Abstract
Data-intensive research depends on tools that manage multi-dimensional, heterogeneous data sets. We have built OME Remote Objects (OMERO), a software platform that enables access to and use of a wide range of biological data. OMERO uses a server-based middleware application to provide a unified interface for images, matrices, and tables. OMERO’s design and flexibility have enabled its use for light microscopy, high content screening, electron microscopy, and even non-image genotype data. OMERO is open source software and available at http://openmicroscopy.org.
Introduction
Modern biology depends on the collection, management, and analysis of comprehensive datasets. Unlike the concerted efforts that drove the completion of the first genome projects, most current datasets derive from specific experiments designed to reveal molecular mechanisms, networks, interactions or phenotypes during a biological transition or after some perturbation. Although these experiments are diverse, they all require facilities for managing and analysing data. In many cases, the available data management resources are limiting, and thus ultimately define what is discovered.
Imaging in biology is a classic example of this. Years of development have delivered automated digital imaging systems that rapidly acquire time-lapse, multispectral and spatially resolved image data to reveal the structure, molecular composition and dynamics of biological cells, tissues, and organisms. The resulting datasets are often written in closed proprietary file formats, and viewed using specific hardware and software solutions, usually on a closed commercial platform1. The metadata that defines the experiment, the structure of the image dataset, the acquisition system settings, and results from analysis of the acquired data are usually stored in distinct, unlinked, heterogeneous data formats. Visualization and analysis tools can be built to use these data, but ‘hard linking’ these tools to a specific data format results in a susceptibility to change in the original format, which limits portability to other formats and data types. While there have been many calls for a single, standardized biological image file format, the heterogeneity of image data and the science it supports make this very difficult. Moreover, the rapid development of imaging methods means that new data types are constantly appearing, and these cannot be hindered by limitations of a standardized file format or access mechanism1,2. Thus the fundamental challenge is not one of standardization of data, but the absence of principles and methods that standardise data access, so that a scientist can choose the tools she or he wishes for data visualization and analysis. Standardising access also helps enable ‘interoperability’, the ability of software tools to interact with data and other software, even though they are not explicitly designed to do so.
Since 2000, the Open Microscopy Environment (OME) has been building open source data access and interoperability tools for biological image data. OME is an international collaborative effort across academic laboratories and commercial entities that produces open specifications and software tools that enable access to data, regardless of file format, programming environment, or geographical location. The OME Data Model is an open and maintained specification that defines the relationship between different data elements that describe the process of image acquisition in a microscope3. Bio-Formats is an image translation library that reads >100 PFFs and translates them into this common data model that can then be accessed by other software tools1. With the OME Data Model and Bio-Formats as a foundation, it is possible to build a system that can manage comprehensive and heterogeneous imaging datasets. Such a system would provide access for any visualization or analysis tool regardless of programming environment, linkage of heterogeneous data types that include metadata and analytic results, data search and query, and an interface to large, distributed computational resources when necessary for analysis and processing. If at all possible, this solution should be adaptable to other kinds of data, even non-image data and provide secure, remote access and mechanisms for sharing with other scientists. Finally, the solution must be maintainable and scalable. Maintenance refers to the need to keep datasets and software current, aware of updates to the data itself, but also filesystem and operating system versions. Scalability strictly refers to an increase in size (i.e., more data records, more users, etc.). However, in scientific settings, a more expansive definition of scalability is required where analysis tools useful for one type of data can easily adapt to significantly larger or more complex datasets. The addition of new searches and queries that express unanticipated scientific questions, new analysis tools, and linkage with new data types must also be considered, and designed into any solution.
To provide a single unified data management platform for image data generators and users we created OME Remote Objects (OMERO), a software platform that includes a number of storage mechanisms, remoting middleware to facilitate access to stored data through a single application programming interface (API), and client applications for biological image data management4. We demonstrate the use of OMERO in the analysis and processing of complex multidimensional image data, scientific image repositories, and extension to imputing of human SNP data.
Architecture and Design
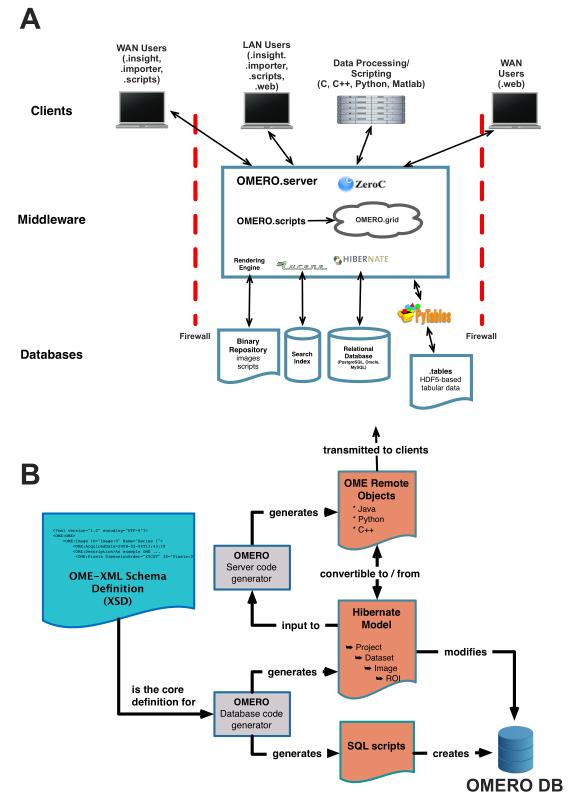
OME Remote Objects (OMERO) is a multi-component data management platform comprised of servers and clients written in Python, Java, and C++. Briefly, OMERO is built as a tiered application of databases, middleware, and remote client applications. The core middleware component, OMERO.server, is a Java application that connects a number of databases that store and manage different data types and uses a single, cross-platform application programming interface (API) to allow access to stored data (Fig. 1A). Client applications connect to OMERO.server’s API through standard internet connections for data access and processing. A schematic outlining the architecture of OMERO is provided in Figure 1A and the technical description can be found in Box 1. Descriptions of some technical terms used in this article are provided in Box 2.
