Abstract
Bacteria display considerable variation in their overall base compositions, which range from 13% to over 75% G+C. This variation in genomic base compositions has long been considered to be a strictly neutral character, due solely to differences in the mutational process; however, recent sequence comparisons indicate that mutational input alone cannot produce the observed base compositions, implying a role for natural selection. Because bacterial genomes have high gene content, forces that operate on the base composition of individual genes could help shape the overall genomic base composition. To explore this possibility, we tested whether genes that encode the same protein but vary only in their base compositions at synonymous sites have effects on bacterial fitness. Escherichia coli strains harboring G+C-rich versions of genes display higher growth rates, indicating that despite a pervasive mutational bias toward A+T, a selective force, independent of adaptive codon use, is driving genes toward higher G+C contents.
Keywords: bacterial adaptation, genome evolution, mutational patterns
Bacterial genomes are highly variable in their overall base compositions, with sequenced genomes ranging from 13% to 75% G+C (1, 2). Genomic base composition in bacteria was the first genetic character to be considered selectively neutral, in the sense that the variation is not adaptive but instead due solely to interspecies differences in the mutational process (3, 4). This view was bolstered by sequence information showing that the base compositional differences among bacteria were most pronounced at synonymous and noncoding positions, sites that are thought to be under the least selective constraints (5). Furthermore, repeated attempts to link genomic base composition with an environmental factor or other selective processes have met with limited success (6–10).
Recently, however, studies based on the comparative analyses of gene sequences have challenged the notion that base composition is driven solely by mutational biases. These studies revealed that mutation is universally biased toward A+T, suggesting selection as an agent that maintains the contemporary base compositions in bacterial genomes (11–13). A difficulty with ascribing a role for selection in determining genomic base composition is that the selective force must be sufficiently strong to operate on a single-nucleotide change despite the fact that each change makes only a tiny contribution toward the overall G+C content of the genome. However, because bacterial genomes are composed primarily of protein-coding genes (14), a selective force that acts on each gene to increase its G+C content can cumulatively influence the overall genomic base composition. To test this possibility, we examined the fitness effects of protein-coding gene variants that differed only in their base composition at synonymous codon positions. For two