Abstract
The elongation of transcription is a highly regulated process that requires negative and positive effectors. By binding the double-stranded stem in the transactivation response (TAR) element, RD protein from the negative transcription elongation factor (NELF) inhibits basal transcription from the long terminal repeat of the human immunodeficiency virus type 1 (HIVLTR). Tat and its cellular cofactor, the positive transcription elongation factor b (P-TEFb), overcome this negative effect. Cdk9 in P-TEFb also phosphorylates RD at sites next to its RNA recognition motif. A mutant RD protein that mimics its phosphorylated form no longer binds TAR nor represses HIV transcription. In sharp contrast, a mutant RD protein that cannot be phosphorylated by P-TEFb functions as a dominant-negative effector and inhibits Tat transactivation. These results better define the transition from abortive to productive transcription and thus replication of HIV.
The elongation of transcription from the human immunodeficiency virus type 1 long terminal repeat (HIVLTR) is regulated negatively and positively by cellular factors and the viral transactivator Tat (22). In the absence of Tat, the elongating RNA polymerase II (RNAPII) is arrested by the negative transcriptional elongation factor (N-TEF), which includes the DRB sensitivity-inducing factor (DSIF) and the negative elongation factor (NELF), resulting in the accumulation of short transcripts (12, 26, 31). However, in the presence of Tat, the positive transcription elongation factor b (P-TEFb), consisting of the cyclin-dependent kinase 9 (Cdk9) and cyclin T1 (CycT1), is recruited to the transactivation response (TAR) element, which forms a stable RNA stem-loop at the 5′ end of all viral transcripts (15, 22, 28, 32). Cdk9 then phosphorylates DSIF and the C-terminal domain (CTD) of RNAPII, which is essential for the productive elongation of transcription (19, 23).
Both DSIF and NELF are found on the HIVLTR after the initiation of viral transcription (18). DSIF is composed of Spt4 and Spt5 (26). Spt5 binds the unphosphorylated but not the phosphorylated form of the CTD (CTDa of RNAPIIa but not CTDo from RNAPIIo) (10, 27). Thus, P-TEFb directly regulates the interaction between DSIF and RNAPII. NELF is comprised of four subunits, NELF-A or WHSC, NELF-B, alternatively spliced NELF-C/D, and NELF-E or RD (17, 29, 31). NELF-A and RD contain RNA recognition motifs (RRM) and bind a number of RNA elements, which are required for the inhibitory effect of NELF on transcription (17, 29, 31). Of importance, RD binds TAR via its RRM (30). This interaction could contribute to low basal levels of viral transcription, and therefore, to the proviral transcriptional latency in infected cells (1, 14). Although P-TEFb can alleviate negative effects of NELF in vitro (25, 27), no mechanism exists for this transition from negative to positive regulation of transcriptional elongation.
In this study, we provide such a mechanism, taken from HIV. First, by binding the bottom stem in TAR, RD from NELF and Spt5 from DSIF cooperatively help to arrest RNAPII on the HIVLTR. Next, the complex between P-TEFb and Tat is recruited to the 5′ bulge and central loop in TAR. Finally, Cdk9 phosphorylates RD, Spt5, and RNAPII, thus removing N-TEF from TAR. As a consequence, productive elongation of HIV transcription ensues.
MATERIALS AND METHODS
Plasmid constructions.
The plasmid targets pHIVCAT, pSLIIBCAT, and pEIAVCAT and the plasmid effectors pcDNA.Tat (pTat), pRev, and pRev.Tat were described elsewhere (6, 21). pSPT5 was a generous gift from Hiroshi Handa (Tokyo Institute of Technology, Tokyo, Japan). To construct glutathione S-transferase-RD (GST.RD) fusion proteins, the DNA fragment corresponding to the full-length RD protein was amplified by PCR using appropriate primers with sites from pBS-RD (Hiroshi Handa). Amplified fragments were inserted into BamHI and EcoRI sites of pGEX-4T1 (Pharmacia, Piscataway, N.J.). The fragments corresponding to RD were also subcloned into a mammalian expression vector, pEF-BOS, using BamHI and EcoRI sites. All RD constructs for mammalian expression contained the c-Myc epitope tag at the C terminus of RD. Western blotting using the polyclonal anti-Myc antibody A14 (Santa Cruz Biotechnology, Santa Cruz, Calif.) confirmed the expression of the proteins in cells. Mutant GST.RD proteins were constructed using QuikChange XL site-directed mutagenesis kit (Stratagene, Carlsbad, Calif.) with appropriate sets of primers. Mutations were verified by DNA sequencing. The sequences of oligonucleotide primers are available upon request.
Transfections and CAT assays.