Figure 1. Architecture and Data Modeling in OMERO.
(A) OMERO Architecture. OMERO is built from a series of database, middleware and client applications. A number of open source libraries provide critical functionality: Hibernate is used for object-relational mapping, Lucene for indexing, Pytables for linking to HDF5 files, and ICE for remoting.
(B) Use of OME Data Model for Code Generation. OME-XML is used by OMERO code generators to generate the OMERO relational database, the object-relational mapping model and the ICE-based remoting system.
Box 1. OMERO architecture.
Databases
OMERO uses a series of databases to efficiently store and retrieve different data types. The Binary Repository is a simple flat file store that holds the binary data, thumbnails, archived images, and any files used for data annotation. For convenience, any other submitted data, including scripts or other uploaded data are stored there as well. The Search Index stores the indices used by Lucene (http://lucene.apache.org) for text indexing. The OMERO relational database, normally run by PostgreSQL (http://www.postgresql.org/), holds all metadata associated with the binary images, all user info, most simple annotations, and records all write transactions within the OMERO installation. The OMERO relational database is constructed using code generation (Fig. 1B). The OMERO tables store is an array of HDF5 files (http://www.hdf5group.org) that contain any tabular data stored within the OMERO installation.
Middleware
The OMERO.server application provides access to the underlying storage facilities and processes data for delivery to client applications. The Rendering Engine reads binary image data, scales it according to parameters stored within the OMERO relational database or instructions from OMERO clients. This can include projections, overlays, and/or compression. Lucene is used for text indexing and running and returning search queries on these indices. Hibernate is used for object-relational mapping between OMERO.server and the OMERO relational database and filters all results according to permissions stored on each object. OMERO.scripts is a scripting service for any process in Python. Scripts are run within a service in OMERO, and passed to a grid of processors ( “OMERO.grid”, based on ZeroC’s IceGrid) where they are executed, and any results returned to the calling script. OMERO.grid processors can exist within OMERO.server or on remote systems. Access to remote clients is through ZeroC’s Internet Communications Engine (ICE; http://www.zeroc.com) framework, allowing communication with a wide variety of client environments using a single API. Hibernate objects and ICE definitions in Python, Java, and C++ are created through code generation (Fig. 1B).
Clients
OMERO provides a Java-based data upload (OMERO.importer) and a Java-based and web browser-based data visualization and analysis clients (OMERO.insight and OMERO.web, respectively). OMERO.importer uses Bio-Formats to read image data and metadata from all formats it supports. OMERO.insight and OMERO.web support visualization, annotation, analysis, and sharing of data in an OMERO installation.
Box 2. Computer and OME terminology.
Application Programming Interface (API)
A software function that provides access to functionality and processing without exposing the underlying complexity in the function. An API ’abstracts’ the technical details of a function but still allows access to it. API’s are usually accompanied by extensive documentation to define how to call a function, what parameters to use, etc. Documentation for the OMERO API is at http://trac.openmicroscopy.org.uk/ome/wiki/OmeroApi.
Client-server application
A two-part application, where one application (the server) stores data and delivers an easy-to-view version of the data to a separate, usually less powerful, application (the client). Data also flows from the client to the server. In most cases, data transfer occurs through an internet connection. A web server and web browser are one example.
Code generators
Software applications that read a specification or model and generate computer code that uses the specification.
Command line interface
A simple, non-graphical tool that allows access to an application (often through an API) by typed commands.
HDF5
A data storage mechanism that allows many different data types to be stored in a single file. Often described as a “whole filesystem in a single file”.
GNU General Public License (GPL)
One of a series of standard open source licenses. Allows users to make any changes to software, but requires any distribution of any derived work to be licensed under the GPL.
Interoperability
The ability of a software tool to interact with data and other software, even though it was not explicitly designed to do so.
Key-value pairs
A simple storage mechanism for data in a computer, where an attribute’s name and value are stored as a pair. A number of mechanisms exist to store key-value pairs. A key-value store is easy to extend—new values are just added to the bottom on the list.