genes, both of which encode proteins that are neither native nor physiologically relevant to Escherichia coli, we detected a strong and significant association between G+C contents and bacterial fitness. G+C-enrichment of mRNAs is observed in a majority of bacterial species, and the G+C content of functionless, nontranscribed regions in the E. coli genome are decidedly lower than that of fourfold degenerate sites in protein-coding genes. Taken together, these data indicate that selection operating on the base composition of individual coding regions guides genomic base composition in bacteria.
Results
Base Composition of an Expressed Gene Impacts E. coli Growth Rate.
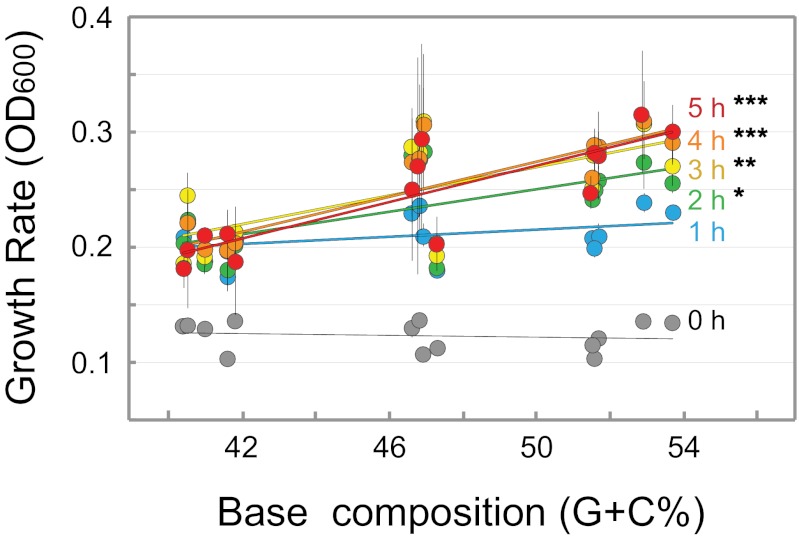
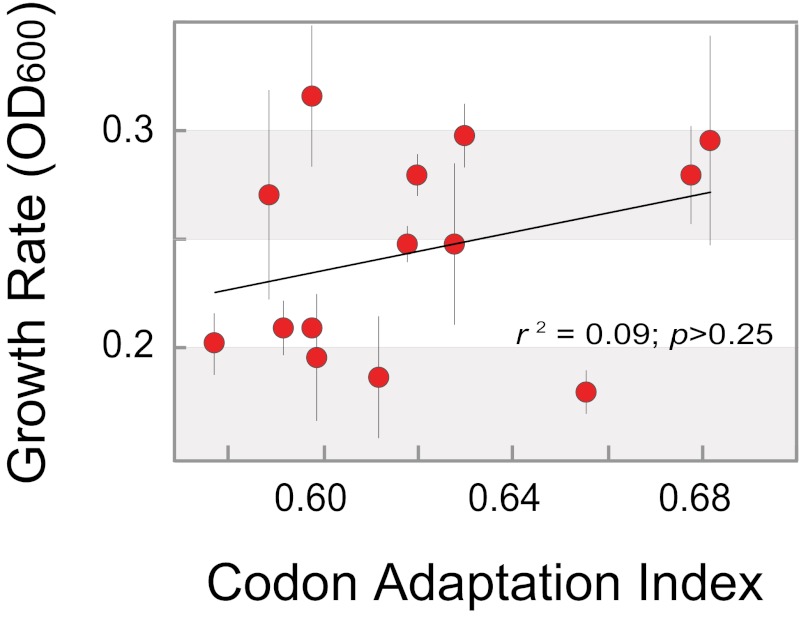
E. coli strains containing variants of a plasmid-borne green fluorescent protein (GFP) gene that differ in base composition (40.4–53.7% G+C; averaging 126 substitutions between gene pairs) were tested for growth at various time points after GFP induction. At 2 h postinduction, there is a significant association between the G+C composition of the expressed GFP gene and bacterial generation time, with strains expressing genes of higher G+C contents exhibiting significantly higher growth rates; the effect becomes more pronounced at later time points (Fig. 1). Performing the same experiment without IPTG induction of the GFP genes abolished the association between bacterial growth rate and the G+C contents of the GFP genes (Fig. S1), indicating that the effect of base composition on growth rate requires gene expression. Additionally, there is no association between the base composition of a GFP gene and its level of protein production (as measured by GFP fluorescence) at any point during the growth experiment (Fig. S2), nor is there an association between codon adaptation index (CAI) of the GFP gene variants and growth rates (Fig. 2).
Fig. 1.
Growth rates of isogenic strains expressing GFP genes of different G+C contents. Growth rate (OD600) was measured hourly for 5 h after induction with 1 mM IPTG. A significant association between %G+C and growth rate is observed at 2 h after induction and is evident at all later time points (0 h, r2 = 0.03; 1 h, r2 = 0.15; 2 h, r2 = 0.40; 3 h, r2 = 0.47; 4 h, r2 = 0.67; 5 h, r2 = 0.72). Asterisks denote level of significance: *P < 0.05, **P < 0.01, ***P < 0.001. Values represent the mean ± SD of three replicates. For detailed results of regressions, see Table S1.
Fig. 2.
No significant association between codon adaptation index (CAI) and growth rate. Growth of E. coli strains expressing GFP genes with CAI values ranging from 0.58 to 0.68 (calculated using CAIcal; ref. 15) were monitored by measuring OD600 every hour. Values shown were measured at 5 h after gene induction and represent the mean ± SD of three replicates.
Association Between Base Composition and Growth Rate Is Observed for Multiple Genes.
