Abstract
RNA silencing in Caenorhabditis elegans is transmitted between cells by the transport of double-stranded RNA (dsRNA). The efficiency of such transmission, however, depends on both the cell type and the environment. Here, we identify systemic RNAi defective-3 (SID-3) as a conserved tyrosine kinase required for the efficient import of dsRNA. Without SID-3, cells perform RNA silencing well but import dsRNA poorly. Upon overexpression of SID-3, cells import dsRNA more efficiently than do wild-type cells and such efficient import of dsRNA requires an intact SID-3 kinase domain. The mammalian homolog of SID-3, activated cdc-42–associated kinase (ACK), acts in many signaling pathways that respond to environmental changes and is known to directly associate with endocytic vesicles, which have been implicated in dsRNA transport. Therefore, our results suggest that the SID-3/ACK tyrosine kinase acts as a regulator of RNA import into animal cells.
Intercellular signaling can occur through the transport of RNA between cells (reviewed in ref. 1). In some animals such transport occurs in response to the expression or ingestion of double-stranded RNA (dsRNA) and in plants the transport of endogenous RNAs between cells can mediate epigenetic effects during development. The import of dsRNA and mobile silencing RNA into Caenorhabditis elegans cells occurs via a conserved dsRNA transporter called systemic RNAi defective-1 (SID-1) (2) and mammalian homologs of SID-1 have been reported to import dsRNA into cells (3, 4). Despite this conservation of import mechanisms and the detection of the transfer of RNAs between cultured mammalian cells (5), the intercellular transport of RNA has not been robustly detected in mammals. In C. elegans, however, transport of silencing RNAs is easily observed from neurons, muscles, and the gut to other tissues (6). However, the efficiency of such transport varies between cell types (6) and is influenced by environmental conditions, such as starvation (2) or the presence of excess environmental dsRNA (7). Furthermore, this variation may underlie the inability to detect RNA transport between C. elegans cells in some studies (8, 9). Similar differences have also been reported in Drosophila: silencing triggered by injected dsRNA spreads throughout the fly (10) but silencing triggered by the expression of dsRNA in specific tissues does not (11). A possible explanation for this variation in the detection of RNA transport between cells is that there are mechanisms that interfere with such transport. Therefore, the identification of signaling pathways that control the efficiency of RNA transport could both enable the discovery of mechanisms that modulate RNA transport in response to the environment and enhance the reliable detection of RNA transport between cells in animals.
Results
sid-3 Encodes the C. elegans Ortholog of Mammalian Activated cdc-42–Associated Kinase.
We previously used a genetic screen in C. elegans to identify genes that control the transport of RNA between cells (2). These systemic RNAi defective (sid) mutants comprise three large complementation groups: sid-1, sid-2, and sid-3. The sid-1 gene encodes a conserved membrane protein with multiple transmembrane domains that imports silencing RNAs into cells (2, 6, 12, 13); sid-2 encodes a single-pass transmembrane domain protein that is required for the import of ingested dsRNA across the gut luminal membrane (14). In this study, we present the analysis of sid-3.
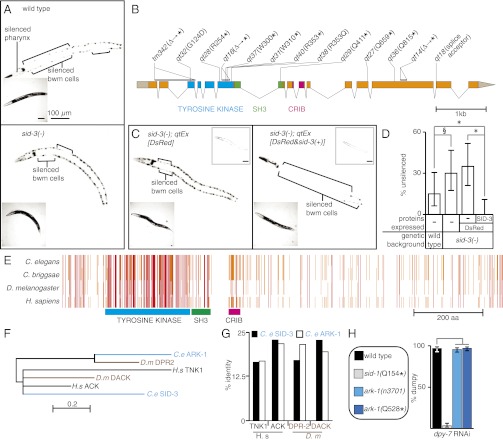
For the sid screen (2), we created a transgenic strain that allows for rapid discrimination between mutants simply defective for RNAi and mutants defective in the transport of RNA silencing signals. Animals of this strain express GFP in two tissues, the pharynx and the body wall muscle (bwm) cells, and also express gfp-dsRNA in the pharynx. In wild-type animals the pharyngeally expressed gfp-dsRNA silences pharyngeal GFP expression and, via the transport of silencing signals, also silences some anterior bwm cell GFP expression. When these animals are grown on bacteria that express gfp-dsRNA, then all anterior and posterior bwm cells are also silenced (Fig. 1A, Upper), making the strain suitable for large-scale phenotypic screening. In RNAi-defective mutants, silencing is not observed in any tissue, but in mutants defective in the uptake of dsRNA from the environment (e.g., sid-2), strong silencing of GFP expression is observed in pharyngeal and some anterior bwm cells but not in posterior bwm cells. In systemic RNAi mutants (e.g., sid-1) the strong silencing of pharyngeal GFP remains intact, but silencing in bwm cells, which is dependent on mobile silencing signals, is not observed. All isolated sid-3 mutants (14 alleles) appeared similar to sid-1 mutants in that silencing of the bwm cell GFP expression was greatly reduced but, unlike sid-1–null mutants, was not eliminated (Fig. 1A, Lower). In addition, GFP expression in the pharynx was silenced to a greater extent in sid-3(−) animals than in wild-type animals (Fig. 1A). Taken together, our results suggest that sid-3 mutants are defective in the transport of silencing RNAs.