Metadata
Often referred to as “data about the data”, this refers to additional information that, while not strictly part of the recorded data defines, how the data were recorded. In some cases, metadata are absolutely necessary to work with the associated data, e.g., the height and width dimensions of an image.
Middleware
Software that mediates communication between many data sources (sometimes referred to as the “front-end”) and one or more databases (the “back-end”).
OME Data Model & OME-XML
An open specification for microscope image data that defines the elements and relationships for microscope image metadata3. The OME Data Model is expressed in Extensible Markup Language (XML), to generate OME-XML, which can be used by software tools as a data specification to generate databases and other applications. OME-XML can be easily stored in file headers to include a metadata specification in image files, e.g., OME-TIFF. Use of the OME Data Model and OME-XML is described at http://www.openmicroscopy.org/site/support/file-formats
OMERO.server, .tables, .importer, .dropbox, .insight, .web
These are components of the OMERO platform. OMERO.server is a middleware server application that provides access to image data and metadata stored in a series of databases. OMERO.tables is one of the databases used by OMERO.server and is an HDF5-based facility for storing large tabular arrays. OMERO.dropbox is a filesystem monitoring application that automatically recognises new files in a specific folder and then imports them into OMERO. OMERO.insight and OMERO.web are Java and web browser-based clients of OMERO.server, respectively, that provide access to data stored inside an OMERO installation.
OME-TIFF
An open scientific image file format that stories binary pixel data in the well-known TIFF format and stores OME-XML describing the image metadata in the TIFF header.
netCDF (network Common Data Form)
A series of open software libraries and protocols for storing scientific data in a programming language and operating system independent format.
Proprietary file formats
Closed, usually commercial data formats that contain recorded image data and instrument configuration metadata.
Relational database
A computer application that stores data in a series of tables, and uses declared relationships between those tables to calculate relationships in the data. A standard language used to query relational databases is Structured Query Language, or SQL.
Data Flow in OMERO
Data is imported into OMERO using Bio-Formats1. All metadata are read, and where possible, mapped to the OME Data Model3. Metadata is stored in the OMERO relational database, and binary pixel data is converted to an efficient binary-only file and stored within a file repository owned by the OMERO installation. To ensure data integrity Bio-Formats also converts all proprietary file format metadata into a table of key-value pairs that is then stored as an annotation on the imported image in OMERO. OMERO supports import using a desktop application (OMERO.importer), a command line interface, or a filesystem monitoring tool (OMERO.Dropbox). Alternatively, third parties can write their own, specialized importers. Any data written to an OMERO database is accompanied by an entry in an “Event” table, which can be used for auditing changes to an OMERO system.
Data Rendering and Display
As most modern biological images consist of multiple image frames recorded at different channels, focal positions, or timepoints, a fast, scalable image rendering tool is needed to display modern image data sets. OMERO.server contains a multi-threaded image-rendering engine that can rapidly render planes for display. These can be transferred, via the API to clients. Clients can choose different levels of on-the-fly JPEG compression to reduce image data size, apply overlays to an image before compression, or specify projection parameters such as maximum or mean intensity to provide a summary view of the image. Users can thus produce multiple views of the data without modifying the acquired data; the default and any user-defined values are stored in OMERO’s database. The rendering settings for other users are also stored, so that the owner of an image can see how co-workers have visualized the same data.
The Importance of a Common API
A standard desktop image processing application like ImageJ normally accesses individual image datasets on a filesystem and provides none of the data management facilities described above. Most commercial desktop imaging applications also access files on a filesystem, and some enable a single user to search for his or her images based on filename, acquisition date, tagging, etc. This level of data management is sufficient for a small research team who work with a limited range of data types. As the complexity and size of data grows, the software that accesses the data becomes more heterogeneous, the requirements for controlling access grows, and the complexity of derived and associated data increases. Building and maintaining the custom linkages between the various applications that directly access files and the results they generate becomes ever more difficult. The computational cost of search grows as well: scanning many data files for metadata values is inefficient because of the overhead of opening and closing each file. While sophisticated file storage and access mechanisms like HDF5 and netCDF5, so called ‘nascent databases’6, are now available, access to individual files by multiple clients is limited, and conflicts between simultaneous reading and writing will inevitably arise. These are problems that databases have been designed to solve, so it makes sense to use them when datasets grow and become more complex. Moreover, middleware applications that add value to data— access control, organization, annotation, indexing, rendering, and processing—will always be necessary.
In OMERO, our general strategy is to use whatever storage mechanism is appropriate and to record its location in the OMERO relational database for easy access and query. OMERO uses the advantages of HDF5 files, relational databases, and filesystems for different data types and exposes them all through a single, defined interface—the OMERO API. For large, complex datasets, this single interface provides a coherent point of access for client applications, regardless of their structure or design. As long as applications use the OMERO API, they can access and operate on the same data, whether or not they are explicitly designed to do so.
Supporting New Data Types in OMERO
In science, rapid developments in technology mean that the types and complexity of data are constantly evolving. Updating the whole OMERO application by hand each time a new mode of imaging appeared would be difficult and error-prone. We therefore constructed code generators, software tools that read the XML version of the OME Data Model (‘OME-XML’) and convert it into scripts that generate the OMERO database, the linkages between the OMERO database and the OMERO.server application (object-relational mapping, see Box 1) and the specifications for the OMERO API used by remote Python, Java, or C++ applications (Fig. 1B). The generated code is then compiled and used to store metadata in the OMERO relational database and to transmit the metadata to remote clients.
