Abstract
African trypanosomes are protozoan parasites transmitted by a tsetse fly vector to a mammalian host. The life cycle includes highly proliferative forms and quiescent forms, the latter being adapted to host transmission. The signaling pathways controlling the developmental switch between the two forms remain unknown. Trypanosoma brucei contains two target of rapamycin (TOR) kinases, TbTOR1 and TbTOR2, and two TOR complexes, TbTORC1 and TbTORC2. Surprisingly, two additional TOR kinases are encoded in the T. brucei genome. We report that TbTOR4 associates with an Armadillo domain-containing protein (TbArmtor), a major vault protein, and LST8 to form a unique TOR complex, TbTORC4. Depletion of TbTOR4 caused irreversible differentiation of the parasite into the quiescent form. AMP and hydrolysable analogs of cAMP inhibited TbTOR4 expression and induced the stumpy quiescent form. Our results reveal unexpected complexity in TOR signaling and show that TbTORC4 negatively regulates differentiation of the proliferative form into the quiescent form.
Keywords: parasitology, cell biology, kinetoplastida
The adaptation of cell metabolism to environmental cues is essential for survival of free-living organisms. Obligate parasites need to adapt cell growth and proliferation rates to conditions encountered in distinct hosts and distinct compartments within a host. The life cycle of Trypanosoma brucei, the etiological agent of sub-Saharan human African trypanosomiasis, alternates between the tsetse fly and the mammal host. In the bloodstream, the parasite decreases cell proliferation to avoid overwhelming the host and to preadapt for transmission to the tsetse fly (1). This differentiation process occurs via quorum sensing in response to the Stumpy inductor factor (SIF), a chemically uncharacterized signal secreted by trypanosomes. Upon high parasitemia, SIF triggers differentiation of the proliferating “slender” bloodstream form to the cell-cycle–arrested “stumpy” form. Laboratory-adapted monomorphic strains are insensitive to SIF and unable to differentiate into the quiescent stumpy form to reduce cell proliferation, causing premature host death (2). The stumpy form is the insect-preadapted quiescent stage, competent for survival in the tsetse gut and irreversibly committed to differentiation to the proliferative procyclic insect form (3).
The highly conserved protein kinase target of rapamycin (TOR) is a master regulator of cell growth, energy homeostasis, and stress resistance in eukaryotes (4). We previously characterized the kinases Trypanosoma brucei TOR1 (TbTOR1) and TbTOR2 (5), which are functionally orthologous to TOR kinases described in other invertebrates (4). Surprisingly, Trypanosoma and the related parasite Leishmania are the only eukaryotes whose genomes encode two additional TOR paralogues, TbTOR3 and TbTOR4 (previously TbTOR-like 1 and 2, respectively) (5) in trypanosomes. Although TbTOR3 is involved in the control of acidocalcisome and polyphosphate metabolism (6), and the Leishmania counterpart is involved in virulence (7), the function of TbTOR4 remains unknown. Our results suggest that TbTOR4 assembles into a structurally and functionally unique TOR complex (TbTORC4) that plays a crucial role in the T. brucei life cycle.
TbTOR4 contains characteristic TOR kinase domains, including HEAT, FAT, and FATC domains, but lacks a rapamycin-binding site (RBS). The RBSs in TbTOR1 and TbTOR3 also are poorly conserved and do not interact with FKBP2-rapamycin (5). Multiple-alignment analysis of TbTOR4 with other members of the PI3K-related kinase (PIKK) superfamily indicates that TbTOR4 clusters with the TOR family (Fig. S1).
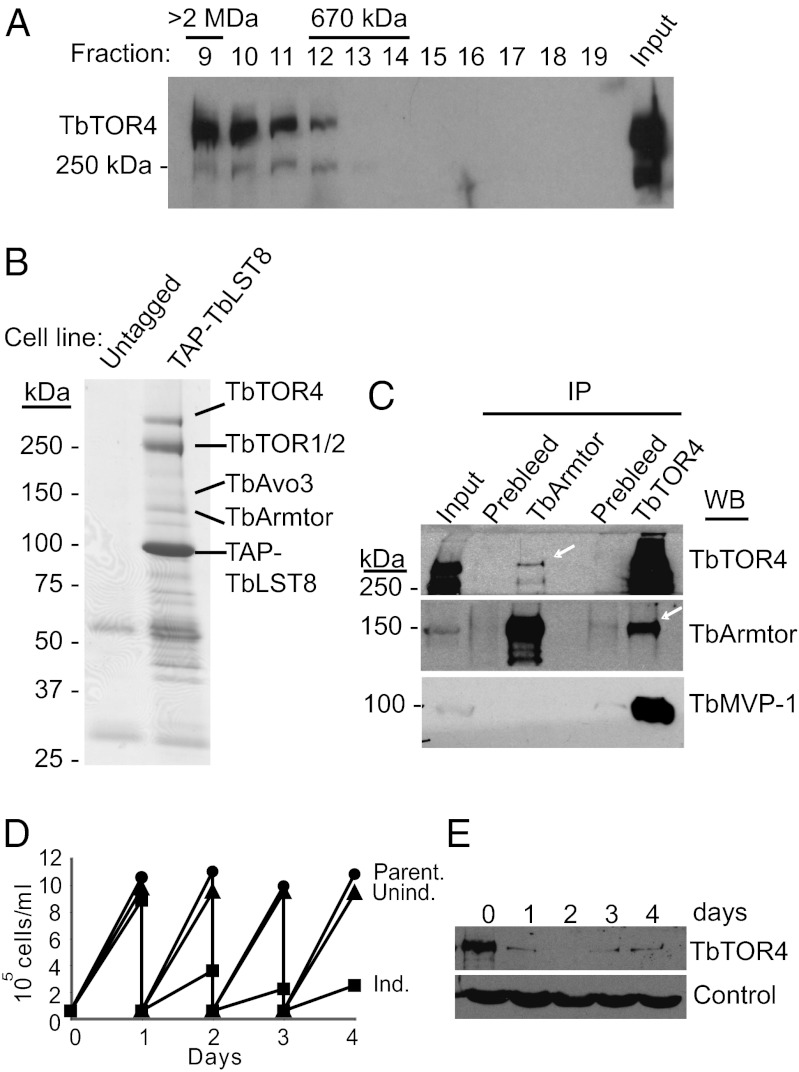
