero R, Manogue KR, Mitchell MD, Wu YK, Oyarzun E, Hobbins JC, Cerami A. Infection and labor. IV. Cachectin-tumor necrosis factor in the amniotic fluid of women with intraamniotic infection and preterm labor. American Journal of Obstetrics and Gynecology. 1989;161:336–341. doi: 10.1016/0002-9378(89)90515-2. [DOI] [PubMed] [Google Scholar]
- 78.Romero R, Brody DT, Oyarzun E, Mazor M, Wu YK, Hobbins JC, Durum SK. Infection and labor. III. Interleukin-1: a signal for the onset of parturition. Am J Obstet Gynecol. 1989;160:1117–1123. doi: 10.1016/0002-9378(89)90172-5. [DOI] [PubMed] [Google Scholar]
- 79.Romero R, Avila C, Santhanam U, Sehgal PB. Amniotic fluid interleukin 6 in preterm labor. Association with infection. J Clin Invest. 1990;85:1392–1400. doi: 10.1172/JCI114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Romero R, Ceska M, Avila C, Mazor M, Behnke E, Lindley I. Neutrophil attractant/activating peptide-1/interleukin-8 in term and preterm parturition. Am J Obstet Gynecol. 1991;165:813–820. doi: 10.1016/0002-9378(91)90422-n. [DOI] [PubMed] [Google Scholar]
- 81.Romero R, Sepulveda W, Mazor M, Brandt F, Cotton DB, Dinarello CA, Mitchell MD. The natural interleukin-1 receptor antagonist in term and preterm parturition. Am J Obstet Gynecol. 1992;167:863–872. doi: 10.1016/s0002-9378(12)80003-2. [DOI] [PubMed] [Google Scholar]
- 82.Romero R, Mazor M, Brandt F, Sepulveda W, Avila C, Cotton DB, Dinarello CA. Interleukin-1 alpha and interleukin-1 beta in preterm and term human parturition. Am J Reprod Immunol. 1992;27:117–123. doi: 10.1111/j.1600-0897.1992.tb00737.x. [DOI] [PubMed] [Google Scholar]
- 83.Romero R, Sepulveda W, Kenney JS, Archer LE, Allison AC, Sehgal PB. Interleukin 6 determination in the detection of microbial invasion of the amniotic cavity. Ciba Found Symp. 1992;167:205–220. doi: 10.1002/9780470514269.ch13. [DOI] [PubMed] [Google Scholar]
- 84.Romero R, Yoon BH, Kenney JS, Gomez R, Allison AC, Sehgal PB. Amniotic fluid interleukin-6 determinations are of diagnostic and prognostic value in preterm labor. Am J Reprod Immunol. 1993;30:167–183. doi: 10.1111/j.1600-0897.1993.tb00618.x. [DOI] [PubMed] [Google Scholar]
- 85.Romero R, Gomez R, Galasso M, Mazor M, Berry SM, Quintero RA, Cotton DB. The natural interleukin-1 receptor antagonist in the fetal, maternal, and amniotic fluid compartments: the effect of gestational age, fetal gender, and intrauterine infection. Am J Obstet Gynecol. 1994;171:912–921. doi: 10.1016/s0002-9378(94)70058-3. [DOI] [PubMed] [Google Scholar]
- 86.Romero R, Gomez R, Galasso M, Munoz H, Acosta L, Yoon BH, Svinarich D, Cotton DB. Macrophage inflammatory protein-1 alpha in term and preterm parturition: effect of microbial invasion of the amniotic cavity. Am J Reprod Immunol. 1994;32:108–113. doi: 10.1111/j.1600-0897.1994.tb01101.x. [DOI] [PubMed] [Google Scholar]
- 87.Maymon E, Ghezzi F, Edwin SS, Mazor M, Yoon BH, Gomez R, Romero R. The tumor necrosis factor alpha and its soluble receptor profile in term and preterm parturition. Am J Obstet Gynecol. 1999;181:1142–1148. doi: 10.1016/s0002-9378(99)70097-9. [DOI] [PubMed] [Google Scholar]
- 88.Keelan JA, Wang K, Chaiworapongsa T, Romero R, Mitchell MD, Sato TA, Brown DA, Fairlie WD, Breit SN. Macrophage inhibitory cytokine 1 in fetal membranes and amniotic fluid from pregnancies with and without preterm labour and premature rupture of membranes. Mol Hum Reprod. 2003;9:535–540. doi: 10.1093/molehr/gag068. [DOI] [PubMed] [Google Scholar]
- 89.Lonergan M, Aponso D, Marvin KW, Helliwell RJ, Sato TA, Mitchell MD, Chaiwaropongsa T, Romero R, Keelan JA. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), TRAIL receptors, and the soluble receptor osteoprotegerin in human gestational membranes and amniotic fluid during pregnancy and labor at term and preterm. J Clin Endocrinol Metab. 2003;88:3835–3844. doi: 10.1210/jc.2002-021905. [DOI] [PubMed] [Google Scholar]