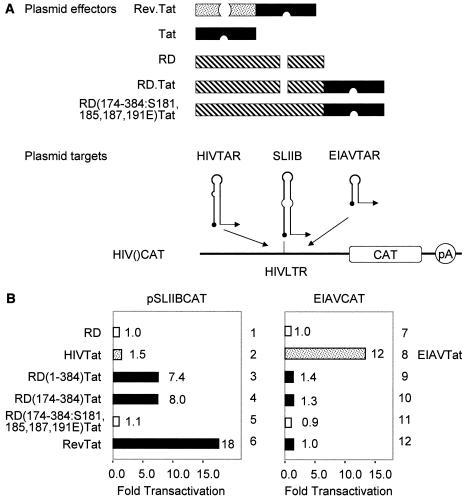
HeLa or HeLa/LTR-luc (generous gift of Jonathan Karn, Case Western Reserve University, Cleveland, Ohio) cells were cotransfected with 0.5 μg of pEF.RD (wild-type and mutant constructions) and/or pCMV.SPT5 and pHIVCAT or pEIAVCAT (0.1 μg), in the absence or presence of pTat (0.1 μg) or pEIAV-Tat (0.1 μg) using Lipofectamine according to the manufacturer's instruction (GIBCO/BRL, Gaithersburg, Md.). Forty-eight hours after transfection, the chloramphenicol acetyltransferase (CAT) and luciferase activities in the cell lysate were measured by using a Lumitech ReportaLight Bioassay kit (Cambrex Bioscience, Baltimore, Md.) and Lumimark Plus microplate reader (Bio-Rad, Hercules, Calif.) by standard procedures described elsewhere (6). For heterologous RNA-tethering assays (see Fig. 4), HeLa cells were cotransfected with 0.5 μg of plasmid effectors (pRev.Tat, pTat, pRD, pRD.Tat, and pRD(174-384:S181,185,187,191E).Tat, as illustrated below [see Fig. 4A]) and 0.2 μg of plasmid targets (pSLIIBCAT and pEIAVCAT, as illustrated below [see Fig. 4A]) using Lipofectamine. Forty-eight hours after transfection, CAT activities in the cell lysate were measured as described above.
FIG. 4.
RD binds TAR in vivo. (A) Schematic presentation of plasmid effectors and targets. pA, polyadenylation site. (B) RD can recruit Tat in the RD.Tat chimera to TAR. pEF.RD, pEF.Tat, pEF.RD.Tat, or mutant pEF.RD.Tat plasmid (0.5 μg) was cotransfected with pSLIIBCAT (left) or pEIAVCAT (right) into HeLa cells. CAT activities were measured as described in the legend to Fig. 1A. Results are presented as fold transactivation over the value obtained with the empty plasmid vector.
EMSAs.
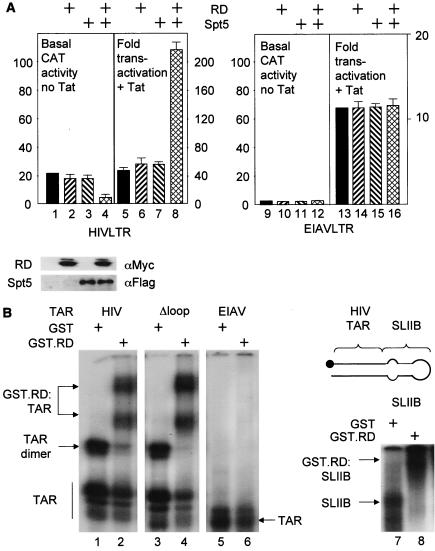
32P-labeled RNA probes were prepared by in vitro transcription of linearized plasmid templates using T7 MAXiscript kit (Ambion, Austin, Tex.). The labeled RNA probes were purified from 5% denaturing polyacrylamide gels containing 7 M urea as described previously (8). The labeled RNA probes were incubated with 0.5 μg of GST fusion proteins in electrophoretic mobility shift assay (EMSA) buffer [30 mM Tris-HCl, 70 mM KCl, 0.01% Nonidet P-40 [NP-40], 5.5 mM MgCl2, 1 mM dithiothreitol [DTT], 20 μg of poly(dI) · poly(dC) per ml, 20 μg of poly(I) · poly(C) per ml] for 20 min at 30°C. RNA-protein complexes were separated on 5% nondenaturing polyacrylamide gels (3 W; 2 h at 4°C). Gels were dried and analyzed by autoradiography (see Fig. 1 and 3) or with a phosphorimager (Bio-Rad) (see Fig. 2).
FIG. 1.
RD and Spt5 inhibit basal transcription and increase Tat transactivation from the HIVLTR but not EIAVLTR. (A) Effects of RD and Spt5 on the HIVLTR in cells. pEF.RD (0.5 μg) and/or pCMV.SPT5 was cotransfected with pHIVCAT (0.1 μg) (lanes 1 to 8) or pEIAVCAT (0.1 μg) (lanes 9 to 12) in the absence or presence of pTat (0.1 μg) or pEIAV-Tat (0.1 μg) into HeLa cells. Two days later, CAT activities were measured. For basal CAT activities, results are presented as raw numbers. For the effects of Tat, results are presented as fold transactivation relative to the CAT activity obtained with the empty plasmid vector (lanes 1 and 9). The expression of RD and Spt5 was visualized by Western blotting using anti-Myc (αMyc) and anti-FLAG (αFlag) antibodies, respectively. The values are means ± standard errors of the mean (error bars) from three independent experiments performed in duplicate. (B) RD binds HIVTAR but not EIAVTAR. 32P-labeled TAR from HIV (lanes 1 and 2), mutant TAR lacking the central loop (Δloop) (lanes 3 and 4), TAR from EIAV (lanes 5 and 6) and SLIIB grafted onto HIVTAR (schematic and lanes 7 and 8) were incubated with (+) purified GST or the GST.RD chimera. The reaction mixture was then separated on a 5% nondenaturing polyacrylamide gel at 4°C. RNA-protein-RNA complexes were visualized by autoradiography. The arrows point to informative RNA-protein complexes, whose composition is given. The position of free probes is indicated by the vertical line at the bottom of the leftmost gel.