Fig. 1.
sid-3 encodes a tyrosine kinase that regulates mobile RNA silencing. (A) sid-3(−) animals are defective in systemic RNAi. Representative wild-type (Upper) and sid-3(−) (Lower) animals where GFP expression in the pharynx and bwm cells is silenced (brackets) because of both pharynx-expressed and ingested gfp-dsRNA. Insets are brightfield images. (B) Gene structure of sid-3 showing mutations in isolated sid-3(−) alleles and the deduced changes in the SID-3 protein. ∆, deletion; *, stop codon. sid-3(tm342) was obtained from the C. elegans stock center. The tyrosine kinase domain (blue), SH3 domain (green), and CRIB domain (pink) are shown. (C) The silencing defect in sid-3(−) animals is rescued by sid-3(+) expression. Unlike expression of the fluorescent protein DsRed alone (Left, qtEx[DsRed]), coexpression of sid-3(+) (Right, qtEx[DsRed&sid-3(+)]) results in robust silencing in the bwm cells of sid-3(−) animals. Left Insets are brightfield images and Right Insets are red channel images. (D) Penetrance of silencing depicted in A and C. For each strain, the proportions of animals that lack detectable silencing are shown. Error bars indicate 95% confidence intervals. n = 40; *P < 0.05; §P = 0.054. (E) Schematic of multiple sequence alignment of SID-3 with its orthologs from Caenorhabditis briggsae, Drosophila melanogaster, and Homo sapiens. Known domains as in B and amino acid residues identical in three (orange) or in four (red) species are shown. (F) Phylogenetic relationship between the kinase domains of SID-3 and that of its paralog and homologs. Scale indicates amino acid substitution rate. (G) Similarity across the whole protein between SID-3, its paralog, and homologs in other species. SID-3 (black) shows greater identity than ARK-1 (white) to the human and fly ACK homolgs. (H) ark-1 mutants do not have a significant defect in silencing the skin gene dpy-7 by feeding RNAi. See Fig 2 for details of the assay.
We used two-factor mapping to narrow the location of the sid-3 mutation to a small region of the C. elegans genome. To determine whether these recombinants were Sid, we used the highly penetrant resistance of sid-3 mutants to feeding RNAi of GFP (Fig. 1A) or of the germ-line and embryonic gene pal-1 (15). This mapping placed sid-3 to the right of the visible marker unc-3 on the X chromosome (Fig. S1A) in a region with significantly reduced recombination (16). To enable the isolation of recombinants in this region, we used a dpy-28(−)/dpy-28(+) background, which increases the crossover frequency in this region (16). Using this approach, we isolated 286 recombinants between the N2 and CB4856 polymorphic strains that had recombined to the right of unc-3 (Fig. S1A). Analysis of these recombinants narrowed the region containing sid-3 to the terminal 588 kb of the X chromosome (Fig. S1A). To identify DNA sequence changes within the sid-3 region associated with the sid-3(qt40) allele, we sequenced the genomic DNA from sid-3(qt40) animals using whole-genome sequencing (17). We identified a point mutation that results in a premature stop codon in the gene B0302.1 (Fig. S1 B and C), the exon-intron structure of which (Fig. 1B) is well supported by RNA-Seq data for each developmental stage (Fig. S2). We next sequenced DNA corresponding to the exons and exon-intron junctions of B0302.1 from 12 additional sid-3 mutant alleles. In all, 9 of the 13 alleles had point mutations or small deletions that result in premature stop codons in the corresponding protein; one had a splicing acceptor mutation; and two others had missense mutations that drastically altered conserved residues (Gly to Asp and Arg to Gln). (Fig. 1B and Figs. S3, S4, and S5). These observations suggest that severe disruption of the SID-3 protein is required to cause a readily detectable defect in mobile RNA silencing in our genetic screen. Confirming the gene identity of sid-3, we found that overexpression of genomic DNA containing B0302.1 along with its promoter and 3′ UTR regions robustly rescued the silencing defect in sid-3(−) animals (Fig. 1C, compare Left and Right). Interestingly, compared with wild-type animals, sid-3(−) animals that overexpress sid-3(+) showed silencing of GFP expression in a greater number of bwm cells (expressivity) and showed robust silencing in a larger fraction of the animals (penetrance) (Fig. 1D). Thus, B0302.1 encodes SID-3 and the extent of silencing because of mobile silencing RNA depends on the level of SID-3 expression.