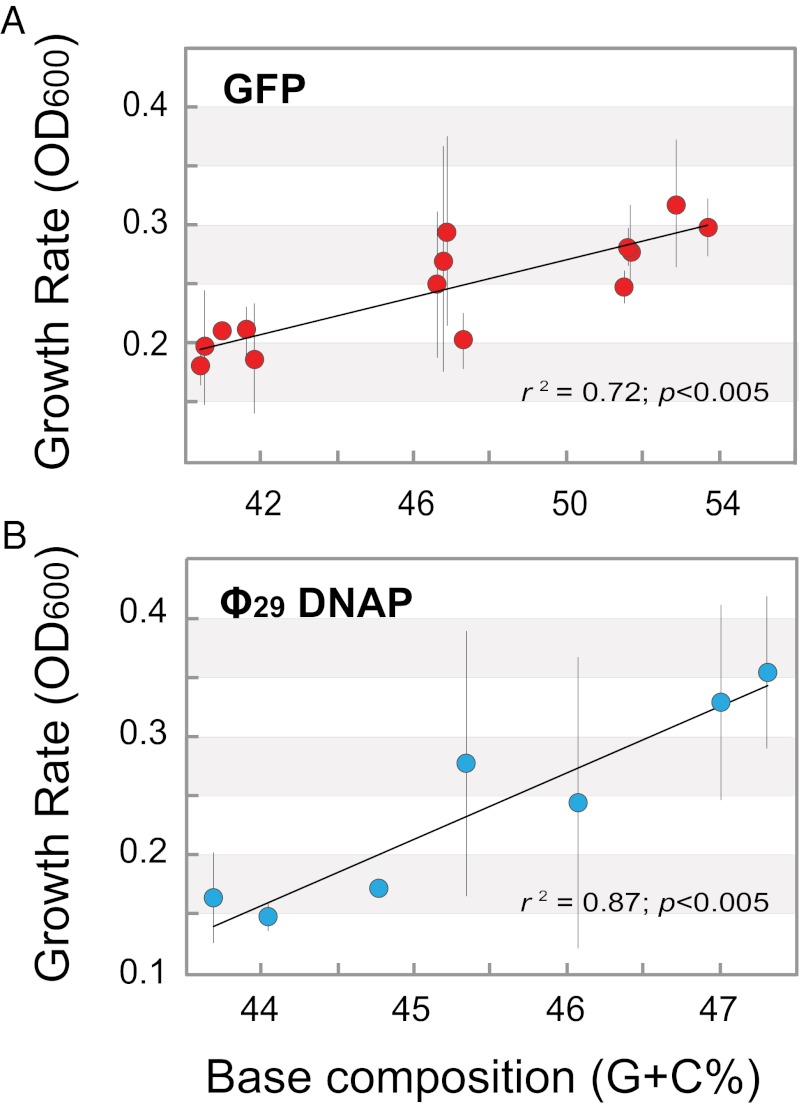
To determine whether the association between base composition and E. coli growth rate was in some way limited to the GFP constructs, we tested a set of Bacillus phage ϕ29 DNA polymerase genes that varied over a more confined range of G+C contents (43.7–47.2% G+C; averaging 213 substitutions between gene pairs). Again, the sequence variants of this gene have similar CAI values, and each encodes the identical protein. As observed with the GFP constructs, there is a significant association between the base compositions of the ϕ29 DNA polymerase gene and bacterial growth rate (Fig. 3), and the association is only apparent when the gene is induced (Fig. S3).
Fig. 3.
Significant association between bacterial growth rate and base composition for genes spanning different ranges of G+C contents. Growth rates for E. coli strains expressing GFP gene variants with base compositions ranging from 40.4–53.7% G+C (A) and Bacillus phage ϕ29 DNA polymerase gene variants with base compositions ranging from 43.7–47.2% G+C (B), measured at 5 h after induction with 1mM IPTG. Values represent the mean ± SD of three replicates.
Obstruction of Translation Diminishes the Effects of Base Composition on Growth Rate.