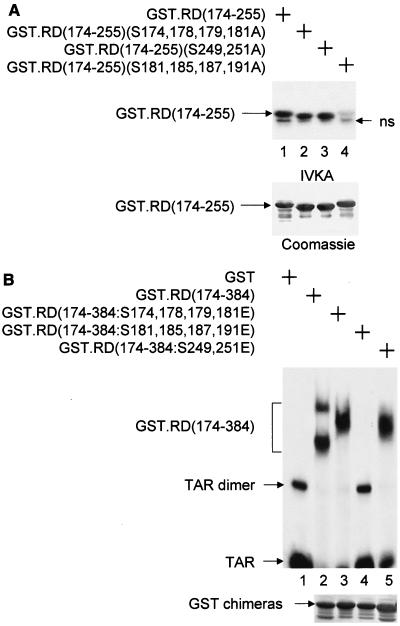
FIG. 3.
Mapping of the phosphorylation site(s) in RD that is required for dissociating RD and TAR. (A) Mutant RD proteins that were examined in this experiment. IVKA was performed as described in the legend to Fig. 2A using the mutant GST.RD chimeras and P-TEFb. ns, nonspecific band. (B) The abilities of the mutant GST.RD fusion proteins to bind TAR were examined by EMSAs. See the legend to Fig. 2B.
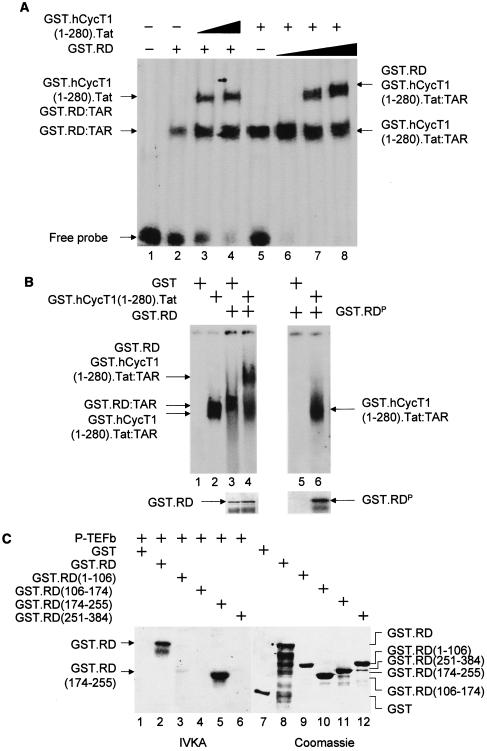
FIG. 2.
Unphosphorylated RD and the complex between Tat and CycT1 (P-TEFb) bind TAR. (A) RD and the complex between CycT1 and Tat bind TAR simultaneously. In lanes 1 to 4, increasing amounts (0.1 and 0.2 μg for lanes 3 and 4, respectively) of the purified GST.hCycT.Tat chimera (hCycT, human CycT) (7) was added to the reaction mixture containing the 32P-labeled HIVTAR and the GST.RD fusion protein. Purified GST protein (0.2 μg) was used as the negative control (lane 2). In lanes 5 to 8, increasing amounts (0.2, 0.5, and 1.0 μg for lanes 2, 3, and 4, respectively) of the purified GST.RD chimera was added to the reaction mixture containing the 32P-labeled HIVTAR and the GST.hCycT.Tat fusion protein (0.1 μg). Purified GST protein (0.2 μg) was used as the negative control (lane 5). The composition of RNA-protein complexes is given next to the arrows pointing to singly shifted and supershifted bands. This EMSA was visualized by the phosphorimager. (B) Phosphorylated RD does not bind TAR in vitro. GST and the GST.RD chimera were phosphorylated by P-TEFb using cold ATP in vitro. As described above, 0.5 μg of unphosphorylated (lanes 1 to 4) and phosphorylated GST.RD (GST.RDP) chimeras (lanes 5 and 6) were used in the absence or presence (+) of the GST.hCycT.Tat fusion protein (0.1 μg) for the EMSA. Phosphorylation was monitored by a parallel experiment using [γ-32P]ATP (lower right panel). After SDS-PAGE, gels were stained with Coomassie brilliant blue to reveal the levels of input proteins (lower left panel). (C) Sequences N terminal to the RRM in RD are phosphorylated by P-TEFb. (Left) RD is phosphorylated by P-TEFb. GST, GST.RD chimera, and its mutant truncated counterparts [GST.RD(1-106), GST.RD(106-174), GST.RD(174-255), and GST.RD(251-384)] were incubated with P-TEFb and [γ-32P]ATP. Proteins were separated by SDS-PAGE, followed by autoradiography. (Right) The input levels of proteins were verified as described for panel B.