To determine if TbTOR4 assembles into a high-molecular-weight complex like other TORs (8), we examined the size of TbTOR4 by gel filtration. The elution profile revealed that TbTOR4 is part of a large complex with an apparent molecular mass >2 MDa (Fig. 1A). To characterize the potential TbTOR4-containing complex further, we performed tandem affinity purification (TAP) on the trypanosome LST8/GβL ortholog, a subunit present in all TOR complexes described so far (9–13). The T. brucei LST8 ortholog (Tb10.61.0700) shares domains with yeast and mammalian LST8, but these domains are separated by insertions, resulting in an unusually large protein (73 kDa) (Fig. S2). TAP-TbLST8 copurifying proteins, identified by tandem mass spectrometry, included TbTOR1 and TbTOR2, confirming that TbLST8 is a mammalian LST8/LST8 ortholog (Fig. 1B). TbLST8 also interacted with TbTOR4. This interaction corroborates that TbTOR4 belongs to the TOR kinase family, because others members within the PIKK superfamily do not interact with LST8. Coimmunoprecipitation experiments failed to detect an interaction between TbTOR4 and TbRaptor (TORC1) or TbAVO3 (TORC2) (Fig. S3A). However, proteomic analysis revealed several TAP-TbTOR4–associated proteins that previously were found by copurification with TAP-TbLST8, some of which had no homology to previously characterized mammalian TOR (mTOR)- or Saccharomyces cerevisiae TOR-interacting proteins. One such protein contained an Armadillo domain involved in protein–protein interactions. We named this protein “TbArmtor” (for “Armadillo-containing TOR-interacting protein”) (Tb927.4.2470). We confirmed that endogenous TbTOR4 interacts with TbArmtor by coimmunoprecipitation experiments using both anti-TbTOR4 and anti-TbArmtor antibodies (Fig. 1C). In addition, a major vault protein (MVP) ortholog, Tb927.5.4460, consistently copurified with TAP-TbTOR4. This interaction was confirmed by proteomic analysis of MVP1–associated proteins and coimmunoprecipitation experiments (Fig. 1C). Although the functions of TbArmtor and MVP proteins in trypanosomes are unknown, it has been shown previously that an MVP complex serves as a kinase scaffold (14, 15). These results suggest that TbTOR4 forms a structurally unique complex that we named “TbTORC4.” Because structurally distinct known TORCs are functionally different, TbTORC4 function may be distinct from that of TORC1 and TORC2.
Fig. 1.
TbTOR4 associates with both TbLST8 and Armtor. (A) Gel filtration elution profile of TbTOR4. Whole-cell lysates prepared in lysis buffer containing 0.3% CHAPS from wild-type bloodstream cells were loaded onto a Superose 6 sizing column. One-milliliter fractions were collected and processed for TbTOR4 immunoblotting. (B). Detection of TbLST8-interacting proteins in affinity purifications of TAP-LST8. Copurified material was resolved by SDS PAGE and visualized using Coomassie Brilliant Blue staining. An untagged cell line was used as negative control. (C) Endogenous TbTOR4 interacts with TbArmtor. Co-IP experiments were carried out using anti-TbTOR4 and anti-TbArmtor antibodies and conditions previously described (5). Western blotting of coimmunoprecipitated material revealed the binding of TbArmtor to TbTOR4. (D) TbTOR4 depletion halts cell proliferation. Growth curves of TbTOR4-depleted bloodstream trypanosomes after RNAi induction with doxycycline (Ind.) is compared with either uninduced (Unind.) or parental cell lines (Parent.) as controls. Cultures were diluted daily to 2.5 × 104 cells/mL to maintain cell density within a range that supports exponential growth. (E) Western blot analysis of TbTOR4 expression in total cell extracts (5 ×106 cells per lane) upon RNAi induction. Tubulin was used as a loading control.