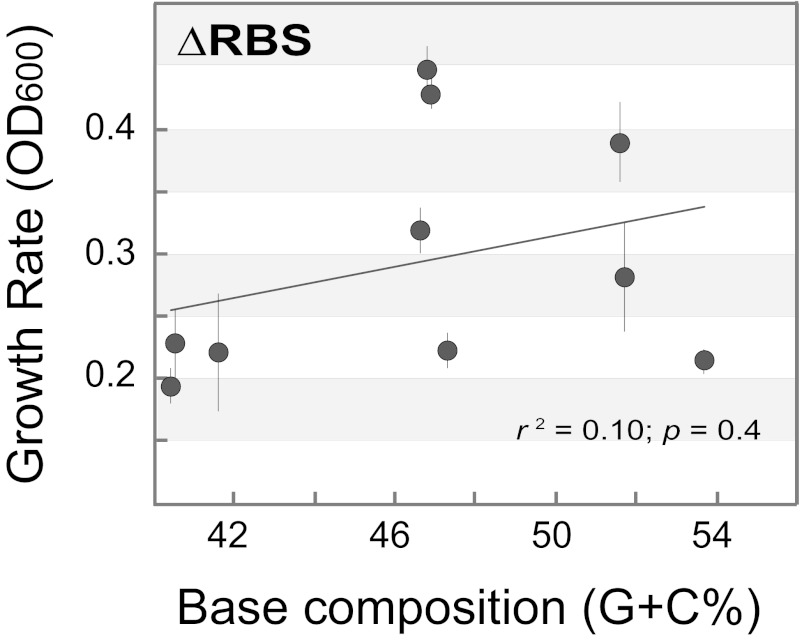
To establish whether the effect of transcript base composition on bacterial growth rate requires translation, we tested the growth rates of E. coli strains that harbor constructs in which translation was prevented through the removal of the ribosome-binding site and start codon, and the introduction of stop codons at the 5′ ends, of the GFP gene. When transformed with these constructs, an association between the base composition of induced genes and bacterial growth rate remains, but it is no longer significant (Fig. 4). This trend suggests that there are other unrecognized selective pressures that counteract the A+T-biased mutational process.
Fig. 4.
Effect of translation on the association between base composition and bacterial growth rates. Growth of E. coli strains expressing GFP gene variants of different base compositions in which translation of the GFP gene was prevented by removing the ribosome binding sites (ΔRBS) and start codons. Growth rates measured at 5 h after induction and represent the mean ± SD of three replicates.
G+C Enrichment of mRNAs Occurs in the Majority of Bacterial Species.
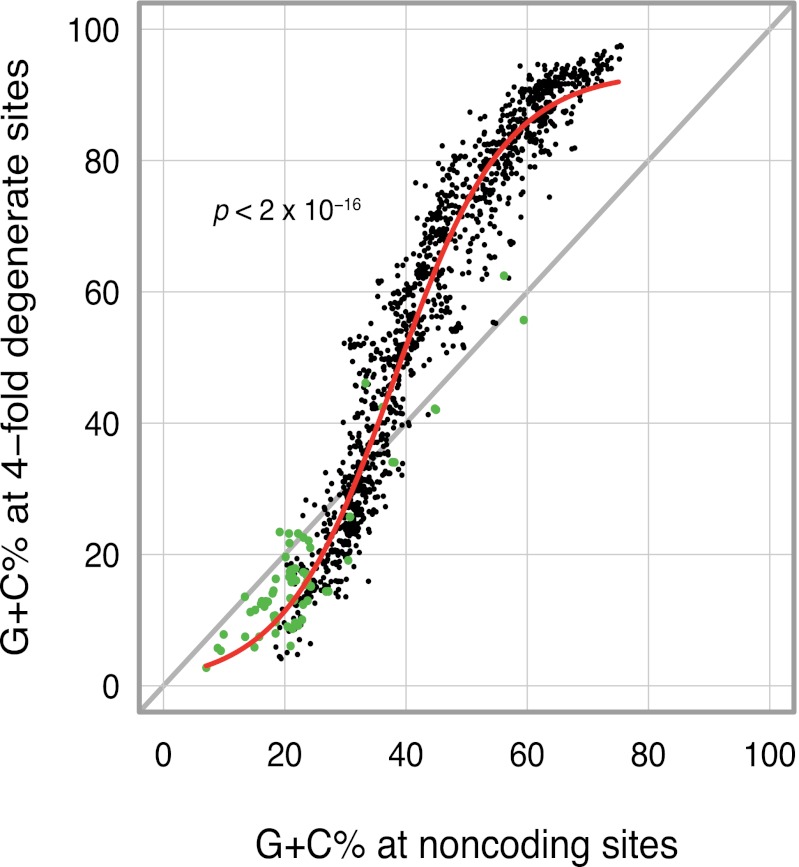
If there is widespread selection across bacterial species for G+C-rich transcripts, then synonymous sites can no longer be considered as evolving in a neutral manner and their G+C contents are expected to be higher than those of noncoding regions within the corresponding genome. We examined the association between nucleotide compositions of fourfold degenerate sites (GC4) and noncoding genomic regions (GCnc) in a phylogenetically diverse sample of sequenced bacterial genomes and found a positive nonlinear relationship in which GC4 was higher than GCnc in the vast majority of genomes with base compositions over 40% G+C (Fig. 5), suggesting that selection acts to enrich transcripts with G/C nucleotides. The converse was detected (i.e., GC4 < GCnc) in most bacterial genomes with base compositions less than 40% G+C.
Fig. 5.
Relationship between the average G+C content at fourfold degenerate sites of all protein-coding genes and the average G+C content of noncoding intergenic regions in fully sequenced bacterial genomes (n = 1430). Green circles denote bacteria with genome size of less than one megabase (n = 67), and the red line indicates the logistic regression model fitted to the data.