This approach is important as the core OME-XML model can be frequently updated to support new and more complex imaging modalities. Moreover, developers of OMERO client applications can easily identify changes and adapt to them—OME-XML specifies the data supported by an OMERO application. This approach balances a commitment to using commonly agreed and defined data types found in a community-based data specification and the need to adapt to newly emerging data types3 in the rapidly developing field of biological imaging.
We recently took advantage of this flexibility to add support for high-content screening (HCS) experiments to OMERO. These experiments, involving treatment of cells with small molecule or genome-wide RNAi libraries and measuring the effects by high-throughput imaging, generate hundreds or thousands of images, each with their own metadata. After consultation with many HCS labs, we revised OME-XML to include a specification for HCS data and used the new version of OME-XML to build a new version of OMERO. Additional development was still necessary to properly display the new data types in OMERO clients (e.g., displays of thumbnails as HCS plates), but manual changes to the underlying server application were minimised. As an example, support for HCS data was recently added to the OMERO-based Dataviewer application that the Journal of Cell Biology uses to provide readers access to raw image data associated with published papers (see below). With the rate that new imaging modes are developed and used, model-based code generation helps keep OMERO’s capabilities aligned with technical advances in the community and ensures that the community can easily determine the data types OMERO supports.
OMERO Clients—Data Access and Interoperability
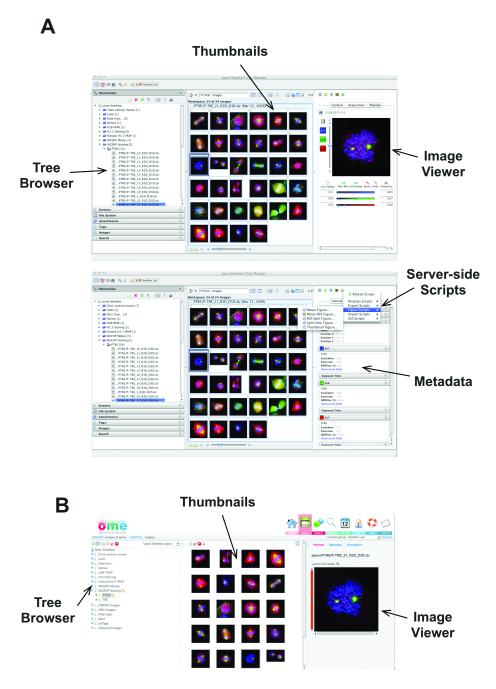
OMERO.server’s architecture delivers two capabilities that are critical for modern science, but rarely available for complex, multi-dimensional image data—remote access and interoperability. Harnessing this functionality requires client applications that run on a desktop computer, a laptop, or even a mobile device and communicate with an installation of OMERO.server. We have built a number of OMERO clients that enable the functionality we require for our research and have included them in the open source, released OMERO software. OMERO.importer and OMERO.insight are two Java-based desktop applications that enable image data import using Bio-Formats and data management, annotation, visualization and analysis (Fig. 2A). OMERO.web, a Django-based web application uses the OMERO Python API and provides access to OMERO’s data hierarchies, and enables annotation and visualization of data. OMERO.web exposes the annotation-based data sharing mechanism where user-defined sets of data can be shared and discussed with other users on the OMERO system (Fig. 2B). A full list of the functionality in these clients is available at the OME web site. OMERO clients can be used as they are, customised using the available source code, or used as examples to guide development of new OMERO client applications developed by others.
Figure 2. OMERO Clients.
(A) Different views of OMERO.insight showing the tree-based data browser, image thumbnails, and a view of a single image (top) and image metadata and annotations and access to server-side analysis scripts (bottom).
(B) OMERO.web client viewing same data as in (A).
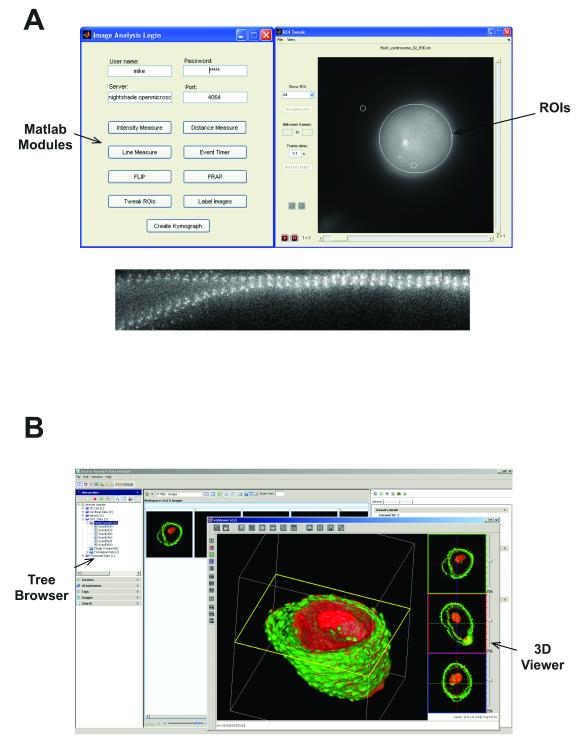
We have also demonstrated integration and re-use of other, third party software tools with specific bridges between OMERO and ImageJ and CellProfiler 1.07, two popular open source image processing tools, thereby making these tools clients of OMERO. Moreover, we have used an existing Java gateway in Matlab to read and write data through the OMERO API (see Supplemental Note). As an example, Figure 3A shows a series of image segmentation and analysis tools implemented in Matlab, analysing data in OMERO. We have also integrated VolViewer, an open source GPU-based 3D volume rendering tool8 with OMERO (Fig. 3B), enabling a powerful image rendering facility to access any data stored in OMERO (see Supplemental Note). All of these facilities are available within the released, open source version of OMERO.