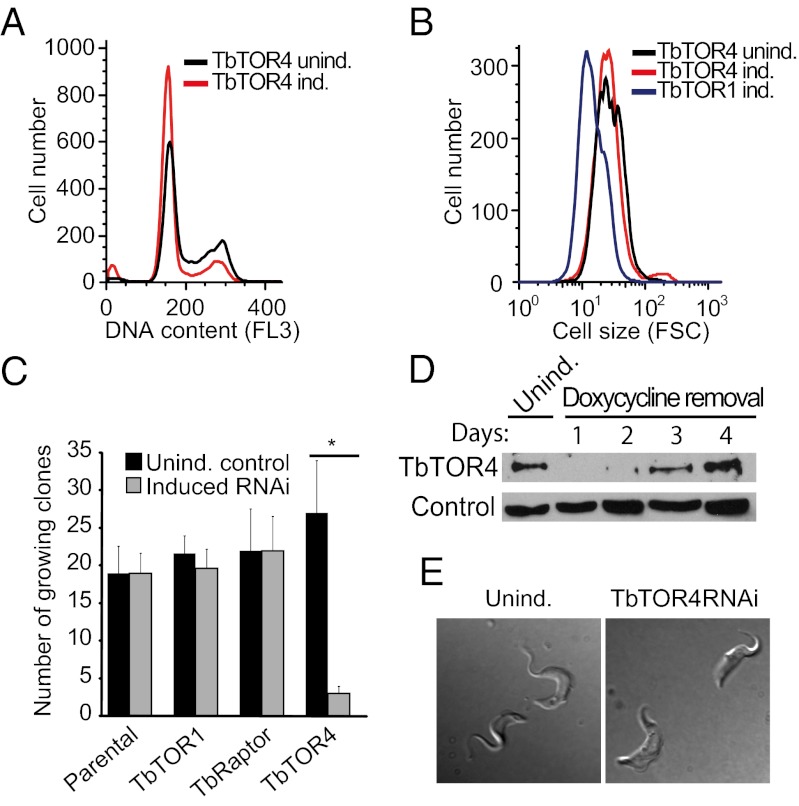
To investigate TbTORC4 function, we knocked down TbTOR4 in bloodstream trypanosomes (SI Materials and Methods). Cells depleted of TbTOR4 showed a reduced rate of proliferation (Fig. 1 D and E), as evidenced by an increased percentage of cells in G1 phase (Fig. 2A). However, the cells remained motile and did not show significant morphological defects. Reduced TbArmtor expression also halted cell-cycle progression at G1 phase (Fig. S3B). In other eukaryotes, TORC1 inhibition impedes cell growth, leading to both G1 arrest and a reduced cell size (4). TbTOR4 depletion did not cause a significant reduction in cell size, in contrast to TbTOR1 depletion (Fig. 2B), suggesting that TbTORC4 regulates cell proliferation rather than cell growth.
Fig. 2.
TbTOR4 knockdown induces irreversible cell-cycle arrest. (A) FACS analysis of DNA content in TbTOR4-depleted cells (TbTOR4 ind.) and in the uninduced control (TbTOR4 unind.) 48 h after RNAi induction with 1μg/mL doxycycline. (B) FACS analysis showing forward scatter of cells (FSC) with reduced expression of TbTOR4 (TbTOR4 ind.), TbTOR1 (TbTOR1 ind.), and the uninduced control (TbTOR4 unind). (C) Relative plating efficiency measured after induction of TbTOR1, TbRaptor, and TbTOR4 RNAi, compared with uninduced and parental cell lines. RNAi was induced by adding doxycycline to the medium for 48 h. Later, cells were washed to recover protein expression, and wells positive for cell growth were counted (SI Materials and Methods). (D) Western blotting analysis using anti-TbTOR4 antisera shows recovery of TbTOR4 expression after doxycycline removal. Cytosolic marker was used as loading control (31). (E) Cell morphology remodeling in bloodstream trypanosomes upon TbTOR4 loss of function. A stumpy morphology is visualized by DIC optics on live cells 96 h after TbTOR4 RNAi induction.
A cellular response to a stimulus can be reversible or, as in the case of development, largely irreversible (16). We sought to investigate the reversibility of the responses triggered upon TbTOR1 or TbTOR4 depletion by examining whether cells arrested in G1 phase can re-enter the cell cycle after recovery from the knockdown. Cells transiently depleted for TbTOR4 showed a significant reduction in plating efficiency (Fig. 2C). In contrast, TbTOR1- and TbRaptor-depleted cells re-entered the cell cycle and proliferated upon restoration of protein expression (Fig. 2C). To rule out the possibility that incomplete recovery from TbTOR4 depletion could account for this observation, we confirmed that TbTOR4 expression was restored to normal levels (Fig. 2D). These results suggest that TbTOR4 depletion triggers a cellular response similar to an irreversible G0 state and that TbTORC4 is functionally distinct from other TbTOR complexes.
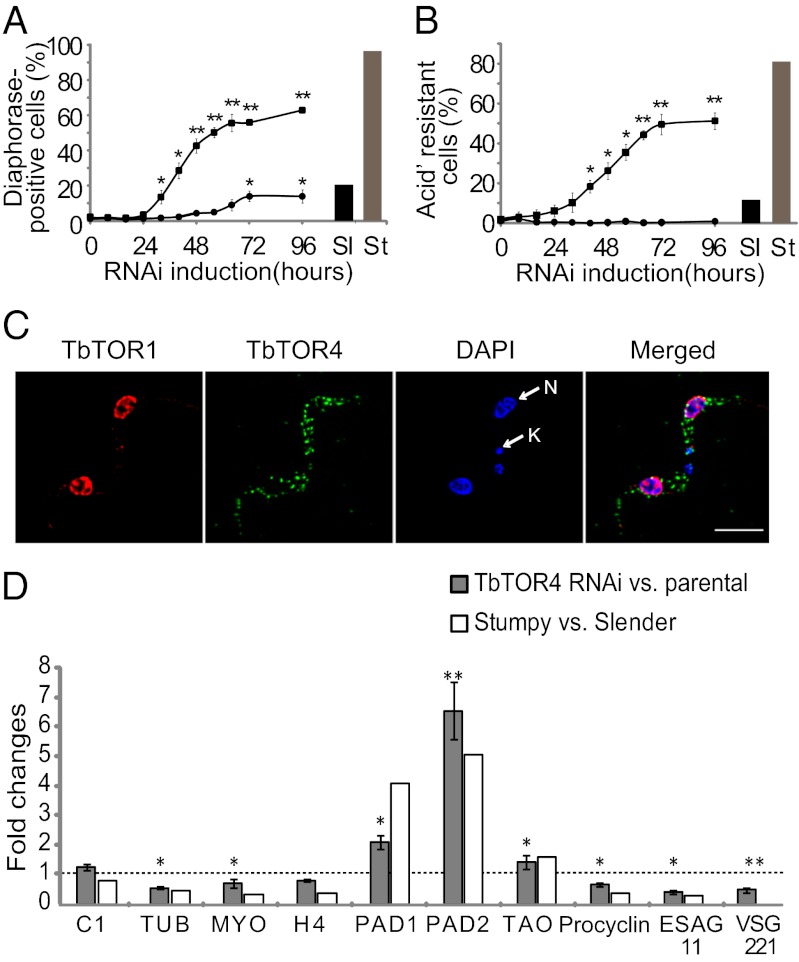
Upon high parasitemia, bloodstream trypanosomes undergo differentiation from a proliferative slender form to a quiescent stumpy form that involves transition to a G0 state. The cell-cycle arrest triggered upon TbTOR4 depletion (Fig. 2C) suggested that TbTORC4 might regulate this cell-differentiation program negatively (17). To investigate this possibility, we examined whether TbTOR4 depletion triggers physiological and morphological changes that normally occur during slender-to-stumpy form differentiation. TbTOR4 knockdown cells exhibited shortening of the flagella and rounded posterior ends (Fig. 2E), as well as reduced motility (Movies S1 and S2), suggesting the appearance of the classical stumpy shape. Mitochondrial activation, enabling use of proline as a source of ATP, is crucial for survival of the stumpy form in the tsetse fly midgut. Cytochemical analysis of NADH-diaphorase activity (18) revealed that TbTOR4 depletion increased mitochondrial oxidative capacity, reaching a maximum 96 h after RNAi induction (Fig. 3A and Fig. S4). Thus, TbTOR4 negatively regulates mitochondrial activity in the proliferative bloodstream trypanosome. We next analyzed whether reduction in TbTOR4 signaling leads to increased resistance to pH fluctuations, as previously described for the stumpy form (19). TbTOR4-depleted cells were resistant to mild acidic pH (Fig. 3B), whereas TbTOR1-depleted and controls cells were sensitive. Using 3D microscopy, we found that TbTOR4 was distributed within the cell cytoplasm, but TbTOR1 was restricted to the nucleus (Fig. 3C). Double indirect immunofluorescence analysis using anti-TbTOR4 antibody suggested that a fraction of TbTOR4 is associated with organelle membranes (Fig. S5 A–E). Moreover, TbTOR4 was found in insoluble fractions of T. brucei homogenates in a urea-sensitive manner, suggesting that TbTOR4 is a peripheral membrane protein (Fig. S5F).
Fig. 3.
TbTOR4 regulates processes involved in slender-to-stumpy differentiation including transcription remodeling. (A) The percentage of NADH dehydrogenase (diaphorase)-positive cells after the induction of TbTOR1 and TbTOR4 RNAi was determined at 8-h intervals for 96 h. Cells were visualized using DIC optics (Fig. S4). (B) The effect of mild acidic extracellular conditions on cell survival was assayed in TbTOR4- and TbTOR1-depleted cells. Cells were harvested at 8-h intervals for 96 h and incubated for 2 h at pH 5.5 and 37 °C. The number of remaining motile cells after the incubation was assessed with a hemocytometer. *P < 0.05, **P < 0.001, unpaired Student’s t test. (C) TbTOR4 and TbTOR1 are localized in distinct subcellular compartments. 3D microscopy localization was performed using anti-TbTOR1 rabbit antiserum and anti-TOR4 monoclonal antibody (Materials and Methods). Cells were counterstained with DAPI to locate the nuclear (N) and kinetoplast (K) mitochondrial DNA. (D) TbTOR4 loss of function induces changes in the transcriptome similar to stumpy differentiation. qRT-PCR analysis of TbTOR4-depleted cells versus parental cells was compared with slender-versus-stumpy RNA samples as a control. The genes analyzed by qRT-PCR were ESAG 11 (Tb427.BES40.19), histone H4 (H4) (Tb927.5.4240), TAO (Tb10.6k15.3640), PAD8 (Tb927.7.6000), control (C1) (Tb10.389.0540), PAD1 (Tb927.7.5930), VSG 221 (MITat 1.2/VSG2), procyclin (Tb927.6.480), myosin (MYO) (Tb11.01.7990), tubulin (TUB) (Tb927.1.2370), and PAD 2 (Tb927.7.5940). The histogram shows gene expression levels expressed as fold changes based on comparative cycle threshold (Ct) calculations using PIK-related (Tb927.2.2260) as house-keeping gene.
These results suggest that TbTOR4 function negatively regulates the slender-to-stumpy transition. Differentiation to the stumpy form also is characterized by specific changes in the transcriptome (20). The transcript profiles of TbTOR4-depleted and parental cells were compared with those of proliferative slender and quiescent stumpy trypanosomes (Fig. 3D). Transcripts known to be selectively regulated in the stumpy form(20) were analyzed by quantitative RT-PCR (qRT-PCR). The abundance of tubulin, myosin, and histone 4 mRNA decreased upon TbTOR4 depletion, but TAO mRNA was increased, as previously described (20). Transcription of the variant surface glycoprotein (VSG) gene, which is responsible for antigenic variation, was reduced markedly upon TbTOR4 depletion (Fig. 3D). VSG cotranscribed expression site-associated gene 11 (ESAG11) was down-regulated also (Fig. 3D), as previously described for the stumpy form (20). Thus, the transcript profile of TbTOR4-depleted cells resembles that of the stumpy form (Fig. 3D and Fig. S6).