To further understand the influence of transcription on genomic nucleotide composition, we measured the G+C content of nontranscribed intergenic regions situated between the transcription start sites (TSSs) of 222 divergently transcribed adjacent genes in E. coli. We found that the base composition of these intergenic regions averaged only 44.1% G+C, even after accounting for (and removing) the A+T-rich −10 and −35 promoters contained within these regions (Fig. S4). This base composition is significantly lower than GC4 (P < 10−16, two-sample t test; Fig. S4) but higher than the equilibrium G+C content of 32% (0.28–0.37; 95% CI) predicted for E. coli (16).
Discussion
We demonstrate that the overall base composition of highly expressed genes affects bacterial fitness: Strains expressing equivalent genes of higher G+C contents have significantly faster doubling times. These findings shed light on the evidence that G+C content of many bacterial genomes is higher than that predicted on the basis of mutational patterns, but only to the extent that selection is operating at the level of individual protein-coding sequences. The interplay between mRNA phenotype and the translation machinery partially explains the observed phenomenon based on several observations: (i) The association between increasing G+C contents and growth rates is observed only in genes that are both transcribed and translated (Fig. 4); (ii) there is no correspondence between the base composition of a GFP gene and its level of protein production (Fig. S2); (iii) mRNAs with higher G+C contents exhibited higher stability (Fig. S5); and (iv) mRNAs are G+C-enriched at fourfold degenerate sites relative to noncoding sites in the majority of bacteria (Fig. 5) and to regions shown experimentally to be nontranscribed in the E. coli genome (Fig. S4).
The degree of secondary structure near the 5′-end of mRNAs has been shown to negatively influence the levels of protein expression (17–19), which is also observed for the GFP sequence variants that we tested (Fig. S6A). However, the stability of mRNA secondary structures near their 5′ ends (-4 to +37) was not correlated with bacterial growth rates (Fig. S6B), ruling out the possibility that the higher fitness of G+C-rich GFP variants results from suppressing the translation of wasteful proteins. Other features of a coding region, aside from those that specify mRNA stability and the amino acid sequence of its encoded protein, may influence its nucleotide sequence, as shown recently for the tendency of bacterial genes to avoid internal Shine–Delgarno sequences (20).
