Abstract
The pHLIP peptide has three states: (I) soluble in aqueous buffer, (II) bound to the bilayer surface at neutral pH, and (III) inserted as a transmembrane (TM) helix at acidic pH. The membrane insertion of pHLIP at low pH can be used to target the acidic tissues characteristic of different diseases, such as cancer. We find that the α-helix content of state II depends on lipid acyl chain length but not cholesterol, suggesting the helicity of the bound state may be controlled by the bilayer elastic bending modulus. Experiments with the P20G variant show the proline residue in pHLIP reduces the α-helix content of both states II and III. We also observe that the membrane insertion pKa is influenced by membrane physical properties, following a biphasic pattern similar to the membrane thickness optima observed for the function of eukaryotic membrane proteins. Because tumor cells exhibit altered membrane fluidity, we suggest this might influence pHLIP tumor targeting. We used a cell insertion assay to determine the pKa in live cells, observing that the properties in liposomes held in the more complex plasma membrane. Our results show that the formation of a TM helix is modulated by both the conformational propensities of the peptide and the physical properties of the bilayer. These results suggest a physical role for helix-membrane interactions in optimizing the function of more complex TM proteins.
Keywords: alpha helix, peptide folding, tumor acidity, bilayer thickness, vesicle properties
The use of peptides that target the cell membrane holds great promise for the diagnostic and treatment of disease (1, 2). However, the human plasma membrane is complex and heterogeneous, containing hundreds of different lipid types (3) that impart specific physical properties to the bilayer. In fact, the function of peptides and proteins interacting with the membrane is often modulated by the bilayer physical properties (4, 5). In particular, membrane parameters such as thickness, fluidity, charge, and curvature have been found to regulate the folding, activity and stability of membrane proteins (6, 7). However, the influence played by the overall physical properties of the bilayer on membrane-targeting therapeutic peptides remains largely unknown. Here we investigate how specific peptide properties, such as helicity, and membrane physical parameters control the membrane insertion of one such peptide, pHLIP®.
The pHLIP peptide (GGEQNPIYWARYADWLFTTPLLLLDLALLVDADEGTCG) is monomeric at low concentrations in three different environments (states): State I, soluble in aqueous buffer, where it remains unstructured; state II, bound to the lipid-water interface in a largely unfolded conformation; and state III, inserted and folded as a transmembrane helix via the translocation of the peptide C-terminal end across the bilayer (8, 9). The transition between states II and III is controlled by the pH: At neutral pH the peptide remains at the bilayer surface (state II), while at acidic pH it inserts across the membrane (state III). The pH-controlled membrane insertion of pHLIP may be useful for cancer diagnosis and therapy (10–13), because tumors are characterized by localized acidity. Interestingly, altered physical membrane properties have been found in cancer. In particular, the cell membranes of several tumor types (lymphoma, leukemia, lung, cervix, and neural cancers) are more fluid than the membranes of normal cells (14–19). Increased fluidity is also correlated with poorer prognosis and increased metastasis rate (17–19). Consequently, we have explored the influence of membrane fluidity on pHLIP insertion.
Results
P20 Reduces Helix Formation in pHLIP.