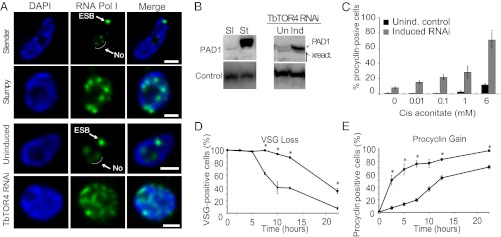
Although ribosomal DNA transcription occurs in the nucleolus, VSG and ESAG genes are transcribed by RNA polymerase I in a nuclear compartment named the “expression site body” (ESB) (21). In the quiescent stumpy form, RNA polymerase I delocalizes from the nucleolus to the nucleoplasm (Fig. 4A). Upon TbTOR4 depletion, RNA polymerase I was dispersed in the nucleoplasm, suggesting that TbTOR4 controls RNA polymerase I-mediated transcription and polymerase I nuclear localization upon development of the stumpy form (Fig. 4A and statistical analysis in Fig. S7). These findings confirm that TbTOR4 inhibits a program that mediates differentiation of the slender proliferative form into the stumpy quiescent form.
Fig. 4.
TbTOR4 loss of function triggers the quiescent stumpy form and preadapts bloodstream trypanosomes for differentiation to the insect form. (A) In the proliferative slender form, TbRPA1 (the largest subunit or RNA polymerase I) (21) is restricted to the nucleolus (No) and the ESB, whereas in the quiescent stumpy form TbRPA1 nuclear localization is scattered in multiple foci within the nucleoplasm. TbTOR4 depletion induced RNA polymerase I delocalization from the nucleolus and the ESB similar that detected in the stumpy form. Maximum intensity projections of three-channel 3D representative stacks show the anti-TbRPA1 signal in green and DAPI staining in blue. (Scale bars, 1 μm.) (B) PAD1 protein expression specific for the stumpy form (St) also is up-regulated upon TbTOR4 depletion as assessed by Western blotting using specific antibodies (22). Cytosolic marker was used as loading control (31). (C) Responsiveness to cis-aconitate during bloodstream-to-procyclic differentiation was analyzed in TbTOR4-depleted cells vs. uninduced control. Differentiation to the procyclic form was triggered in DTM medium at 28 °C using increasing concentrations of cis-aconitate (SI Materials and Methods). (D and E) TbTOR4 loss of function promotes a rapid differentiation to the procyclic form. Shown are the results of a time-course experiment following changes in the expression of stage-specific markers during in vitro bloodstream-to-procyclic differentiation. The expression on the surface of the procyclin and VSG was followed by indirect immunofluorescence (SI Materials and Methods).
Next, we investigated whether loss of TbTOR4 induces commitment to the stumpy form, the form preadapted to differentiation to the insect procyclic form. A recent study (22) identified a family of carboxylate transporters named “proteins associated with differentiation” (PADs) whose expression is stumpy-specific and required for increased responsiveness to cis-aconitate during bloodstream stumpy-to-procyclic differentiation. Upon TbTOR4 depletion, PAD1 expression increased both at the mRNA and protein levels (Figs. 3D and 4B). The expression of PADs in TbTOR4-depleted cells may confer increased potential for differentiation to the procyclic insect form, as suggested previously for the stumpy form (22). Bloodstream-to-procyclic differentiation was triggered with increasing concentrations of cis-aconitate. TbTOR4-depleted cells were hypersensitive to cis-aconitate as compared with the uninduced control (Fig. 4C). Moreover, TbTOR4-depleted cells rapidly released VSG from the membrane (Fig. 4D) and expressed procyclin during the course of differentiation (Fig. 4E). This finding suggests that the loss of TbTOR4 function preadapts the bloodstream trypanosome to differentiate rapidly to the insect form. Conversely, TbTOR1-depleted cells showed a reduced rate of differentiation into procyclic forms (Fig. S8), similar to that previously described with TORC1 inhibition during differentiation processes in metazoans (23). Collectively, these results suggest that TbTOR4 depletion is necessary for bloodstream-to-procyclic differentiation to occur rapidly and efficiently through the development of the preadapted stumpy form.