Examined across bacterial genomes, it has been recorded by us and by others (e.g., refs. 5, 13, 21, and 22) that the G+C contents of synonymous sites are often higher than those of noncoding regions (Fig. 5). Because synonymous sites were previously considered to be evolving neutrally in all but the most highly expressed bacterial genes, this pattern has been taken as evidence that noncoding regions are under some form of selection for lower G+C contents. However, our results indicate the converse: Selection instead serves to increase the G+C contents of synonymous sites. Because this selective force for higher G+C contents is operating on expressed sequences, we examined the base composition of spacers situated between divergent TSSs, regions with very low probability of expression (Fig. S4). The G+C content of this set of nontranscribed regions is substantially lower than GC4 but higher than the predicted equilibrium G+C% of 0.32 (16), suggesting that although the region may be evolving in a predominately neutral manner, it might still contain regulatory sequences that are under selection.
It is curious that the G+C content of noncoding sequences (GCnc) is higher than that of synonymous sites (GC4) in the most A+T-rich genomes (Fig. 5). Because A/T-to-G/C mutations at synonymous sites are rare in A+T-rich genomes (12, 13, 23), it is possible that the enrichment of A and U in mRNA transcripts is caused by the formation of secondary structures that are stabilized by A:U pairings, which occur much more readily than mutations leading to G:C pairings. Although the association between GC4 and GCnc is expected to become nonlinear at the limits of the distribution, a logistic curve best fits the relationship between GC4 and GCnc suggesting that there is cooperativity in the occurrence of complementary A:T or G:C pairings, as might be expected if there were compensatory changes that serve to stabilize mRNAs (24).
In sum, for two genes, neither of which encodes a physiologically relevant protein, we detected a strong and significant association between G+C content and bacterial fitness, indicating that a selective force within genes compensates for mutations that are naturally A+T-biased in bacteria. The elevated fitness of strains expressing mRNAs with high G+C content was most pronounced when genes are expressed at very high levels, suggesting that the selective forces responsible for this effect are low (12, 13), perhaps on par with those operating on adaptive codon bias (25). However, because bacterial genomes consist primarily of protein-coding regions, even a small selective effect that operates to change the base composition of individual genes in a common direction will have the effect of altering overall genomic base composition. Under this view, the extreme A+T richness of highly reduced genomes of host-restricted bacteria would result from the reduced efficacy of selection in these species (11, 26–28) rather than from changes in mutational patterns. Thus, the broad variation in genomic base composition among bacterial species reflects, in part, differences in population-level parameters that affect the efficacy of selection.
Materials and Methods
A set of 14 clones containing GFP genes that varied in their base compositions from 40.4% to 53.7% G+C was selected from those reported in Kudla et al. (18). A second set of plasmids containing the Bacillus phage ϕ29 DNA polymerase gene with base compositions ranging from 43.7% to 47.2% G+C were selected from those reported in Welch et al. (29). Within each set, the amino acid sequence of the protein encoded by all constructs was identical, and clones in each set were selected to represent a narrow range of CAI values for the E. coli host (CAI of GFP constructs = 0.58–0.68; CAI of ϕ29 constructs = 0.53–0.59) to abate the effects of selection on translational optimization (30). To further analyze the sole effect of base composition on bacterial fitness, we selected constructs with very different nucleotide sequences, even among those of similar base composition. For example, among the GFP constructs of low G+C content (40.4–41.8%), there are, on average, 58 substitutions; among those of medium G+C (46.6–47.3%), 121 substitutions; and among those of high G+C (51.5–53.7%), 99 substitutions. There are, on average, 126 synonymous substitutions between pairs of GFP constructs (240 codons), and 213 differences between the ϕ29 DNA polymerase constructs (575 codons). Genes were supplied in similar pET vectors (Novagen), with the gene of interest linked to a T7 promoter. To test constructs in which translation is prevented, we designed GFP genes in which the start codon was removed and stop codons introduced into their 5′ ends. These amplicons were cloned into a pET11a vector (Novagen) that was engineered, by XbaI and BamHI digestion, to lack its ribosome-binding site.