Figure 3. External applications as OMERO Clients.
(A) Screenshots of custom applications written in Matlab that define and measure features of ROIs based on Otsu thresholding or calculate kymographs based on a user-defined region. Any processing or analysis facility within Matlab can be used for data stored within OMERO.
(B) VolViewer, a 3D GPU-based multi-channel volume rendering engine8 viewing image data obtained from an OMERO installation. VolViewer requires a GPU for processing, but implementations using VolViewer as a client of OMERO, or running as an OMERO service on a server with a GPU are available.
Supporting Analytic Data
Analysis of large primary datasets inevitably generates new sets of data. Acquired images may be processed to improve contrast and generate a second set of derived images, or analysed to generate calculated features derived from objects in the image—segmented objects, fitted functions, etc. In OMERO, generated images can be annotated and stored together with the original image. However, many analyses generate derived measurements that are separate from the images. In OMERO, these files (.xml, .xls, etc.) can be stored as ‘StructuredAnnotations’ in an OMERO file repository, which is linked from the OMERO relational database. The OMERO API exposes this linkage, allowing any OMERO client to access the original data and any derived measurements.
One use of OMERO’s StructuredAnnotations involves annotating data with specific ontological terms. Biological image data is still heterogeneous, and there are not yet defined specifications for annotating microscope image data. To help begin this process, we have implemented support for defined terms of the Open Biological and Biomedical Ontologies (OBO)9 within OMERO.server, and accessed them through the OMERO API as StructuredAnnotations. This implementation was used for the ASCB CELL Image Library (see below). We expect that as the use of these ontologies develops and software tools for image annotation improve, the specific ontologies used can be harmonised to start to enforce common annotations across all images in different domains. An approach similar to OMERO’s StructuredAnnotations is followed in Bisque, an alternative open source image management application10. In the future, it may be possible to standardise the annotations used in these and other image management applications to allow seamless communication between them.
In contrast to individual terms, many derived measurements are of the form of large tabular arrays and are often stored as “comma-separated value” (CSV) or spreadsheet files. In principle these data would be well-suited for inclusion in OMERO’s database, but since each lab or institute may use its own algorithms and structures for data storage, derived measurements often vary in structure and form. They are also often quite large-- a standard HCS use-case is the calculation of >1M measurements per plate (384 wells × 10 images/well × 25 cells/image × 20 calculated features/image). Importing many millions of calculated values into a relational database, in the absence of any specialised hardware or database configuration, imposes an unacceptable performance burden. In addition, dynamic updates to the database are possible11, but risk database corruption by anyone not fully adept at data modelling. For these reasons, we developed a simple flexible framework for storing tabular data and derived measurements known as OMERO.tables (see Supplemental Note). This facility uses an HDF5-based data store accessed through the OMERO API to provide a flexible structure for naming, storing and accessing data stored in tables within an OMERO.server installation (see Supplemental Note).
In some cases, it may be appropriate to store data in completely defined, relational models. One example is regions-of-interest (ROIs) that describe the boundaries of objects defined by manual or automatic object definition or segmentation methods. As ROIs are used in many imaging applications, OMERO includes support for 5D ROIs (space, time, and channel) in its core model, and thus within the OMERO database. Using this facility, any tools that define objects can save them to the OMERO database, making them available to any other tool that can analyse or process these objects.