- 90.Keelan JA, Yang J, Romero RJ, Chaiworapongsa T, Marvin KW, Sato TA, Mitchell MD. Epithelial cell-derived neutrophil-activating peptide-78 is present in fetal membranes and amniotic fluid at increased concentrations with intra-amniotic infection and preterm delivery. Biol Reprod. 2004;70:253–259. doi: 10.1095/biolreprod.103.016204. [DOI] [PubMed] [Google Scholar]
- 91.Esplin MS, Romero R, Chaiworapongsa T, Kim YM, Edwin S, Gomez R, Mazor M, Adashi EY. Monocyte chemotactic protein-1 is increased in the amniotic fluid of women who deliver preterm in the presence or absence of intra-amniotic infection. J Matern Fetal Neonatal Med. 2005;17:365–373. doi: 10.1080/14767050500141329. [DOI] [PubMed] [Google Scholar]
- 92.Mittal P, Romero R, Kusanovic JP, Edwin SS, Gotsch F, Mazaki-Tovi S, Espinoza J, Erez O, Nhan-Chang CL, Than NG, Vaisbuch E, Hassan SS. CXCL6 (granulocyte chemotactic protein-2): a novel chemokine involved in the innate immune response of the amniotic cavity. Am J Reprod Immunol. 2008;60:246–257. doi: 10.1111/j.1600-0897.2008.00620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hassett S, Moynagh P, Reen D. TNF-alpha is a mediator of the anti-inflammatory response in a human neonatal model of the non-septic shock syndrome. Pediatr Surg Int. 2006;22:24–30. doi: 10.1007/s00383-005-1574-7. [DOI] [PubMed] [Google Scholar]
- 94.Pettersson A, Richter J, Owman C. Flow cytometric mapping of the leukotriene B4 receptor, BLT1, in human bone marrow and peripheral blood using specific monoclonal antibodies. Int Immunopharmacol. 2003;3:1467–1475. doi: 10.1016/S1567-5769(03)00145-0. [DOI] [PubMed] [Google Scholar]
- 95.Tager AM, Luster AD. BLT1 and BLT2: the leukotriene B(4) receptors. Prostaglandins Leukot Essent Fatty Acids. 2003;69:123–134. doi: 10.1016/s0952-3278(03)00073-5. [DOI] [PubMed] [Google Scholar]
- 96.Pettersson A, Sabirsh A, Bristulf J, Kidd-Ljunggren K, Ljungberg B, Owman C, Karlsson U. Pro- and anti-inflammatory substances modulate expression of the leukotriene B4 receptor, BLT1, in human monocytes. J Leukoc Biol. 2005;77:1018–1025. doi: 10.1189/jlb.1204740. [DOI] [PubMed] [Google Scholar]
- 97.Plante H, Picard S, Mancini J, Borgeat P. 5-Lipoxygenase-activating protein homodimer in human neutrophils: evidence for a role in leukotriene biosynthesis. Biochem J. 2006;393:211–218. doi: 10.1042/BJ20060669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Benjamim CF, Canetti C, Cunha FQ, Kunkel SL, Peters-Golden M. Opposing and hierarchical roles of leukotrienes in local innate immune versus vascular responses in a model of sepsis. J Immunol. 2005;174:1616–1620. doi: 10.4049/jimmunol.174.3.1616. [DOI] [PubMed] [Google Scholar]
- 99.Espinoza J, Chaiworapongsa T, Romero R, Edwin S, Rathnasabapathy C, Gomez R, Bujold E, Camacho N, Kim YM, Hassan S, Blackwell S, Whitty J, Berman S, Redman M, Yoon BH, Sorokin Y. Antimicrobial peptides in amniotic fluid: defensins, calprotectin and bacterial/permeability-increasing protein in patients with microbial invasion of the amniotic cavity, intra-amniotic inflammation, preterm labor and premature rupture of membranes. J Matern Fetal Neonatal Med. 2003;13:2–21. doi: 10.1080/jmf.13.1.2.21. [DOI] [PubMed] [Google Scholar]
- 100.Soto E, Espinoza J, Nien JK, Kusanovic JP, Erez O, Richani K, Santolaya-Forgas J, Romero R. Human beta-defensin-2: a natural antimicrobial peptide present in amniotic fluid participates in the host response to microbial invasion of the amniotic cavity. J Matern Fetal Neonatal Med. 2007;20:15–22. doi: 10.1080/14767050601036212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cole AM, Cole AL. Antimicrobial polypeptides are key anti-HIV-1 effector molecules of cervicovaginal host defense. Am J Reprod Immunol. 2008;59:27–34. doi: 10.1111/j.1600-0897.2007.00561.x. [DOI] [PubMed] [Google Scholar]