IVKAs.
In vitro kinase assay (IVKA) was performed as described elsewhere (4). Briefly, purified GST, GST.RD, and its truncation mutants [GST.RD(1-106), GST.RD(106-174), GST.RD(174-255), and GST.RD(251-380)] bound to glutathione-Sepharose 4B beads (Pharmacia) were incubated with purified P-TEFb and [γ-32P]ATP in IVKA buffer (50 mM Tris-HCl, 0.1 M KCl, 1 mM MgCl2, 1 mM DTT [pH 7.5]) for 30 min at room temperature. After the proteins were washed twice with IVKA buffer without MgCl2, the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by autoradiography. To prepare phosphorylated RD proteins for EMSAs, IVKA was performed as described above except that cold ATP was used instead of [γ-32P]ATP. Unphosphorylated GST and GST.RD chimera were prepared by omitting P-TEFb and ATP from the reaction mixture. The proteins were then purified using glutathione-Sepharose beads. Phosphorylation was monitored by a parallel experiment using [γ-32P]ATP.
Coimmunoprecipitations.
Myc epitope-tagged wild-type and mutant RD proteins were expressed in 293T cells, which were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, Calif.). Forty-eight hours later, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, 0.15 M NaCl, 1 mM EDTA, 1% sodium deoxycholate, 1% NP-40, 0.1% SDS, 1 mM DTT [pH 7.4]) in the presence of protease inhibitors (Sigma-Aldridge, St. Louis, Mo.). Cell lysates were incubated with 1 μg of monoclonal antibody against NELF-A subunit (provided by Hiroshi Handa) overnight at 4°C. After the cell lysates were allowed to bind to the antibody, reaction mixtures were incubated with protein G-agarose beads (Santa Cruz Biotechnology) at 4°C. Immunoprecipitated proteins were washed extensively with RIPA buffer and subjected to SDS-PAGE, followed by Western blotting with anti-Myc polyclonal antibody (A14; Santa Cruz Biotechnology).
RESULTS
RD and Spt5 cooperatively decrease basal levels of transcription and increase Tat transactivation from the HIVLTR.
To investigate the mechanism whereby P-TEFb alleviates the effects of NELF on viral transcription, first we examined RD and Spt5 activities on the HIVLTR in HeLa cells. In these cells, RD was coexpressed with a plasmid target that encodes the CAT reporter gene under control of the HIVLTR (pHIVCAT) in the absence or presence of Tat (Fig. 1A). Equine infectious anemia virus long terminal repeat (EIAVLTR) linked to CAT (pEIAVCAT) was used as the control (see below). As presented in Fig. 1A, the overexpression of RD alone had little effect on basal HIV transcription (compare lanes 1 and 2). However, although Spt5 alone had no effect, the coexpression of RD and Spt5 had a synergistic negative effect on the HIVLTR (Fig. 1A, lanes 3 and 4). In sharp contrast, Tat activated HIV transcription to maximal levels, regardless of the expression of these negative factors, resulting in the highest transactivation in the presence of RD and Spt5 (Fig. 1A, lane 8). Importantly, the expression of RD or Spt5 alone or the coexpression of these proteins had no effect on the levels of basal transcription from the EIAVLTR (Fig. 1A, lanes 9 to 12). Therefore, no differences were observed in Tat transactivation in the EIAV system (Fig. 1A, lanes 13 to 16). Since HIV replication requires the recruitment of P-TEFb to TAR by Tat, we conclude that DSIF, NELF, and TAR decrease basal levels of viral transcription, which is overcome by Tat and P-TEFb.
RD, Tat, and CycT1 bind the bottom stem, 5′ bulge, and central loop in TAR, respectively.
Consistent with the observation of Yamaguchi et al. (30), purified RD protein bound TAR in the EMSA (Fig. 1B, lane 2). RD also bound the mutant TAR RNA species containing deletions in the central loop (Fig. 1B, lane 4) and bulge sequences (data not presented) and the chimeric SLIIB, in which stem-loop II (SLII) from the Rev response element (RRE) was grafted onto the lower double-stranded stem of TAR (Fig. 1B, lanes 7 and 8). In sharp contrast, RD did not bind EIAV TAR (Fig. 1B, lane 6), which is consistent with the coexpression data in cells (Fig. 1A). Since TAR from EIAV bears a shorter stem sequence than TAR from HIV (10 and 20 bp in EIAV and HIV, respectively), these results suggest that RD binds the double-stranded RNA stem-loop in HIVTAR. Moreover, RD supershifted the complex between CycT1, Tat, and TAR and vice versa in an EMSA (Fig. 2A, lanes 2 to 4 and 6 to 8), which completes mapping of the binding between RD and the stem in HIVTAR. We conclude that NELF decreases the basal HIV transcription via the specific interaction between RD and TAR.