The sid-3 gene encodes a conserved protein with a tyrosine kinase domain, a Src homology 3 (SH3) domain, and a Cdc42/Rac interactive binding (CRIB) domain (Fig. 1 B and E, and Fig. S4). This domain composition is present in the activated Cdc-42–associated kinase (Ack) family of cytoplasmic tyrosine kinases (Fig. 1E and Fig. S4) (reviewed in ref. 18). C. elegans contains two members of this broadly conserved family of tyrosine kinases: SID-3 and Ack related kinase-1 (ARK-1). Phylogenetic analysis suggests that the kinase domain of SID-3, rather than that of ARK-1, is more closely related to human ACK (Fig. 1F). Although both ARK-1 and SID-3 are more similar to ACK than the other human Ack-family protein TNK1 (Fig. 1G), ark-1(−) animals do not have a defect in silencing because of mobile RNA (Fig. 1H). To test whether ACK can rescue the silencing defects in sid-3(−) animals, we transformed sid-3(−) animals with a mouse ACK cDNA under the control of either the sid-3 promoter or a bwm promoter (see SI Materials and Methods for details). In both cases, we failed to obtain viable transgenic lines but observed larval and embryonic lethality in the progeny of injected animals. Thus, although it remains unclear whether ACK can functionally replace SID-3 in systemic RNAi, the sequence similarity and analysis of ark-1(−) animals suggest that SID-3 is the only C. elegans ortholog of the mammalian ACK protein with a defect in the transport of mobile RNA.
SID-3 Is Required for Efficient Feeding RNAi in all Tested Tissues and Is Localized in the Cytoplasm of Many Tissues.
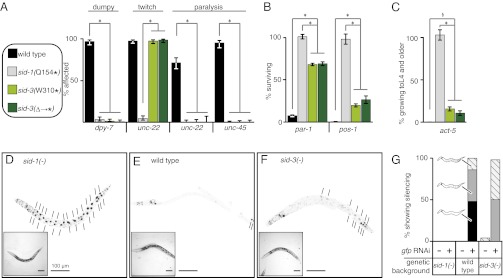
To test whether sid-3(−) animals have defects in RNA silencing beyond the inability to silence GFP expression in bwm cells in response to mobile RNA, we examined their response to feeding RNAi of genes expressed in multiple tissues. Using two outcrossed sid-3 null mutants [sid-3(W310Stop) and the sid-3(tm342) deletion that leads to a premature stop codon], we tested the requirement for sid-3 to silence skin- (dpy-7), muscle- (unc-22 and unc-45), germ line- (par-1 and pos-1), and intestine-expressed (act-5) genes (Fig. 2 A–C). In all cases, sid-3 null mutants were measurably defective in silencing the target gene, although the severity of the defect varied among tissues and was often less penetrant than sid-1–null mutants (e.g., germ-line and intestinal genes). In particular, sid-3(−) animals were only mildly defective in silencing the intestine-expressed gene act-5. This observation suggested that, unlike SID-1 or SID-2, the SID-3 protein may not be required for the import of ingested dsRNA into intestinal cells but rather is required for the subsequent export of dsRNA from intestinal cells to internal tissues. Because the sur-5::gfp transgene expresses nuclear-localized GFP in all somatic cells, which is easily observed in the large intestinal nuclei, we measured the extent of GFP silencing in intestinal nuclei of individual sid-3(−);sur-5::gfp animals in response to gfp feeding RNAi. We found that sid-3(−) animals clearly showed a measurable defect in silencing GFP expression in intestinal cells, although they were more silenced than the completely defective sid-1(−) animals (Fig. 2 D–G). In summary, these results suggest that because SID-3 is required to fully silence genes expressed in many tissues, including intestinal cells in response to ingested dsRNA, sid-3 is likely to function broadly to control silencing in most tissues.