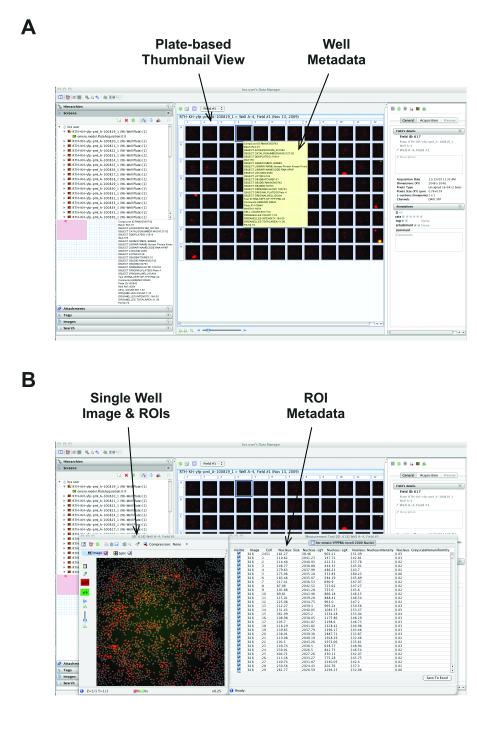
An example of a combined use of the OMERO database, StructuredAnnotations, and OMERO.tables occurs in support for HCS data. Figure 4 shows a view of a plate from a targeted siRNA screen focusing on modifiers of SUMOylation. The acquired image data was analysed using custom third-party software that generated tabular output in separate CSV files, which are stored in OMERO as StructuredAnnotations on the Screen. In addition, the data identifying gene name, treatment, etc. for each well are stored in OMERO.tables, and viewable by mousing over each individual well (Fig. 4A).
Figure 4. Experimental, heterogeneous data stored and viewed in OMERO.
(A) Screenshot of OMERO.insight showing data from a targeted siRNA screen recorded on an InCell 1000 platform. Images are stored in OMERO’s file store and acquisition metadata are stored in the OMERO relational database. Experimental metadata defining gene name, siRNA and other experimental parameters were originally stored in CSV files and imported into OMERO using the Annotation service, and stored in the OMERO.tables HDF5-based store.
(B) Screenshot of OMERO.insight showing view of data of single field from single well from dataset in (A). Choosing any well thumbnail opens an image view window (Fig. 4B), and viewing the measurements in that image reveals ROIs calculated by proprietary software on the InCell platform, stored in the OMERO DB and visualised as overlays on the displayed image. Calculated features from each ROI are stored in the OMERO.tables HDF5-based store, and displayed alongside the ROIs.
Foundation for Flexible Large-scale Processing
A useful data management system must provide a mechanism to initiate, run, and monitor analyses of the data it holds. OMERO provides this facility through its Scripting Service. Python scripts are loaded on the server, registered in the OMERO relational database, and then exposed to the clients through the OMERO API. Scripts can be called from a client and run either within the server or distributed to external compute resources. Scripts can run functions themselves, or wrap libraries written in other environments (e.g., C++ and Java). The current OMERO distribution ships with demonstrator scripts for image export, projection, and stitching.
As the size and complexity of analysis applications grows, the efficient identification and recall of metadata becomes limiting in large-scale calculations. The case of HCS provides one example of this problem. One HCS plate commonly consists of 3840 images, each 2 MB in size (1024 × 1024 pixels × 2bytes/pixel). Distributing the segmentation and feature calculations for all the images in one plate across a similar number of cores is possible, but the simultaneous I/O calls for thousands of image files can cause performance issues. Some throttling of job distribution is thus necessary, and this requires access to metadata since throttling strategies will depend on image size and calculation type. Meta-calculations are therefore required to define the structure and the performance of the main calculation. This can only be achieved if metadata are linked to the original data and are easily retrievable, as in OMERO.
Supporting Non-Image Data
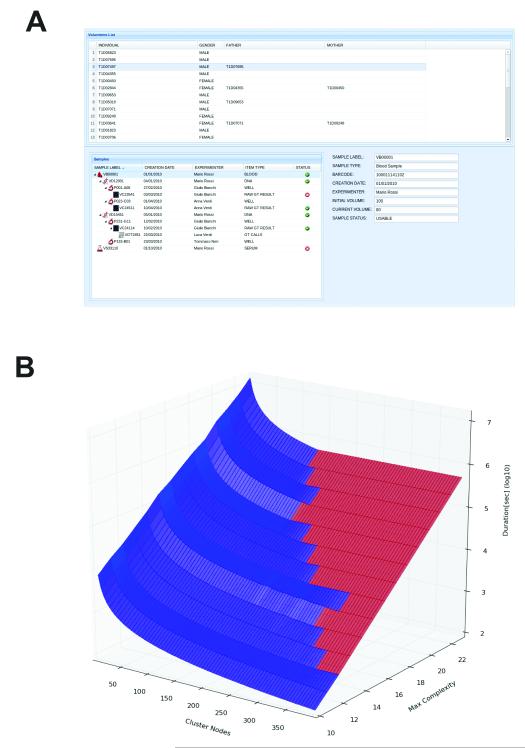
Thus far, we have focussed on solutions for image data and the range of data types associated with image analysis. However, the data management and remoting facilities in OMERO are required across life sciences research for other data types. As an example, OMERO has been customised to support large scale, high-resolution Genome Wide Association Studies (GWAS) of human autoimmune diseases12 simply by adding specialized model types pertaining to genotyping and clinical health records (Fig. 5A; see Supplemental Note). We chose this implementation to solve a critical data processing bottleneck that occurs when extrapolating experimentally obtained genotyping data13 to combine and impute missing genotypes for a set of individuals by using information available on their relatives14. The duration of a single imputation run depends strongly – from minutes to weeks on a standard computing node -- on the complexity of the family tree structure. In a large-scale population study it is common to have batches of hundred of thousands of runs for an imputation calculation. As most compute resources are shared, the efficient distribution and monitoring of compute jobs are crucial. Without accurate planning of compute resource allocation, large numbers of jobs can be started on a compute cluster with no improvement in the total run time. Figure 5B shows how the expected time needed for an imputation computation for 6,863 genotypes depends on the number of computational nodes N used and the maximal family “bit complexity” (BC, a measure of family pedigree complexity). The boundary between blue and red data points represents the set of optimal pairs of BC and N. For a given BC, increasing the cluster size beyond the relative optimal N value does not improve overall run time. Using the pedigree metadata is absolutely critical for efficient use of computational resources. Processing metadata and setting up a calculation within OMERO takes minutes at most, whereas directly reading metadata from the raw data files (ped files, ~20 GB each) takes many hours.