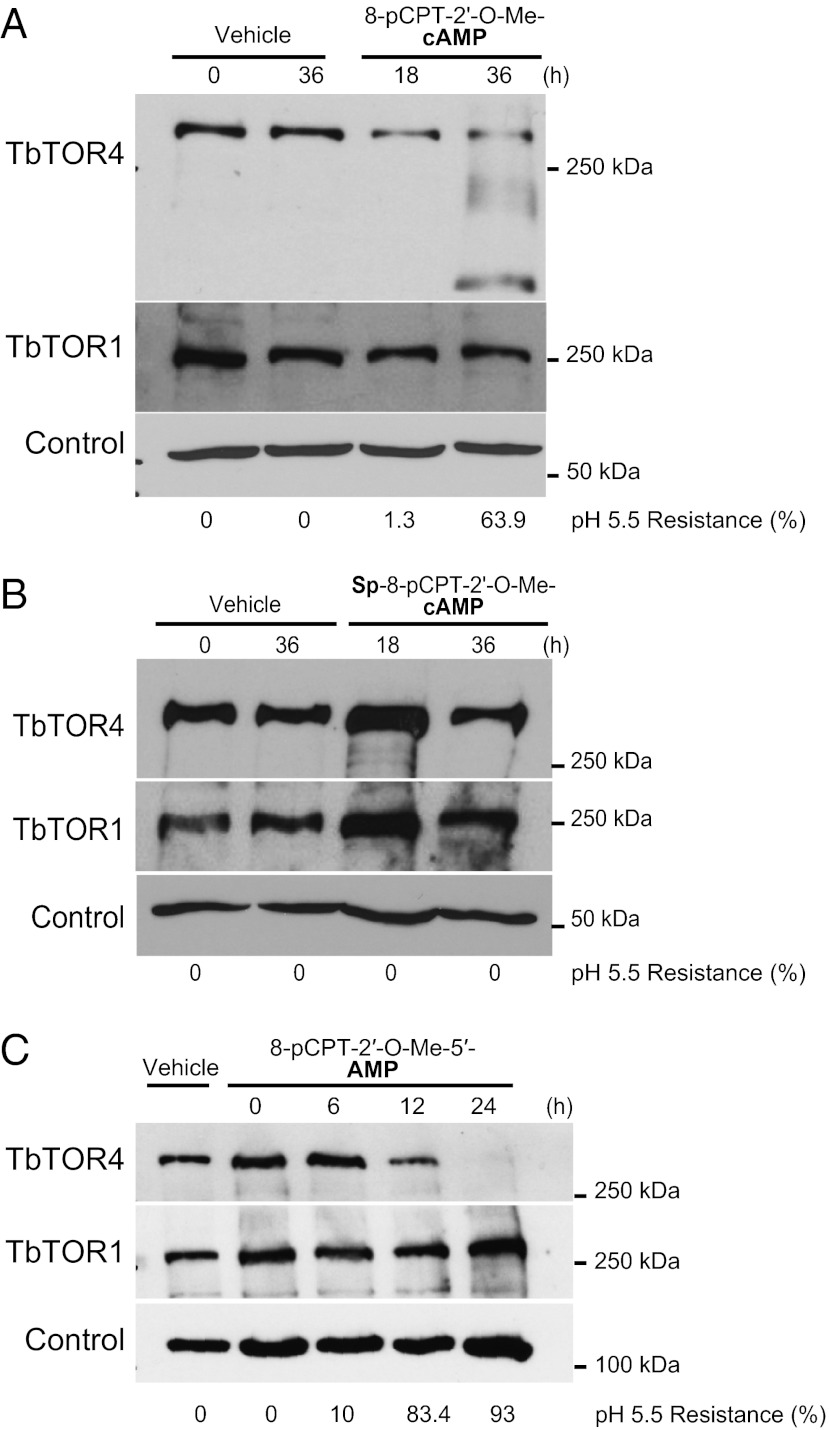
To gain insight into the regulation of the signaling pathway controlled by TbTORC4, we analyzed TbTOR4 expression in stumpy-like monomorphic trypanosomes. Hydrolysable cAMP analogs promote differentiation to stumpy-like trypanosomes (24). Time-course analysis of cells grown in the presence of the hydrolysable cAMP analog 8-pCPT-2′-O-Me-cAMP showed a progressive decrease in TbTOR4 protein, but TbTOR1 remained unaltered (Fig. 5A). The hydrolysable cAMP analog induced stumpy-like cells, as confirmed by the acquisition of resistance to mild acidic pH (Fig. 5A). Conversely, the hydrolysis-resistant cAMP analog Sp-8-pCPT-2′-O-Me-cAMPS did not reduce TbTOR4 protein levels or induce stumpy-like cells (Fig. 5B). Next, we investigated whether the AMP analog 8-pCPT-2′-O-Me-5′-AMP induces differentiation into the stumpy form. The AMP analog not only induced stumpy differentiation but also promoted rapid TbTOR4 down-regulation but did not affect TbTOR1 expression (Fig. 5C). Thus, AMP appears to be necessary to down-regulate TbTOR4 and thereby to trigger stumpy differentiation. Furthermore, these data argue against a cAMP-dependent pathway underlying slender-to-stumpy differentiation as suggested previously (24) and point to AMP as the signal that promotes the differentiation process. These results suggest a model in which reduced cellular energy (high AMP:ATP ratio) inactivates TbTORC4 to trigger differentiation into the quiescent stumpy form.
Fig. 5.
AMP and hydrolysable analogs of cAMP inhibited TbTOR4 expression and induced the stumpy-like quiescent form. (A) The membrane-permeable and hydrolysable cAMP analog (8-pCPT-2′-O-Me-cAMP) (10 μM) down-regulates TbTOR4 expression and induces stumpy-like (pH5.5-resistant) cells in monomorphic trypanosomes; TbTOR1 expression is unaffected. Anti-tubulin was used as a loading control. (B) The hydrolysis-resistant cAMP analog (Sp-8-pCPT-2′-O-Me-cAMPS) (10 μM) does not affect TbTOR4 expression levels or induction of stumpy-like cells. Anti-tubulin was used as a loading control. (C) Membrane permeable 5′-AMP (8-pCPT-2′-O-Me-5′-AMP) (5 μM) promotes rapid TbTOR4 down-regulation and induces stumpy-like monomorphic trypanosomes. Anti-TbRRP4 was used as a loading control (31).
In this study, we describe a TOR kinase, TbTOR4, that forms a structurally and functionally distinct complex, TbTORC4. TbTORC4 contains TbLST8, MVP, and TbArmtor. TbTORC4 governs a signaling pathway distinct from those controlled by the conventional TOR complexes TORC1 and TORC2. The TbTORC4 pathway regulates a developmental switch between the slender and stumpy bloodstream forms of T. brucei. As in yeast, multiple TOR kinases might have arisen in trypanosomes through duplication followed by functional diversification in Kinetoplastidae (25). Most likely, the novel TORs provided new functions and conferred selective advantages to the complex life cycles of these early eukaryotes.