Fig. 2.
sid-3 mutants are defective for feeding RNAi in all tested tissues. (A–C) Feeding RNAi of endogenous genes. Fourth larval stage (L4) animals of wild-type, sid-1(−), and two sid-3(−) strains with mutations that result in early stop codons [sid-3(W310*) and the tm342 deletion sid-3(∆→*)] were fed bacteria expressing dsRNA that target skin (dpy-7), muscle (unc-22 and unc-45) (A), embryonic (par-1 and pos-1) (B), or intestinal (act-5) genes (C). Percentage of affected (for skin and muscle genes) or surviving (for embryonic and intestinal genes) progeny are shown. Error bars indicate 95% confidence intervals. n ≥ 100; *P < 0.05; §sid-3(W310*) vs. wild-type (P = 0.02) and sid-3(∆→*) vs. wild-type (P = 0.053). (D–G) Feeding RNAi of gfp expression in transgenic animals. L4 animals that express GFP in all somatic nuclei (sur-5::gfp) in sid-1(−) (D), wild-type (E), or sid-3(tm342) (F) backgrounds were fed bacteria expressing gfp-dsRNA. Short lines indicate unsilenced gut nuclei (black). Proportions of progeny that show increasing extents of silencing (hatch < gray < black) are indicated along with representative schematics (G). n = 25–50 worms; Insets are brightfield images. (Scale bars, 100 μm.)
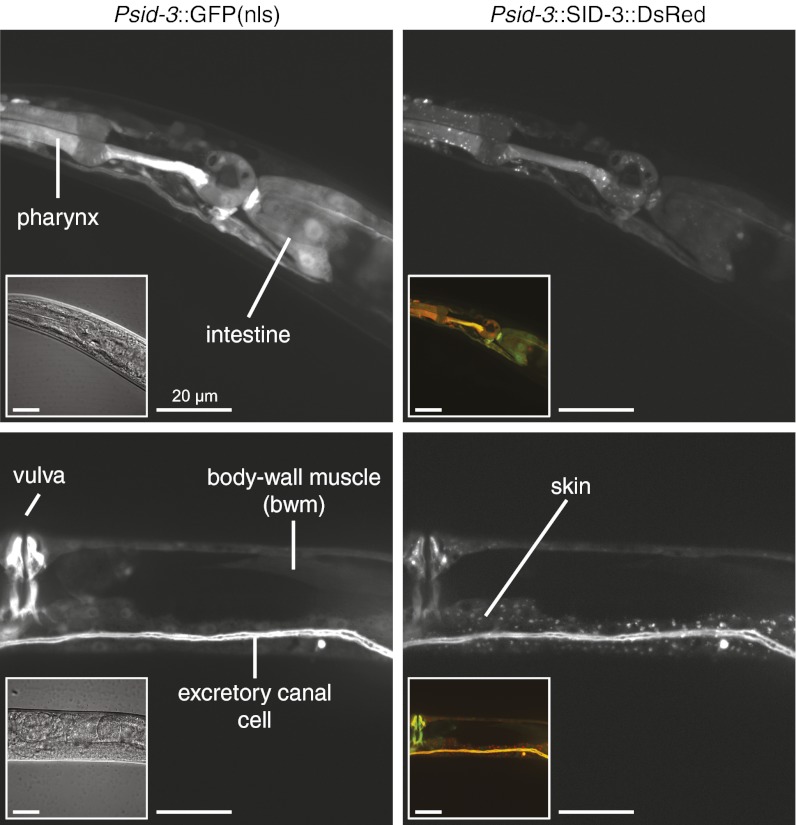
To determine the tissues that express the sid-3 gene, we constructed reporters designed to express either GFP or DsRed under the control of the sid-3 promoter and 3′ UTR regions that were used to rescue sid-3(−) in Fig. 1. These transcriptional reporters were expressed in multiple distinct cell types, including the gut, pharynx, bwm, skin, and excretory canal cells (Fig. 3). To examine the subcellular localization of the SID-3 protein, we generated translational reporter constructs designed to express a SID-3::DsRed fusion protein under the control of the same sid-3 promoter and 3′ UTR regions used above. The SID-3::DsRed fusion protein rescued the silencing defects in sid-3(−) animals (100% of animals expressing SID-3::DsRed showed silencing upon dpy-7 feeding RNAi) and, similar to the mammalian Ack proteins (19), was localized within the cytoplasm of cells in a diffuse as well as punctate pattern in all examined tissues (Fig. 3). To ensure that the punctate localization of SID-3::DsRed was not a result of nonspecific aggregation induced by fusion to the DsRed protein, we similarly generated a SID-3::GFP fusion protein. This fusion protein was also broadly expressed and localized to the cytoplasm of cells in a diffuse as well as punctate pattern (Fig. S6).
Fig. 3.