To express the plasmid libraries and monitor growth rates, we transformed constructs into E. coli BL21(DE3) (NEB), which has a chromosomal copy of T7 RNA polymerase under the control of a lacUV5 promoter. E. coli strain Lemo21(DE3) (NEB), which is similar to BL21(DE3) but can express T7 lysozyme under a rhamnose promoter, was used to measure mRNA stability.
For growth rate experiments, 10 μL of an overnight culture were inoculated into 140 μL of LB medium containing 100 μg/mL ampicillin into 96-well plates and grown at 37 °C with constant shaking. After 1 h, 1 mM IPTG was added, and GFP fluorescence and optical density (OD) were measured each hour for 5 h on a Victor3 microplate reader (PerkinElmer). Regression analyses of bacterial growth rates and GFP fluorescence were performed on values averaged from three independent experiments.
For measurements of mRNA stability, Lemo21(DE3) cells were grown as above but induced with 20 μM IPTG for 2 h, followed by the induction of T7 lysozyme with 2 mM l-rhamnose. After 15 min, samples were collected and processed for quantification as described above. Quantitative PCR was performed with primers that target the 3′-end common to all of the GFP mRNAs, and transcript abundance was calculated from threshold cycle (Ct) after normalizing to the Ct obtained with 16S rRNA primers (dCt).
Genome sequences and annotations of 1,430 fully sequenced bacterial genomes were obtained from NCBI FTP server (ftp.ncbi.nih.gov). For each genome, noncoding regions were identified as those not encoding proteins, ribosomal RNA, and transfer RNA, and the fourfold degenerate sites of protein-coding genes were identified based on the standard genetic code. The G+C content of fourfold degenerate sites (GC4) was calculated for each genome as the proportion of all fourfold degenerate sites in the entire genome that are either G or C, and the G+C content of noncoding regions (GCnc) for each genomes was similarly calculated. The relationship between GC4 and GCnc (Fig. 5 and Fig. S7) was fitted with a logistic function, and the goodness of fit of this regression was measured using analysis of deviance with the nls package in R.
To examine nontranscribed regions that are putatively free of selective constraints, we located intergenic regions situated between experimentally verified, divergent transcription start sites in the E. coli MG1655 genome (31, 32). Any of these intergenic regions that contained noncoding RNAs (33) were removed before analysis. Because many promoter sequences are AT-rich and could bias nucleotide counts in short intergenic regions, the AT-rich σ70 promoter sequences (TATAAT, -10 region and TTGACA, -35 region) were not included in the calculations of G+C contents. RNA free energy was calculated using the mfold web server (34).
Supplementary Material
Acknowledgments
We thank Joshua Plotkin and Grzegorz Kudla for providing the GFP constructs and Mark Welch for the Bacillus phage ϕ29 DNA polymerase plasmids. We also thank Nancy Moran and Adam Eyre-Walker for helpful discussions and comments on the manuscript, Kim Hammond for assistance with the figures, and the staff at the Yale University Faculty of Arts and Sciences High Performance Computing Center. This work was supported in part by National Institutes of Health Grant GM74738 (to H.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205683109/-/DCSupplemental.