P-TEFb phosphorylates RD, which no longer binds TAR.
These results indicated that RD, Tat, and P-TEFb bind TAR simultaneously, which suggested that P-TEFb alleviates negative effects by a mechanism other than the simple displacement of NELF from TAR. Additionally, the entire NELF complex also binds TAR (30). Therefore, we examined whether a posttranslational modification of RD, such as phosphorylation, could affect the ability of RD to bind TAR. IVKAs were performed using the GST.RD fusion protein and P-TEFb, which were purified from Escherichia coli and from insect cells using baculovirus, respectively (4). As presented in Fig. 2B, the GST.RD fusion protein, but not GST alone, was phosphorylated by P-TEFb (Fig. 2B, lower right gel, and Fig. 2C, lane 2). A partial mapping of the phosphorylation sites indicated that only the fragment from positions 174 to 255 was phosphorylated by P-TEFb (Fig. 2C, lanes 3 to 6). Surprisingly, this phosphorylated form of RD bound neither TAR alone nor the complex between Tat, TAR, and P-TEFb in an EMSA (Fig. 2B, compare lanes 3, 4, 5, and 6). Importantly, the region of RD that is phosphorylated by P-TEFb (positions 174 to 255) overlaps its RRM (positions 247 to 342) (see below) (30). When coincubated with both TAR and P-TEFb prior to the EMSA, RD also no longer bound TAR (data not presented). Therefore, the phosphorylation of this portion could have a significant effect on the ability of RRM to bind TAR. We conclude that the phosphorylation of RD by P-TEFb prevents interactions between RD and TAR.
P-TEFb phosphorylates RD next to the RRM.
The fragment of RD that is phosphorylated by P-TEFb [RD(174-255)] contains one serine residue (S181) that conforms to the consensus sequence for phosphorylation by cyclin-dependent kinases (SPXX) and eight other serines at positions 174, 178, 179, 185, 187, 191, 249, and 251. IVKA with a mutant RD protein, in which S181 was mutated to alanine [RD(S181A)], indicated that S181 is a phosphorylation site, but not the only one (data not presented). Therefore, we constructed mutant RD proteins that contained groups of changed serines (S174,178,179,181, S181,185,187,191, and S249,251) to alanines (Fig. 3A). These mutant GST.RD chimeras were examined for their phosphorylation by P-TEFb in IVKAs. As presented in Fig. 3A, only the mutant GST.RD(174-255, S181,185,187,191A) fusion protein was not phosphorylated by P-TEFb (lane 4). Next, to mimic their phosphorylated counterparts, these serines were mutated to glutamates in the RD fragment that contained the RRM [RD(174-384)] (Fig. 3B). As presented in Fig. 3B, the mutant GST.RD(174-384, S181,185,187,191E) chimera did not bind TAR in EMSA (lane 4), which is consistent with the hypothesis that the phosphorylation of these serines by P-TEFb is responsible for the dissociation between RD and TAR.
Only unphosphorylated RD binds TAR in vivo.
Next, we examined whether this phosphorylation-dependent interaction between RD and TAR occurs in vivo. To detect the interaction between RD and TAR in HeLa cells, we used the plasmid reporter in which the TAR sequence in pHIVCAT was replaced by SLIIB (pSLIIBCAT [6, 24]). As described above, SLIIB contains the stem sequence from TAR that is the binding site for RD (Fig. 1B). Thus, as effectors, wild-type and mutant RD proteins were fused in frame with Tat. Since neither Tat nor P-TEFb binds SLIIB, only the Rev.Tat fusion protein but not Tat alone activates this hybrid promoter (6). On the other hand, if RD were to bind SLIIB in cells, the complex between Tat and P-TEFb could be recruited and activate transcription from pSLIIBCAT via this heterologous RNA-tethering system (Fig. 4A). Since RD does not bind TAR from EIAV, pEIAVCAT was used as the negative control (Fig. 1B) (21). Additionally, to confirm the specificity of our chimeras, free RD and Tat proteins were used as negative controls (Fig. 4A). As presented in Fig. 4B, the full-length hybrid RD.Tat [RD(1-384)].Tat protein activated pSLIIBCAT more than sevenfold in HeLa cells (Fig. 4B, lane 3). A truncated mutant RD.Tat fusion protein containing the RRM [mutant RD(174-384).Tat chimera] also activated pSLIIBCAT (Fig. 4B, lane 4). In sharp contrast, the truncated mutant RD(174-384, S181,185,187,191E).Tat chimera, which contained glutamates rather than serines in its phosphorylation domain, had no activity (Fig. 4B, lane 5). These results are consistent with the binding data in vitro (Fig. 3B). The activity obtained with the RD.Tat chimera was somewhat lower than with the hybrid Rev.Tat protein that was used as the positive control (Fig. 4B, 7.4- versus 15-fold, compare lanes 3 and 7). This finding could be due to the multimerization of Rev on SLIIB, which increases the number of P-TEFb molecules that are recruited by Rev.Tat rather than RD.Tat chimeras. Also, since P-TEFb phosphorylates RD, this could shorten the residence of RD on TAR in vivo. Results with the mutant RD(174-384, S181,185,187,191E).Tat chimera support this hypothesis (Fig. 4B, lane 5). As presented in Fig. 4B, the RD.Tat fusion protein and its truncated derivatives did not activate pEIAVCAT, which is also consistent with previous binding data (Fig. 1B). We conclude that RD binds TAR specifically and that the binding depends on the phosphorylation state of RD in vivo.