Loss of TbMAPK5 or zinc finger kinase (ZFK) enhances differentiation to the stumpy form in pleomorphic but not in monomorphic trypanosomes (26, 27). TbTOR4 is the only protein, thus far, whose loss has been shown to trigger this developmental process in a monomorphic strain. TbMAPK5, TbZFK, and TbTOR4 likely participate in a complex signaling pathway that modulates slender-to-stumpy differentiation. Interestingly, the atypical ZFK belongs to the AGC kinase family, well-known TOR substrates in both yeast and mammals, suggesting ZFK might be a downstream effector of TbTOR4.
A TbTORC4 pathway may be unique to Kinetoplastidae. However, there is increasing evidence for mTORC1- and mTORC2-independent functions of TOR in mammals. The TSC–mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in an mTORC1- and mTORC2-independent manner (28), suggesting that a distinct TORC may exist in other eukaryotes. Finally, TbTOR4 represents a valid and promising drug target for the treatment of the disease, given that TOR4 inactivation leads to an irreversible quiescent state of the parasite.
Materials and Methods
Resistance to Mild Acidic pH Assay.
Bloodstream trypanosome survival in mildly acidic conditions was performed as described previously (19). Briefly, 106 cells were resuspended in 1 mL of PBS supplemented with glucose [1% (wt/vol)], leupeptin (20 mg/mL) and FBS (15%) for 2 h at 37 °C. The percentage of cells resistant to mild acidic pH was determined by comparing the number of living and motile cells in pH 5.5 buffer and in pH 7.4 buffer using a Neubauer hemocytometer. Two samples per time point for each experimental condition were analyzed by two independent observers in four independent experiments. Statistical analysis was performed by unpaired t test. Slender- and stumpy-enriched samples were processed as negative and positive controls, respectively.
Coimmunoprecipitation.
Cells (2.5 109) were washed with PBS, lysed in 1 mL lysis buffer B (SI Materials and Methods), and subsequently centrifuged at 13,000 × g for 15 min. The soluble fraction was cleared with 40 μL of Sepharose 4B (Sigma-Aldrich) for 2 h. The suspension was incubated overnight with 15 μL of the specified antiserum or unspecific antibody (prebleed). Immunoprecipitates were washed three times and resuspended in 2× Laemmli buffer. Samples were resolved in a 6–8% SDS/PAGE, and proteins were transferred to nitrocellulose membranes and detected by Western blotting using specific antibodies (SI Materials and Methods).
Microscopy.
Cellular localization analysis was performed by 3D immunofluorescence as described previously (29). Images displayed in the figures are maximum intensity projections from digitally deconvolved multichannel 3D image data sets using Huygens Essential software v. 2.9 (Scientific Volume Imaging). Cells were permeabilized with 1% Nonidet P-40 for 20 min. Immunofluorescence analysis was carried out in 0.5% blocking reagent (Roche) in PBS (Sigma) using the monoclonal anti-TbTOR4 antibody (5H7) (1:2) and rabbit affinity-purified antiserum anti-TbTOR1 (1:400) (5). Affinity purified anti-TbRPA1 (21) (1:600) was used to localize RNA polymerase I in stumpy form and TbTOR4-depleted cell nuclei (SI Materials and Methods). Pseudocoloring and maximum intensity projections were performed using ImageJ software v. 1.43 (National Institutes of Health). Live-cell imaging was performed at 37 °C, and cells were visualized under differential interference contrast (DIC) optics.
Differentiation to Procyclic Form.
Differentiation from the bloodstream form to the insect procyclic form was induced by cis-aconitate, with a temperature shift from 37 °C to 28 °C and switching the medium to Differentiating Trypanosome Medium (DTM). To assess the differentiation process, we monitored the developmental expression of the surface glycoproteins by double immunofluorescence using anti-procyclin and anti-VSG221 antibodies. Importantly, cold shock at 20 °C was not carried out to avoid premature procyclin expression; thus procyclin expression is driven by the availability of cis-aconitate and the sensitivity of the trypanosome to this metabolite (30). The procyclic form obtained after TbTOR4 depletion consistently was maintained for longer periods than in control cells.
Supplementary Material
Acknowledgments
We thank Keith Matthews for supplying anti-PAD antibodies; A. Estevez for the control antibodies anti-CSM and anti-RRP44; J. Bangs for anti-TbBiP; S. Reed for anti-TcP0; D. González Pacanowska for anti-LmMVAK antibodies; and Sabine Hilfiker for discussions. This work was supported by Grant SAF2009-07587 from the Spanish Ministerio de Ciencia e Innovación, Grant CTS-5841 from the Junta de Andalucia, Grant RD06/0001/0010 from Red de Investigacion de Centros de Enfermedades Tropicales (RICET) and Howard Hughes Medical Institute Grant HHMI-55005525. J.-M.B. is supported by Miguel Servet Fellowship CP09/00300. M.N.H. acknowledges the Swiss National Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210465109/-/DCSupplemental.