SID-3 is a widely expressed cytoplasmic protein. Fluorescence images of animals that coexpress nuclear-enriched GFP (Left) and a rescuing SID-3::DsRed fusion protein (Right) under the control of the sid-3 promoter and 3′ UTR. Left Insets are differential interference contrast images and Right Insets are merged red and green channel images. Fluorescence from SID-3::DsRed fusion was detected diffusely throughout the cytoplasm and in cytoplasmic foci. Similar diffuse and focal expression was also observed using a SID-3::GFP fusion protein (Fig. S6). Note that extrachromasomal arrays, which express the fluorescent proteins above, are lost mitotically, resulting in mosaic expression. For the more stable extrachromosomal arrays, the mosaic expression patterns largely match the known cell lineage. (Scale bars, 20 μm.)
These results suggest that the SID-3 protein likely functions in the cytoplasm of most tissues to enable efficient RNAi in response to ingested dsRNA.
SID-3 Is Not Defective in Cell-Autonomous RNAi but Is Defective in the Transport of RNA Between Cells.
This broad distribution of SID-3 could reflect that SID-3 is necessary for the efficient execution of RNAi (cell-autonomous RNAi) in all tissues or that SID-3 is necessary for the efficient transport of dsRNA to all tissues. The lack of a silencing defect in pharyngeal cells that express dsRNA (Fig. 1B) suggests that the ability to execute RNAi is not compromised in sid-3(−) animals. However, because the level of dsRNA expression in the pharynx is unknown, a high level of dsRNA expression in the pharynx could mask a mild defect in the execution of RNAi in sid-3(−) animals.
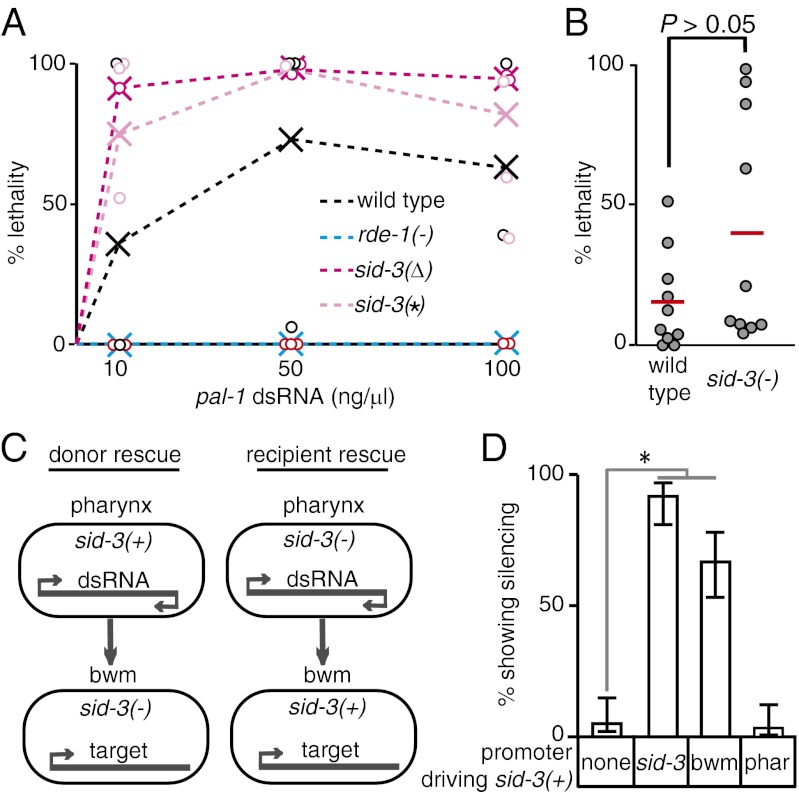
To clearly distinguish defects in the execution of RNAi from defects solely in the transport of dsRNA, we measured the response of sid-3(−) animals to known amounts of dsRNA injected into cells. Specifically, we injected different concentrations of dsRNA targeting an embryonic gene (pal-1) directly into both gonad arms of animals and measured the fraction of progeny that showed embryonic lethality (Fig. 4A). The germ line within the gonad is syncytial and so does not require dsRNA transport for silencing. Wild-type animals injected with increasing concentrations of pal-1-dsRNA laid an increasing proportion of dead progeny. The lethality was specifically caused by RNAi because no lethality was observed at any of the concentrations tested in rde-1(−) animals, which are resistant to RNAi. In this assay, sid-3 mutants laid a greater proportion of dead progeny than did wild-type animals at all dsRNA doses. This finding suggests that cell-autonomous RNAi may be enhanced in sid-3(−) animals. To test this possibility rigorously we used a single needle containing a limiting but identical concentration (10 ng/μL) of pal-1–dsRNA to inject both gonad arms of 10 wild-type worms and 10 sid-3(−) worms. We then measured the proportion of dead progeny laid by each doubly injected animal (Fig. 4B). Although sid-3(−) animals appear marginally more sensitive to silencing than wild-type animals, these results were statistically indistinguishable (P > 0.05 Mann–Whitney U test). Nevertheless, these results clearly demonstrate that sid-3 mutants are not defective in the execution of RNAi. Furthermore, the minor enhanced RNAi observed in response to injection of dsRNA into the germ line and the enhanced silencing of pharyngeal GFP because of gfp-dsRNA expression within the pharynx (Fig. 1A) suggest that sid-3 mutants may even be enhanced for the execution of RNAi. Therefore, our results are consistent with the idea that the partial silencing defect in response to mobile RNA observed in sid-3(−) animals solely reflects a defect in RNA transport between cells.