Mutant glutamate-substituted RD protein and Spt5 no longer decrease cooperatively basal transcription from the HIVLTR.
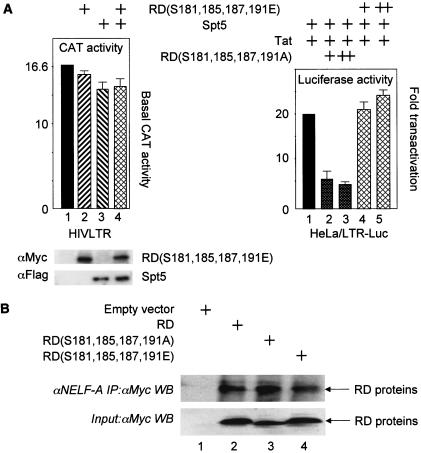
To determine whether this mutant RD(S181,185,187,191E) protein had any residual activity on the HIVLTR, it was coexpressed with Spt5 and pHIVCAT in HeLa cells. As presented in Fig. 5A, this mutant RD(S181,185,187,191E) protein alone or with Spt5 did not inhibit expression from the HIVLTR (compare lanes 1, 2, 3, and 4). The mutant RD(S181,185,187,191E) protein also had no effect on Tat transactivation in the EIAV system (data not presented). Thus, only the wild-type, unphosphorylated RD protein binds TAR and affects levels of transcription from the HIVLTR.
FIG. 5.
Effects of mutant RD proteins on viral transcription and their incorporation into NELF. (A) (Left) The mutant glutamate-substituted RD protein no longer inhibits HIV transcription. pHIVCAT, mutant RD(S181,185,187,191E) and Spt5 were expressed in HeLa cells, and CAT assays were performed as described in the legend to Fig. 1A. (Right) The mutant alanine-substituted RD protein inhibits Tat transactivation. Tat (0.1 μg), Spt5 (0.1 μg), mutant RD(S181,185,187,191E) (0.1 and 0.3 μg) or mutant RD(S181,185,187,191A) (0.1 and 0.3 μg) proteins were expressed in HeLa cells that stably contained the HIVLTR linked to the luciferase reporter gene. Luciferase activities were measured 48 h later. (B) Wild-type and mutant RD proteins are incorporated equivalently into NELF. 293T cells expressed Myc epitope-tagged RD proteins. Immunoprecipitations (IP) with the anti-NELF-A monoclonal antibody (αNELF-A)were followed by Western blotting (WB) with the anti-Myc antiserum (αMyc). The bottom gel contains 5% of input proteins. In lane 1, cells were transfected with the empty plasmid vector. Arrows point to wild-type and mutant RD proteins in lanes 2, 3, and 4.
Mutant alanine-substituted RD protein inhibits Tat transactivation.
Next, we examined whether a mutant RD protein that can no longer be phosphorylated but does not mimic the phosphorylated RD protein could function in a dominant-negative fashion for Tat transactivation. This finding would confirm that native RD protein binds TAR but once phosphorylated, it no longer inhibits viral transcription. Indeed, as presented in Fig. 5A, right panel, whereas the mutant RD(S181,185,187,191E) protein had no effect on Tat transactivation (compare lanes 1, 4, and 5), the alanine-substituted mutant RD(S181,185,187,191A) protein inhibited effects of Tat four- to fivefold in a dose-dependent fashion (compare lanes 1, 2, and 3). We conclude that mutant RD proteins that bind but cannot dissociate from TAR have dominant-negative effects on Tat transactivation and viral transcription.
Mutant RD proteins are incorporated into NELF as efficiently as the wild-type RD protein.
Finally, we wanted to determine whether the mutant RD proteins that had no effect or significantly reduced levels of Tat transactivation were incorporated into NELF in cells. Our Myc epitope-tagged RD proteins were expressed in 293T cells. Following the immunoprecipitation with the anti-NELF-A monoclonal antibody, Western blotting was performed with the anti-Myc polyclonal antiserum. As presented in Fig. 5B, all three RD proteins, the wild-type counterpart and mutant RD(S181,185,187,191E) and RD(S181,185,187,191A) proteins, coimmunoprecipitated with the largest subunit of NELF. Thus, not only were wild-type and mutant RD proteins incorporated efficiently into NELF, but since their effects also required Spt5, they occurred in the context of these larger multiprotein complexes.