References
- 1.Fenn K, Matthews KR. The cell biology of Trypanosoma brucei differentiation. Curr Opin Microbiol. 2007;10:539–546. doi: 10.1016/j.mib.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner CM, Aslam N, Dye C. Replication, differentiation, growth and the virulence of Trypanosoma brucei infections. Parasitology. 1995;111:289–300. doi: 10.1017/s0031182000081841. [DOI] [PubMed] [Google Scholar]
- 3.Matthews KR, Gull K. Commitment to differentiation and cell cycle re-entry are coincident but separable events in the transformation of African trypanosomes from their bloodstream to their insect form. J Cell Sci. 1997;110:2609–2618. doi: 10.1242/jcs.110.20.2609. [DOI] [PubMed] [Google Scholar]
- 4.Soulard A, Cohen A, Hall MN. TOR signaling in invertebrates. Curr Opin Cell Biol. 2009;21:825–836. doi: 10.1016/j.ceb.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Barquilla A, Crespo JL, Navarro M. Rapamycin inhibits trypanosome cell growth by preventing TOR complex 2 formation. Proc Natl Acad Sci USA. 2008;105:14579–14584. doi: 10.1073/pnas.0802668105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Jesus TC, et al. Target of rapamycin (TOR)-like 1 kinase is involved in the control of polyphosphate levels and acidocalcisome maintenance in Trypanosoma brucei. J Biol Chem. 2010;285:24131–24140. doi: 10.1074/jbc.M110.120212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madeira da Silva L, Beverley SM. Expansion of the target of rapamycin (TOR) kinase family and function in Leishmania shows that TOR3 is required for acidocalcisome biogenesis and animal infectivity. Proc Natl Acad Sci USA. 2010;107:11965–11970. doi: 10.1073/pnas.1004599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wullschleger S, Loewith R, Oppliger W, Hall MN. Molecular organization of target of rapamycin complex 2. J Biol Chem. 2005;280:30697–30704. doi: 10.1074/jbc.M505553200. [DOI] [PubMed] [Google Scholar]
- 9.Loewith R, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 10.Kim DH, et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 11.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 12.Matsuo T, Otsubo Y, Urano J, Tamanoi F, Yamamoto M. Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol Cell Biol. 2007;27:3154–3164. doi: 10.1128/MCB.01039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang G, et al. Autophagy induced by Alexander disease-mutant GFAP accumulation is regulated by p38/MAPK and mTOR signaling pathways. Hum Mol Genet. 2008;17:1540–1555. doi: 10.1093/hmg/ddn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolli S, Zito CI, Mossink MH, Wiemer EA, Bennett AM. The major vault protein is a novel substrate for the tyrosine phosphatase SHP-2 and scaffold protein in epidermal growth factor signaling. J Biol Chem. 2004;279:29374–29385. doi: 10.1074/jbc.M313955200. [DOI] [PubMed] [Google Scholar]
- 15.Berger W, Steiner E, Grusch M, Elbling L, Micksche M. Vaults and the major vault protein: Novel roles in signal pathway regulation and immunity. Cell Mol Life Sci. 2009;66:43–61. doi: 10.1007/s00018-008-8364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: Dynamics of regulatory and signaling pathways in the cell. Curr Opin Cell Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 17.Blomen VA, Boonstra J. Cell fate determination during G1 phase progression. Cell Mol Life Sci. 2007;64:3084–3104. doi: 10.1007/s00018-007-7271-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vassella E, Reuner B, Yutzy B, Boshart M. Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. J Cell Sci. 1997;110:2661–2671. doi: 10.1242/jcs.110.21.2661. [DOI] [PubMed] [Google Scholar]
- 19.Nolan DP, Rolin S, Rodriguez JR, Van Den Abbeele J, Pays E. Slender and stumpy bloodstream forms of Trypanosoma brucei display a differential response to extracellular acidic and proteolytic stress. Eur J Biochem. 2000;267:18–27. doi: 10.1046/j.1432-1327.2000.00935.x. [DOI] [PubMed] [Google Scholar]
- 20.Kabani S, et al. Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei. BMC Genomics. 2009;10:427. doi: 10.1186/1471-2164-10-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navarro M, Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414:759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- 22.Dean S, Marchetti R, Kirk K, Matthews KR. A surface transporter family conveys the trypanosome differentiation signal. Nature. 2009;459:213–217. doi: 10.1038/nature07997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyler WA, et al. Activation of the mammalian target of rapamycin (mTOR) is essential for oligodendrocyte differentiation. J Neurosci. 2009;29:6367–6378. doi: 10.1523/JNEUROSCI.0234-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laxman S, Riechers A, Sadilek M, Schwede F, Beavo JA. Hydrolysis products of cAMP analogs cause transformation of Trypanosoma brucei from slender to stumpy-like forms. Proc Natl Acad Sci USA. 2006;103:19194–19199. doi: 10.1073/pnas.0608971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shertz CA, Bastidas RJ, Li W, Heitman J, Cardenas ME. Conservation, duplication, and loss of the Tor signaling pathway in the fungal kingdom. BMC Genomics. 2010;11:510. doi: 10.1186/1471-2164-11-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller IB, Domenicali-Pfister D, Roditi I, Vassella E. Stage-specific requirement of a mitogen-activated protein kinase by Trypanosoma brucei. Mol Biol Cell. 2002;13:3787–3799. doi: 10.1091/mbc.E02-02-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vassella E, et al. Deletion of a novel protein kinase with PX and FYVE-related domains increases the rate of differentiation of Trypanosoma brucei. Mol Microbiol. 2001;41:33–46. doi: 10.1046/j.1365-2958.2001.02471.x. [DOI] [PubMed] [Google Scholar]
- 28.Patursky-Polischuk I, et al. The TSC-mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol Cell Biol. 2009;29:640–649. doi: 10.1128/MCB.00980-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landeira D, Bart JM, Van Tyne D, Navarro M. Cohesin regulates VSG monoallelic expression in trypanosomes. J Cell Biol. 2009;186:243–254. doi: 10.1083/jcb.200902119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engstler M, Boshart M. Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes Dev. 2004;18:2798–2811. doi: 10.1101/gad.323404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Estévez AM, Kempf T, Clayton C. The exosome of Trypanosoma brucei. EMBO J. 2001;20:3831–3839. doi: 10.1093/emboj/20.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.