Figure 5. Non-Image Data and the Use of Metadata for Large-Scale Computations.
(A) Alternative use of OMERO for storing and tracking clinical laboratory specimens and genotype data. Sample type, creation date, processing, assay results, genotyping results, and any familial relationships are all recorded. The modelling for this system is based on OpenEHR archetypes. See Supplemental Note for more details.
(B) Performance of a SNP imputing calculation as a function of pedigree complexity (an index of the number of members of a genetically related family) and compute nodes used to perform the calculation. Low complexity imputation benefits from increasing the number of compute nodes, and only occupies nodes for relatively short periods. High complexity calculations take orders of magnitude longer, and do not benefit from adding more nodes. The point where adding number of nodes provides no improvement in performance is shown by the interface between the blue and red domains. Time and cost-efficient use of compute resources therefore depends on access to metadata, determination of complexity, and planning of processing strategy before initiating the imputing calculation.
OMERO-based Image Repositories
OMERO was originally built to support management of image data at individual sites—laboratory or department imaging facilities that required a data management facility. However, the same architecture can provide the foundation for public image data repositories. OMERO has been used as the foundation for two public repositories, the JCB DataViewer15 and the ASCB’s CELL Image Library. Both applications use a customised import strategy based on Bio-Formats, and use OMERO to store, manage, and provide access to image data. The ASCB CELL Image Library collects images via public submission, and following on well-established methods in the genomics community, uses a team of human annotators to apply terms from a series of defined ontologies to the submitted images. In this case, the ontological terms are represented in XML, stored within OMERO.server, and then exposed within a custom web browser-based user interface.
To demonstrate the flexibility of OMERO and the OME Data Model, we imported data from the Electron Microscopy Data Bank16 into OMERO and extended OMERO.web’s data viewing capabilities to include display of fitted models from single particle reconstructions from electron microscopy (EM) data (see Supplemental Note). A modified version of OpenAstexViewer is used for 3D map viewing, and facilities for search and display of EM metadata in customized frames were also added. No changes were needed to the underlying OMERO relational database or API for this conversion, demonstrating that OMERO’s underlying model is generic enough to handle most types of biological imaging. As we continue to develop the underlying OME Data Model, EM-specific metadata can be directly added to and accessed within the OMERO database.
Discussion
OMERO is a flexible, scalable image data management system that provides storage, access, processing, and visualization of a diverse set of data types. We used model-based code generation techniques to ensure that extension beyond OMERO’s original intended use in light microscopy is straightforward and we adapted it for use in HCS, EM, scientific image repositories and GWAS studies. While this involved changes to the client user interfaces, addition of many new metadata types was enabled with little change to the components of OMERO.server. This flexibility simplifies software maintenance and allows OMERO to serve as the foundation for a range of image management and analysis applications (Figs. 2, 3 and 4), public image repositories, and analysis of genomic data from large populations (Fig. 5).
The goal of OMERO is not to provide a full toolkit of data analysis tools, as there are already many examples of these-- ImageJ17, ITK/VTK18, R, and Matlab are just a few. Instead, OMERO provides common interfaces for these and any other application to access all types of data supported by OMERO and Bio-Formats, independent of the specific programming environment used. OMERO clients enable remote access to large, complex datasets so that data can be viewed and analysed in remote locations, through secure, authenticated connections and allow collaborators with appropriate access privileges to view, annotate, and analyse data as well. We intend the released OMERO clients to serve as a starting point for other clients and utilities developed by others, to suit their own needs.
Using OMERO
OMERO is an open source software application, and available for download at http://openmicroscopy.org. Extensive documentation is available, and support for installation, configuration, implementation and usage is also available using mailing lists and public forums. As of this writing (January 2012), OMERO is installed at 3160 sites worldwide, and in regular use at ~500 sites. The software is available under the GPL license, which allows any customization for local needs.
In many laboratories, the scale of data collected and analysed is small, or the need to share data between users, sites, or even different analysis tools is limited. For these cases, powerful, well-developed commercial and open source desktop processing tools for imaging are available (ImageJ17, Osirix19, etc.) are certainly the preferred solutions—the simple data management capabilities available in desktop applications are sufficient and the burden of running a full enterprise-level data management application is not justified. Enterprise-level data management applications are complex and an experienced informatics staff person and significant computational and data storage capacity are required for production use. However, the level of automation and thus the scale and complexity of data routinely collected in laboratories are rapidly increasing (e.g., machine learning tools to automatically record datasets according to specific criteria20), making the use of appropriate data management applications imperative.