DISCUSSION
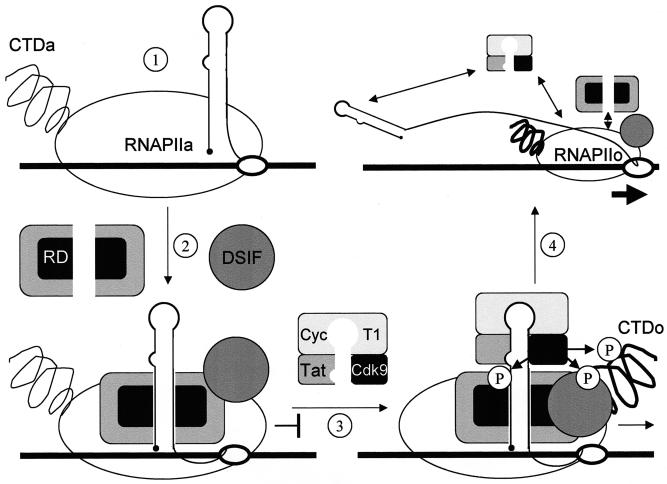
These results reveal a mechanism for the transition from negative to positive regulation of transcriptional elongation on the HIVLTR. First, RD binds TAR in the absence of the complex between Tat and P-TEFb. Second, RD dissociates from TAR upon its phosphorylation by P-TEFb, which is recruited to TAR by Tat. Therefore, during the activation of HIV transcription, Tat and P-TEFb phosphorylate not only the CTD of RNAPII and Spt5 from DSIF but also RD (Fig. 6). This phosphorylation occurs in the region that is just N terminal to and partially overlaps the RRM that binds TAR. The putative three-dimensional structure of this region, which was modeled on that from U1, which contains a similar RRM (16), revealed that these phosphorylation sites reside close to the positively charged α-helix that contacts RNA (data not presented). Therefore, adding negative charges in this region by phosphorylation could displace the α-helix from RNA, resulting in the dissociation of RD from TAR. Importantly, both negative and positive effectors are recruited to TAR, which could be essential for the accumulation of short transcripts (12). Moreover, in latently infected cells from seropositive individuals, short transcripts predominate (1, 14). These findings suggest an active involvement of the negative transcription factors in proviral latency and emphasize the role played by TAR as the switch between abortive and productive viral transcription and replication.
FIG. 6.
A model for negative and positive transcriptional regulation on the HIVLTR. In step 1, the unphosphorylated RNAPIIa clears the viral promoter. In step 2, the transcription complex is arrested near TAR with the help of DSIF and NELF. RD in NELF binds the lower stem in TAR. After the synthesis of Tat, P-TEFb is recruited to the 5′ bulge and central loop in TAR. Cdk9 phosphorylates the CTD of RNAPIIa (RNAPIIo), Spt5 in DSIF, and RD in NELF (step 3). The phosphorylated RD no longer binds TAR. In step 4, negative factors are converted into positive elongation factors, and RNAPIIo leaves the HIVLTR so that the viral genome is copied efficiently and cotranscriptional processing can take place. Altered N-TEF most likely stays associated with RNAPIIo.
NELF can be recruited to the transcriptional complex by different mechanisms. For example, NELF-A also binds RNA and RNAPII (29). RD displays a broad specificity of binding, which suggests that NELF can function on a wide variety of genes (30). In each case, RNA binding is essential for the activity of NELF. Therefore, the recruitment of N-TEF to the elongating RNAPII via interactions between one or more subunits of NELF and nascent transcripts could represent a key step in its mode of action. Supporting this hypothesis is the observation that NELF can also be found in the elongating complex without negative effects, which indicates that anchoring to RNA is critical for its inhibitory activity (18). Moreover, transcriptional pausing is required for P-TEFb to stimulate transcriptional elongation (25). After the release, the phosphorylation of the CTD by P-TEFb is observed (3). In this scenario, N-TEF pauses RNAPII at an early step of transcriptional elongation, whereupon P-TEFb can be recruited by many different activators, e.g., the androgen receptor, CIITA, c-Myc, NF-κB, MyoD, and Tat (2, 5, 11, 13, 20) (Fig. 6). As described above, our results indicate that the interaction between RD and TAR is regulated by the phosphorylation of RD by P-TEFb. This finding provides a mechanism for the removal of negative factors from RNA and therefore for the activation of transcriptional elongation. Of interest, this mechanism of transcriptional arrest and elongation uncannily parallels the termination by Nus proteins and antitermination by N at the NutB RNA stem-loop of bacteriophage λ (9).
Acknowledgments
We thank members of the Peterlin laboratory for help with experiments, Hiroshi Handa for reagents, and Warner Greene, Hiroshi Handa, Jonathan Karn, Eric Verdin, and Keith Yamamoto for comments on the work. We thank Lindsey McGowen for technical assistance.
This study was supported in part by grants from AmFAR, Center for AIDS Research in CWRU, NIH, and UARP.