Fig. 4.
Efficient import of dsRNA requires sid-3. (A) Response to different concentrations of dsRNA injected into the germline. Adult animals of wild-type, rde-1(−), and two sid-3(−) genotypes [sid-3(W310*)–sid-3(*) and the tm342 deletion–sid-3(∆)] were injected with a similar volume of the indicated pal-1 dsRNA concentrations. pal-1-RNAi is embryonic lethal. The proportion of dead embryos laid by each injected animal (circle) and the average pooled proportion of dead embryos for each concentration and genotype (X) is plotted. (B) Response to limiting amounts of dsRNA (10 ng/μL) injected into the germ line of wild-type and sid-3(W310*) animals. Red bars and circles indicate average and individual proportions of dead embryos laid, respectively. P value is based on Mann–Whitney U test. (C) Schematic of experiment to test the role of SID-3 in exporting and importing tissues. (D) SID-3 is not required in the exporting tissue but is required in the importing tissue for silencing because of mobile RNA. sid-3(−) animals that express GFP in the pharynx and in bwm cells but express gfp-dsRNA only in the pharynx were transformed with constructs that express sid-3(+) under the control of its own (sid-3), bwm-specific (bwm), or pharynx-specific (phar) promoter and the percentage of transgenic animals that show gfp silencing in bwm cells was determined. Error bars indicate 95% confidence intervals. n = 100 L4 animals; *P < 0.05.
SID-3 Is Required for the Import of Silencing RNA into Cells but Not for Their Export from Cells.
SID-3 may control the export and the import of dsRNA as well as of mobile silencing RNA, which are also forms of dsRNA (20). To evaluate the roles of SID-3 in RNA transport between cells, we rescued sid-3(−) animals in gfp-dsRNA–expressing donor tissue or in GFP-expressing recipient tissue and measured silencing of GFP in the recipient tissue (Fig. 4 C and D). Specifically, we used sid-3(−) animals that express GFP and gfp-dsRNA in the pharynx (donor) and that express GFP in the bwm cells (recipient). Expression of sid-3(+) under the control of a pharynx-specific promoter failed to rescue GFP silencing in bwm cells, suggesting that SID-3 does not have a detectable effect on the efficiency of export of mobile silencing RNA. In contrast, expression of sid-3(+) under the control of a bwm-specific promoter robustly rescued GFP silencing in bwm cells, suggesting that SID-3 plays a role in controlling the efficiency of mobile silencing RNA import into recipient cells. Thus, these results support the idea that SID-3 is specifically required to ensure the efficient import of mobile silencing RNA and dsRNA into C. elegans cells.
Tyrosine Kinase Domain of SID-3 Is Required for the Efficient Import of dsRNA into Cells.
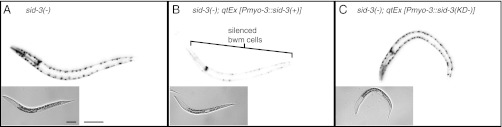
SID-3 contains several conserved protein interaction domains (Fig. 1B) and thus may play a structural rather than a catalytic role to promote the import of dsRNA into cells. Curiously, 10 of 13 recovered mutants contain nonsense, small-deletion, or splice-site mutations that via nonsense-mediated decay would result in substantially reduced mRNA levels and produce at most truncated proteins. In addition, although we isolated two missense mutants, both in the tyrosine kinase domain, modeling the mutations on the known crystal structure of the ACK tyrosine kinase domain (21) shows that these mutations are distant from the catalytic residues important for kinase activity and ATP binding (Fig. S5). To directly evaluate the requirement for an active kinase domain, we tested whether a kinase-dead SID-3 protein can rescue the silencing defect in sid-3 mutants. Specifically, we transformed sid-3 mutant animals with constructs that express either wild-type SID-3 or a SID-3(K139A) mutant protein. This lysine residue corresponds to ACK1(K158), which is required for ATP binding (21, 22). We then evaluated the silencing of GFP expression in bwm cells in response to gfp feeding RNAi and gfp-dsRNA expression in the pharynx. Unlike constructs that express wild-type SID-3, constructs that express SID-3(K139A) failed to rescue bwm silencing (Fig. 5). Thus, the kinase activity of SID-3 is essential to enable efficient import of dsRNA into C. elegans cells.