- 102.Foell D, Frosch M, Sorg C, Roth J. Phagocyte-specific calcium-binding S100 proteins as clinical laboratory markers of inflammation. Clin Chim Acta. 2004;344:37–51. doi: 10.1016/j.cccn.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 103.Buhimschi CS, Buhimschi IA, bdel-Razeq S, Rosenberg VA, Thung SF, Zhao G, Wang E, Bhandari V. Proteomic biomarkers of intra-amniotic inflammation: relationship with funisitis and early-onset sepsis in the premature neonate. Pediatr Res. 2007;61:318–324. doi: 10.1203/01.pdr.0000252439.48564.37. [DOI] [PubMed] [Google Scholar]
- 104.Newton RA, Hogg N. The human S100 protein MRP-14 is a novel activator of the beta 2 integrin Mac-1 on neutrophils. J Immunol. 1998;160:1427–1435. [PubMed] [Google Scholar]
- 105.Chishti AD, Shenton BK, Kirby JA, Baudouin SV. Neutrophil chemotaxis and receptor expression in clinical septic shock. Intensive Care Med. 2004;30:605–611. doi: 10.1007/s00134-004-2175-y. [DOI] [PubMed] [Google Scholar]
- 106.Henneke P, Osmers I, Bauer K, Lamping N, Versmold HT, Schumann RR. Impaired CD14-dependent and independent response of polymorphonuclear leukocytes in preterm infants. J Perinat Med. 2003;31:176–183. doi: 10.1515/JPM.2003.024. [DOI] [PubMed] [Google Scholar]
- 107.Kim SK, Romero R, Chaiworapongsa T, Pedro KJ, Mazaki-Tovi S, Mittal P, Erez O, Vaisbuch E, Gotsch F, Pacora P, Yeo L, Teresa GM, Lamont RF, Yoon BH, Hassan SS. Evidence of changes in the immunophenotype and metabolic characteristics (intracellular reactive oxygen radicals) of fetal, but not maternal, monocytes and granulocytes in the fetal inflammatory response syndrome. J Perinat Med. 2009;37:543–552. doi: 10.1515/JPM.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pinsky MR. Sepsis: a pro- and anti-inflammatory disequilibrium syndrome. Contrib Nephrol. 2001:354–366. doi: 10.1159/000060100. [DOI] [PubMed] [Google Scholar]
- 109.Phan UT, Lackman RL, Cresswell P. Role of the C-terminal propeptide in the activity and maturation of gamma -interferon-inducible lysosomal thiol reductase (GILT) Proc Natl Acad Sci U S A. 2002;99:12298–12303. doi: 10.1073/pnas.182430499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Becker-Herman S, Arie G, Medvedovsky H, Kerem A, Shachar I. CD74 is a member of the regulated intramembrane proteolysis-processed protein family. Mol Biol Cell. 2005;16:5061–5069. doi: 10.1091/mbc.E05-04-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Muller Kobold AC, Tulleken JE, Zijlstra JG, Sluiter W, Hermans J, Kallenberg CG, Tervaert JW. Leukocyte activation in sepsis; correlations with disease state and mortality. Intensive Care Med. 2000;26:883–892. doi: 10.1007/s001340051277. [DOI] [PubMed] [Google Scholar]
- 112.Pachot A, Monneret G, Brion A, Venet F, Bohe J, Bienvenu J, Mougin B, Lepape A. Messenger RNA expression of major histocompatibility complex class II genes in whole blood from septic shock patients. Crit Care Med. 2005;33:31–38. doi: 10.1097/01.ccm.0000150958.20209.a3. [DOI] [PubMed] [Google Scholar]
- 113.Monneret G, Lepape A, Voirin N, Bohe J, Venet F, Debard AL, Thizy H, Bienvenu J, Gueyffier F, Vanhems P. Persisting low monocyte human leukocyte antigen-DR expression predicts mortality in septic shock. Intensive Care Med. 2006;32:1175–1183. doi: 10.1007/s00134-006-0204-8. [DOI] [PubMed] [Google Scholar]
- 114.Fumeaux T, Pugin J. Role of interleukin-10 in the intracellular sequestration of human leukocyte antigen-DR in monocytes during septic shock. Am J Respir Crit Care Med. 2002;166:1475–1482. doi: 10.1164/rccm.200203-217OC. [DOI] [PubMed] [Google Scholar]