REFERENCES
- 1.Adams, M., L. Sharmeen, J. Kimpton, J. M. Romeo, J. V. Garcia, B. M. Peterlin, M. Groudine, and M. Emerman. 1994. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. USA 91:3862-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. [DOI] [PubMed] [Google Scholar]
- 3.Bourgeois, C. F., Y. K. Kim, M. J. Churcher, M. J. West, and J. Karn. 2002. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol. 22:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao, S. H., K. Fujinaga, J. E. Marion, R. Taube, E. A. Sausville, A. M. Senderowicz, B. M. Peterlin, and D. H. Price. 2000. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J. Biol. Chem. 275:28345-28348. [DOI] [PubMed] [Google Scholar]
- 5.Eberhardy, S. R., and P. J. Farnham. 2001. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 276:48562-48571. [DOI] [PubMed] [Google Scholar]
- 6.Fujinaga, K., T. P. Cujec, J. Peng, J. Garriga, D. H. Price, X. Grana, and B. M. Peterlin. 1998. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J. Virol. 72:7154-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujinaga, K., D. Irwin, R. Taube, F. Zhang, M. Geyer, and B. M. Peterlin. 2002. A minimal chimera of human cyclin T1 and Tat binds TAR and activates human immunodeficiency virus transcription in murine cells. J. Virol. 76:12934-12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujinaga, K., R. Taube, J. Wimmer, T. P. Cujec, and B. M. Peterlin. 1999. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc. Natl. Acad. Sci. USA 96:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenblatt, J., J. R. Nodwell, and S. W. Mason. 1993. Transcriptional antitermination. Nature 364:401-406. [DOI] [PubMed] [Google Scholar]
- 10.Ivanov, D., Y. T. Kwak, J. Guo, and R. B. Gaynor. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 20:2970-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanazawa, S., T. Okamoto, and B. M. Peterlin. 2000. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity 12:61-70. [DOI] [PubMed] [Google Scholar]
- 12.Kao, S. Y., A. F. Calman, P. A. Luciw, and B. M. Peterlin. 1987. Antitermination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330:489-493. [DOI] [PubMed] [Google Scholar]
- 13.Lee, D. K., H. O. Duan, and C. Chang. 2001. Androgen receptor interacts with the positive elongation factor P-TEFb and enhances the efficiency of transcriptional elongation. J. Biol. Chem. 276:9978-9984. [DOI] [PubMed] [Google Scholar]
- 14.Lin, X., D. Irwin, S. Kanazawa, L. Huang, J. Romeo, T. S. Yen, and B. M. Peterlin. 2003. Transcriptional profiles of latent human immunodeficiency virus in infected individuals: effects of Tat on the host and reservoir. J. Virol. 77:8227-8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mancebo, H. S., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagai, K., C. Oubridge, T. H. Jessen, J. Li, and P. R. Evans. 1990. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature 348:515-520. [DOI] [PubMed] [Google Scholar]
- 17.Narita, T., Y. Yamaguchi, K. Yano, S. Sugimoto, S. Chanarat, T. Wada, D. K. Kim, J. Hasegawa, M. Omori, N. Inukai, M. Endoh, T. Yamada, and H. Handa. 2003. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 23:1863-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ping, Y. H., and T. M. Rana. 2001. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J. Biol. Chem. 276:12951-12958. [DOI] [PubMed] [Google Scholar]
- 19.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simone, C., P. Stiegler, L. Bagella, B. Pucci, C. Bellan, G. De Falco, A. De Luca, G. Guanti, P. L. Puri, and A. Giordano. 2002. Activation of MyoD-dependent transcription by cdk9/cyclin T2. Oncogene 21:4137-4148. [DOI] [PubMed] [Google Scholar]
- 21.Taube, R., K. Fujinaga, D. Irwin, J. Wimmer, M. Geyer, and B. M. Peterlin. 2000. Interactions between equine cyclin T1, Tat, and TAR are disrupted by a leucine-to-valine substitution found in human cyclin T1. J. Virol. 74:892-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taube, R., K. Fujinaga, J. Wimmer, M. Barboric, and B. M. Peterlin. 1999. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology 264:245-253. [DOI] [PubMed] [Google Scholar]
- 23.Taube, R., X. Lin, D. Irwin, K. Fujinaga, and B. M. Peterlin. 2002. Interaction between P-TEFb and the C-terminal domain of RNA polymerase II activates transcriptional elongation from sites upstream or downstream of target genes. Mol. Cell. Biol. 22:321-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiley, L. S., S. J. Madore, M. H. Malim, and B. R. Cullen. 1992. The VP16 transcription activation domain is functional when targeted to a promoter-proximal RNA sequence. Genes Dev. 6:2077-2087. [DOI] [PubMed] [Google Scholar]
- 25.Wada, T., G. Orphanides, J. Hasegawa, D. K. Kim, D. Shima, Y. Yamaguchi, A. Fukuda, K. Hisatake, S. Oh, D. Reinberg, and H. Handa. 2000. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol. Cell. 5:1067-1072. [DOI] [PubMed] [Google Scholar]
- 26.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi, Y., J. Filipovska, K. Yano, A. Furuya, N. Inukai, T. Narita, T. Wada, S. Sugimoto, M. M. Konarska, and H. Handa. 2001. Stimulation of RNA polymerase II elongation by hepatitis delta antigen. Science 293:124-127. [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi, Y., N. Inukai, T. Narita, T. Wada, and H. Handa. 2002. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 22:2918-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 32.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 11:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]