Fig. 5.
The kinase domain of SID-3 is required for the efficient import of dsRNA into cells. sid-3(−) animals that express GFP in the pharynx and in bwm cells but that express gfp-dsRNA only in the pharynx (A) were transformed with constructs that express either wild-type SID-3 [sid-3(+)] (B) or a kinase-dead version of SID-3 [sid-3(KD−)] (C) in bwm cells. Representative fluorescence images of silencing in response to gfp feeding RNAi in third larval-staged animals of each of the above three genotypes are shown. Only animals that express sid-3(+) in bwm showed silencing of bwm cells (brackets). Insets are brightfield images. (Scale bars, 50 μm.)
Discussion
We have shown that: (i) SID-3 functions in the importing cell to enable silencing by mobile RNA; (ii) SID-3 function requires an intact kinase domain; (iii) cells that lack SID-3 are not defective, but are marginally enhanced for cell-autonomous RNAi; and (iv) overexpression of SID-3 enhances silencing because of mobile RNA. Taken together, these observations suggest that the conserved tyrosine kinase SID-3 promotes dsRNA import into cells.
Three considerations lead us to speculate that SID-3/ACK functions to enhance the endocytic import of dsRNA into animal cells. First, the ACK tyrosine kinase was initially identified as a protein that binds and prolongs the activity of Cdc42 (23), a small GTPase that promotes endocytosis (24). Notably, the CRIB domain required for this binding is highly conserved (56% identical) between the human ACK protein and SID-3 (Fig. 1E). Second, a screen using Drosophila cultured cells revealed a conserved role for clathrin-mediated endocytosis in the import of dsRNA into S2 cells (25). Third, in cultured mammalian cells, activated ACK associates with Cdc42 and invaginated endocytic vesicles (26). Consistent with a possible role for SID-3–dependent tyrosine phosphorylation in endocytosis, the C. elegans dynamin, DYN-1, was recently found to be phosphorylated at a tyrosine (27). Thus, signaling through the SID-3/ACK tyrosine kinase may promote the endocytic uptake of dsRNA for the eventual import into the cytoplasm through the conserved dsRNA transporter SID-1 in C. elegans and in mammals. Signaling through ACK is activated by multiple extracellular stimuli that include growth factors and cell attachment (28). Therefore, the receptivity of animal cells for dsRNA uptake may be sensitive to developmental or environmental signals that activate the tyrosine kinase SID-3/ACK to promote the import of mobile silencing RNA.
Materials and Methods
Worm strains and transgenic animals were generated and maintained using standard methods (6). The position of sid-3 was narrowed using SNP mapping and the corresponding mutation was identified using whole-genome sequencing (17). Rescues of sid-3 mutants were performed using PCR products amplified from genomic DNA and fused with different promoter sequences. Resistance to RNAi was evaluated using feeding RNAi or injection of dsRNA. Detailed procedures and a list of the PCR primers used are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Marie Sutherlin (C.P.H. laboratory) for mapping data; Daniel Schott (C.P.H. laboratory) for advice on phylogeny; Siavash Karimzadegan (O. Hobert laboratory) and Christian Daly (Center for Systems Biology, Harvard University) for advice on Illumina sequencing; Robert Horvitz and members of the C.P.H. laboratory, particularly Kenneth Pang, Daniel Schott, Jacqueline Brooks, and Philip Shiu, for critical comments on the manuscript; and the Caenorhabditis elegans Genetics Center for some of the strains used. This work was funded in part by the National Institutes of Health (A.M.J and C.P.H.) and the National Science Foundation (C.P.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201153109/-/DCSupplemental.