References
- 1.McCutcheon JP, Moran NA. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2010;2:708–718. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas SH, et al. The mosaic genome of Anaeromyxobacter dehalogenans strain 2CP-C suggests an aerobic common ancestor to the delta-proteobacteria. PLoS ONE. 2008;3:e2103. doi: 10.1371/journal.pone.0002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sueoka N. On the genetic basis of variation and heterogeneity of DNA base composition. Proc Natl Acad Sci USA. 1962;48:582–592. doi: 10.1073/pnas.48.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freese E. On the evolution of base composition of DNA. J Theor Biol. 1962;3:82–101. [Google Scholar]
- 5.Muto A, Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci USA. 1987;84:166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurst LD, Merchant AR. High guanine-cytosine content is not an adaptation to high temperature: a comparative analysis amongst prokaryotes. Proc Biol Sci. 2001;268:493–497. doi: 10.1098/rspb.2000.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naya H, Romero H, Zavala A, Alvarez B, Musto H. Aerobiosis increases the genomic guanine plus cytosine content (GC%) in prokaryotes. J Mol Evol. 2002;55:260–264. doi: 10.1007/s00239-002-2323-3. [DOI] [PubMed] [Google Scholar]
- 8.Foerstner KU, von Mering C, Hooper SD, Bork P. Environments shape the nucleotide composition of genomes. EMBO Rep. 2005;6:1208–1213. doi: 10.1038/sj.embor.7400538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musto H, et al. Genomic GC level, optimal growth temperature, and genome size in prokaryotes. Biochem Biophys Res Commun. 2006;347:1–3. doi: 10.1016/j.bbrc.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 10.Wang HC, Susko E, Roger AJ. On the correlation between genomic G+C content and optimal growth temperature in prokaryotes: Data quality and confounding factors. Biochem Biophys Res Commun. 2006;342:681–684. doi: 10.1016/j.bbrc.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 11.Balbi KJ, Rocha EPC, Feil EJ. The temporal dynamics of slightly deleterious mutations in Escherichia coli and Shigella spp. Mol Biol Evol. 2009;26:345–355. doi: 10.1093/molbev/msn252. [DOI] [PubMed] [Google Scholar]
- 12.Hildebrand F, Meyer A, Eyre-Walker A. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 2010;6:e1001107. doi: 10.1371/journal.pgen.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hershberg R, Petrov DA. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 2010;6:e1001115. doi: 10.1371/journal.pgen.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- 15.Puigbò P, Bravo IG, Garcia-Vallve S. CAIcal: A combined set of tools to assess codon usage adaptation. Biol Direct. 2008;3:38. doi: 10.1186/1745-6150-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch M. The origins of genome architecture. Sunderland: Sinauer; 2007. [Google Scholar]
- 17.Eyre-Walker A, Bulmer M. Reduced synonymous substitution rate at the start of enterobacterial genes. Nucleic Acids Res. 1993;21:4599–4603. doi: 10.1093/nar/21.19.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allert M, Cox JC, Hellinga HW. Multifactorial determinants of protein expression in prokaryotic open reading frames. J Mol Biol. 2010;402:905–918. doi: 10.1016/j.jmb.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernardi G, Bernardi G. Compositional constraints and genome evolution. J Mol Evol. 1986;24:1–11. doi: 10.1007/BF02099946. [DOI] [PubMed] [Google Scholar]
- 22.Osawa S, et al. Directional mutation pressure and transfer RNA in choice of the third nucleotide of synonymous two-codon sets. Proc Natl Acad Sci USA. 1988;85:1124–1128. doi: 10.1073/pnas.85.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 24.Chamary JV, Hurst LD. Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol. 2005;6:R75. doi: 10.1186/gb-2005-6-9-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharp PM, Emery LR, Zeng K. Forces that influence the evolution of codon bias. Philos Trans R Soc Lond B Biol Sci. 2010;365:1203–1212. doi: 10.1098/rstb.2009.0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moran NA. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersson SGE, Kurland CG. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- 28.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 29.Welch M, et al. Design parameters to control synthetic gene expression in Escherichia coli. PLoS ONE. 2009;4:e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp PM, Li W-H. The Codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keseler IM, et al. EcoCyc: A comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39(Database issue):D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raghavan R, Sage A, Ochman H. Genome-wide identification of transcription start sites yields a novel thermosensing RNA and new cyclic AMP receptor protein-regulated genes in Escherichia coli. J Bacteriol. 2011;193:2871–2874. doi: 10.1128/JB.00398-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raghavan R, Groisman EA, Ochman H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 2011;21:1487–1497. doi: 10.1101/gr.119370.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.