- 115.Hanna N, Hanna I, Hleb M, Wagner E, Dougherty J, Balkundi D, Padbury J, Sharma S. Gestational age-dependent expression of IL-10 and its receptor in human placental tissues and isolated cytotrophoblasts. J Immunol. 2000;164:5721–5728. doi: 10.4049/jimmunol.164.11.5721. [DOI] [PubMed] [Google Scholar]
- 116.Gotsch F, Romero R, Kusanovic JP, Erez O, Espinoza J, Kim CJ, Vaisbuch E, Than NG, Mazaki-Tovi S, Chaiworapongsa T, Mazor M, Yoon BH, Edwin S, Gomez R, Mittal P, Hassan SS, Sharma S. The anti-inflammatory limb of the immune response in preterm labor, intra-amniotic infection/inflammation, and spontaneous parturition at term: a role for interleukin-10. J Matern Fetal Neonatal Med. 2008;21:529–547. doi: 10.1080/14767050802127349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Than NG, Kim SS, Abbas A, Han YM, Hotra J, Tarca AL, Erez O, Wildman DE, Kusanovi JP, Pineles B, Montenegro D, Edwin SS, Mazaki-Tovi S, Gotsch F, Espinoza J, Hassan SS, Papp Z, Romero R. Chorioamnionitis and increased galectin-1 expression in PPROM --an anti-inflammatory response in the fetal membranes? Am J Reprod Immunol. 2008;60:298–311. doi: 10.1111/j.1600-0897.2008.00624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McGowan EC, Kostadinov S, McLean K, Gotsch F, Venturini D, Romero R, Laptook AR, Sharma S. Placental IL-10 dysregulation and association with bronchopulmonary dysplasia risk. Pediatr Res. 2009 doi: 10.1203/PDR.0b013e3181b3b0fa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Thaxton JE, Romero R, Sharma S. TLR9 activation coupled to IL-10 deficiency induces adverse pregnancy outcomes. J Immunol. 2009;183:1144–1154. doi: 10.4049/jimmunol.0900788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Macardle PJ, Weedon H, Fusco M, Nobbs S, Ridings J, Flego L, Roberton DM, Zola H. The antigen receptor complex on cord B lymphocytes. Immunology. 1997;90:376–382. doi: 10.1111/j.1365-2567.1997.00376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Weinschenk NP, Farina A, Bianchi DW. Premature infants respond to early-onset and late-onset sepsis with leukocyte activation. J Pediatr. 2000;137:345–350. doi: 10.1067/mpd.2000.107846. [DOI] [PubMed] [Google Scholar]
- 122.Thome M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004;4:348–359. doi: 10.1038/nri1352. [DOI] [PubMed] [Google Scholar]
- 123.Karin M. How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene. 1999;18:6867–6874. doi: 10.1038/sj.onc.1203219. [DOI] [PubMed] [Google Scholar]
- 124.Sato S, Miller AS, Inaoki M, Bock CB, Jansen PJ, Tang ML, Tedder TF. CD22 is both a positive and negative regulator of B lymphocyte antigen receptor signal transduction: altered signaling in CD22-deficient mice. Immunity. 1996;5:551–562. doi: 10.1016/s1074-7613(00)80270-8. [DOI] [PubMed] [Google Scholar]
- 125.Yamazaki T, Nagumo H, Hayashi T, Sugane K, Agematsu K. CD72-mediated suppression of human naive B cell differentiation by down-regulating X-box binding protein 1. Eur J Immunol. 2005;35:2325–2334. doi: 10.1002/eji.200425639. [DOI] [PubMed] [Google Scholar]
- 126.Elsmen E, Ley D, Cilio CM, Hansen-Pupp I, Hellstrom-Westas L. Umbilical cord levels of interleukin-1 receptor antagonist and neonatal outcome. Biol Neonate. 2006;89:220–226. doi: 10.1159/000089838. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Globin RNA transcript abundance was assayed in triplicate with TaqMan inventoried gene expression assays (Applied Biosystems) for alpha, beta, and gamma globin. The currently available GlobinClear-Human kit (Ambion) results in >90% depletion of alpha (HBA) and beta (HBB) globin but minimal reduction in gamma globin (HBG, fetal globin) as is demonstrated with the solid bars (representative n=4). With the addition of gamma globin capture oligo, this transcript is also reduced by >90% (hatched bars; representative n=10).