References
- 1.Melnyk CW, Molnar A, Baulcombe DC. Intercellular and systemic movement of RNA silencing signals. EMBO J. 2011;30:3553–3563. doi: 10.1038/emboj.2011.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- 3.Wolfrum C, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 4.Duxbury MS, Ashley SW, Whang EE. RNA interference: A mammalian SID-1 homologue enhances siRNA uptake and gene silencing efficacy in human cells. Biochem Biophys Res Commun. 2005;331:459–463. doi: 10.1016/j.bbrc.2005.03.199. [DOI] [PubMed] [Google Scholar]
- 5.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 6.Jose AM, Smith JJ, Hunter CP. Export of RNA silencing from C. elegans tissues does not require the RNA channel SID-1. Proc Natl Acad Sci USA. 2009;106:2283–2288. doi: 10.1073/pnas.0809760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Timmons L, Tabara H, Mello CC, Fire AZ. Inducible systemic RNA silencing in Caenorhabditis elegans. Mol Biol Cell. 2003;14:2972–2983. doi: 10.1091/mbc.E03-01-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tijsterman M, May RC, Simmer F, Okihara KL, Plasterk RH. Genes required for systemic RNA interference in Caenorhabditis elegans. Curr Biol. 2004;14:111–116. doi: 10.1016/j.cub.2003.12.029. [DOI] [PubMed] [Google Scholar]
- 9.Esposito G, Di Schiavi E, Bergamasco C, Bazzicalupo P. Efficient and cell specific knock-down of gene function in targeted C. elegans neurons. Gene. 2007;395:170–176. doi: 10.1016/j.gene.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Dzitoyeva S, Dimitrijevic N, Manev H. Gamma-aminobutyric acid B receptor 1 mediates behavior-impairing actions of alcohol in Drosophila: Adult RNA interference and pharmacological evidence. Proc Natl Acad Sci USA. 2003;100:5485–5490. doi: 10.1073/pnas.0830111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roignant JY, et al. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA. 2003;9:299–308. doi: 10.1261/rna.2154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- 13.Shih JD, Hunter CP. SID-1 is a dsRNA-selective dsRNA-gated channel. RNA. 2011;17:1057–1065. doi: 10.1261/rna.2596511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winston WM, Sutherlin M, Wright AJ, Feinberg EH, Hunter CP. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc Natl Acad Sci USA. 2007;104:10565–10570. doi: 10.1073/pnas.0611282104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter CP, Kenyon C. Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell. 1996;87:217–226. doi: 10.1016/s0092-8674(00)81340-9. [DOI] [PubMed] [Google Scholar]
- 16.Tsai CJ, et al. Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev. 2008;22:194–211. doi: 10.1101/gad.1618508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarin S, Prabhu S, O’Meara MM, Pe’er I, Hobert O. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat Methods. 2008;5:865–867. doi: 10.1038/nmeth.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prieto-Echagüe V, Miller WT. Regulation of ack-family nonreceptor tyrosine kinases. J Signal Transduct. 2011;2011:742372. doi: 10.1155/2011/742372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grøvdal LM, Johannessen LE, Rødland MS, Madshus IH, Stang E. Dysregulation of Ack1 inhibits down-regulation of the EGF receptor. Exp Cell Res. 2008;314:1292–1300. doi: 10.1016/j.yexcr.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 20.Jose AM, Garcia GA, Hunter CP. Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Nat Struct Mol Biol. 2011;18:1184–1188. doi: 10.1038/nsmb.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lougheed JC, Chen R-H, Mak P, Stout TJ. Crystal structures of the phosphorylated and unphosphorylated kinase domains of the Cdc42-associated tyrosine kinase ACK1. J Biol Chem. 2004;279:44039–44045. doi: 10.1074/jbc.M406703200. [DOI] [PubMed] [Google Scholar]
- 22.Yokoyama N, Miller WT. Biochemical properties of the Cdc42-associated tyrosine kinase ACK1. Substrate specificity, authphosphorylation, and interaction with Hck. J Biol Chem. 2003;278:47713–47723. doi: 10.1074/jbc.M306716200. [DOI] [PubMed] [Google Scholar]
- 23.Manser E, Leung T, Salihuddin H, Tan L, Lim L. A non-receptor tyrosine kinase that inhibits the GTPase activity of p21cdc42. Nature. 1993;363:364–367. doi: 10.1038/363364a0. [DOI] [PubMed] [Google Scholar]
- 24.Harris KP, Tepass U. Cdc42 and vesicle trafficking in polarized cells. Traffic. 2010;11:1272–1279. doi: 10.1111/j.1600-0854.2010.01102.x. [DOI] [PubMed] [Google Scholar]
- 25.Saleh MC, et al. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen H, et al. Constitutive activated Cdc42-associated kinase (Ack) phosphorylation at arrested endocytic clathrin-coated pits of cells that lack dynamin. Mol Biol Cell. 2011;22:493–502. doi: 10.1091/mbc.E10-07-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zielinska DF, Gnad F, Jedrusik-Bode M, Wiśniewski JR, Mann M. Caenorhabditis elegans has a phosphoproteome atypical for metazoans that is enriched in developmental and sex determination proteins. J Proteome Res. 2009;8:4039–4049. doi: 10.1021/pr900384k. [DOI] [PubMed] [Google Scholar]
- 28.Galisteo ML, Yang Y, Ureña J, Schlessinger J. Activation of the nonreceptor protein tyrosine kinase Ack by multiple extracellular stimuli. Proc Natl Acad Sci USA. 2006;103:9796–9801. doi: 10.1073/pnas.0603714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.