OMERO and other applications that follow similar design principles provide open, flexible solutions for these challenges. Open source data management applications for specific domains have been developed, e.g., HCDC for HCS (http://hcdc.ethz.ch) and a number of picture archive systems (“PACS”) for medical imaging (http://www.osirix-viewer.com; http://www.dcm4che.org/). In general, these are technically simpler systems—they use standard web services techniques for remote access to data and store files and metadata on a filesystem. Like OMERO, Bisque uses Bio-Formats to read image data, a relational database for data storage, a tiered architecture like OMERO for its server application, a flexible annotation structure to adapt to complex heterogeneous data, and a web interface for remote client access10. By contrast, OMERO’s ICE-based framework remoting interface is many times faster than web services and avoids scaling problems associated with filesystem-based applications. OMERO is demonstrated to scale to very large multi-terabyte datasets across a wide range of biological applications and is used in a variety of publicly available applications. By combining facilities for large-scale data management and a flexible, model-based architecture, OMERO provides a foundation for many different data management challenges, especially where large, heterogeneous data sets must be accessed, viewed, analysed and shared.
Supplementary Material
Acknowledgements
We thank Dr Emma Hill for helpful comments on the manuscript. Work on OME, Bio-Formats and OMERO in JRS’s lab is supported by the Wellcome Trust (Ref: 085982 & 095931) and the BBSRC (Ref: BB/G022585 & BB/I000755). Work on OMERO EMDB at EBI is supported by the BBSRC (Ref: BB/G022577). Work on OMERO at the John Innes Center, Norwich is supported by JISC (Ref: REXDAT03).
References
- 1.Linkert M, et al. Metadata matters: access to image data in the real world. J. Cell. Biol. 2010;189:777–82. doi: 10.1083/jcb.201004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swedlow JR, Eliceiri KW. Open source bioimage informatics for cell biology. Trends Cell Biol. 2009;19:656–660. doi: 10.1016/j.tcb.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg IG, et al. The Open Microscopy Environment (OME) Data Model and XML File: Open Tools for Informatics and Quantitative Analysis in Biological Imaging. Genome Biol. 2005;6:R47. doi: 10.1186/gb-2005-6-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swedlow JR, Goldberg IG, Eliceiri KW. Bioimage Informatics for Experimental Biology. Annu Rev Biophys. 2009;38:327–346. doi: 10.1146/annurev.biophys.050708.133641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dougherty MT, et al. Unifying biological image formats with HDF5. Commun. ACM. 2009;52:42–47. doi: 10.1145/1562764.1562781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray J, et al. Scientific data management in the coming decade. Microsoft Research, Inc.; Redmond, WA: 2005. [Google Scholar]
- 7.Carpenter A, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee K, et al. Visualizing Plant Development and Gene Expression in Three Dimensions Using Optical Projection Tomography. The Plant Cell Online. 2006;18:2145–2156. doi: 10.1105/tpc.106.043042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith B, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotech. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kvilekval K, Fedorov D, Obara B, Singh A, Manjunath BS. Bisque: a platform for bioimage analysis and management. Bioinformatics. 2010;26:544–52. doi: 10.1093/bioinformatics/btp699. [DOI] [PubMed] [Google Scholar]
- 11.Swedlow JR, Goldberg I, Brauner E, Sorger PK. Informatics and quantitative analysis in biological imaging. Science. 2003;300:100–2. doi: 10.1126/science.1082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanna S, et al. Variants within the immunoregulatory CBLB gene are associated with multiple sclerosis. Nat. Genet. 2010;42:495–497. doi: 10.1038/ng.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Browning SR. Missing data imputation and haplotype phase inference for genome-wide association studies. Human genetics. 2008;124:439–50. doi: 10.1007/s00439-008-0568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Willer C, Sanna S, Abecasis G.a. Genotype Imputation. Ann. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill E. Announcing the JCB DataViewer, a browser-based application for viewing original image files. J. Cell Biol. 2008;183:969–970. [Google Scholar]
- 16.Lawson CL, et al. EMDataBank.org: unified data resource for CryoEM. Nucleic Acids Res. 2011;39:D456–64. doi: 10.1093/nar/gkq880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 18.Yoo TS, et al. Engineering and Algorithm Design for an Image Processing API: A Technical Report on ITK - The Insight Toolkit. In: Westwood J, editor. Proc. of Medicine Meets Virtual Reality. IOS Press; Amsterdam: 2002. pp. 586–592. [PubMed] [Google Scholar]
- 19.Rosset A, Spadola L, Ratib O. OsiriX: An Open-Source Software for Navigating in Multidimensional DICOM Images. Journal of Digital Imaging. 2004;17:205–216. doi: 10.1007/s10278-004-1014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conrad C, et al. Micropilot: automation of fluorescence microscopy-based imaging for systems biology. Nat. Methods. 2011;8:246–9. doi: 10.1038/nmeth.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.