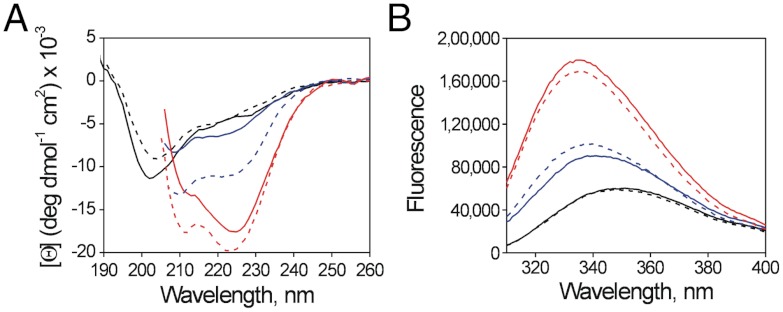
At neutral and basic pH (state II), pHLIP interacts with the surfaces of lipid membranes (8). Instead of forming an interfacial α-helix, as is common for many other membrane-interacting peptides (20), pHLIP remains largely unfolded. pHLIP contains a single proline at sequence position 20 (P20). Prolines influence helical protein secondary structure because they have no hydrogen atom on the backbone amide group to act as a hydrogen bond donor in stabilizing helical backbones. To study whether P20 is responsible for the lack of helicity in state II, we replaced P20 with glycine to make the variant P20G (AAEQNPIYWARYADWLFTTGLLLLDLALLVDADEGT). Analytical ultracentrifugation of P20G in aqueous solution shows a single species (Fig. S1) (9). Circular dichroism (CD) spectra of P20G and Wild Type (WT) pHLIP in buffer at pH 7.8 (state I) have similar minima at approximately 203 nm, characteristic of disordered polypeptides (Fig. 1A). However, the CD for P20G in the presence of liposomes shows major differences from WT pHLIP. For state II (18∶1-PC liposomes at pH 7.8), the CD spectrum of pHLIP has α-helical features (minimum at 208 and shoulder at 222 nm) but the ellipticity is low, suggesting a small content of α-helix. Conversely, P20G exhibits more ellipticity at 208 and 222 nm, indicating that P20G becomes more helical in the presence of lipid at neutral pH. The CD spectrum for state III (18∶1-PC liposomes at pH 4.5) is α-helical for both peptides but the ellipticity is again greater for P20G. The characteristic helical minima at 208 and at 222 nm appear to be slightly red-shifted, as has been observed for some membrane proteins (21). Taken together, these results suggest that P20 inhibits α-helix formation in state II and, to a lesser extent, in state III. The state I and state III fluorescence spectra of the peptides are similar (Fig. 1B), indicating that the environment of the two tryptophan residues, located at the N terminus of the transmembrane domain, does not change. In state II, however, P20G causes a shift in the spectral maximum from 342 to 340 nm, suggesting deeper tryptophan burial in the membrane and consistent with the state II CD changes.
Fig. 1.
Helical structure and tryptophan fluorescence responses to the membrane environment of WT and P20G pHLIP. (A) Circular dichroism experiments of WT (continuous lines) and P20G (dotted lines) in buffer at pH 7.8 (state I; black lines) and 18∶1-PC liposomes, at pH 7.8 (state II; blue lines) and pH 4.5 (state III; red lines). (B) Emission fluorescence spectra, with lines as in panel A.
State II Helicity Is Altered by Lipid Acyl Chain Length.
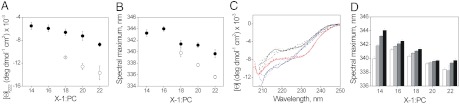
The conformational flexibility observed for the surface bound (state II) pHLIP prompted us to investigate whether its secondary structure could be influenced by lipid composition. We studied the interaction of P20G and WT pHLIP with a series of cis-9 di-monounsaturated phosphatidylcholine (PC) lipids of 14, 16, 18, 20, and 22 acyl chain carbon length (X∶1-PC, being X length), at liquid phase temperatures and neutral pH. We observed that the ellipticity at 222 nm increases progressively with acyl chain length for both peptides (Fig. 2A), with more ellipticity for P20G than for WT. For reasons not well understood, the CD spectra of P20G in 14∶1-PC and 16∶1-PC showed strong minima at 228 nm (Fig. S2), suggesting an aromatic residue exciton contribution (9) which precluded secondary structure analysis. The observed increases in helicity for longer lipids in both peptides were accompanied by decreases of the wavelength of maximum emission (Fig. 2B), indicating the tryptophan environment is less polar for the more helical states.
Fig. 2.
State II pHLIP helicity is sensitive to lipid acyl chain length but not cholesterol. (A) Ellipticity at 222 nm of WT (closed symbols) and P20G (open symbols) for PC lipids of different acyl chain length. (B) Fluorescence emission maxima for WT (closed symbols) and P20G (open symbols) incubated with PC lipids of different length. Data for 14∶1-PC and 16∶1-PC have been omitted, as discussed in the main text. (C) CD spectra of WT pHLIP in liposomes of different acyl chain length and cholesterol content. Red lines: 14∶1-PC; black lines: 18∶1-PC; blue lines: 22∶1-PC. Solid lines: 0%, dashed lines: 10%, dash/dot lines: 20% and dash lines: 30% cholesterol. (D) Fluorescence spectral maximum of WT pHLIP in liposomes of different acyl chain length and cholesterol content (white: 0%, light gray: 10%, dark gray: 20% and black: 30% cholesterol). All experiments were performed at pH 7.8.
Cholesterol Does Not Alter State II Helicity.
To provide a deeper analysis of how membrane composition can affect pHLIP’s state II at the bilayer surface, we incubated pHLIP at pH 7.8 with 14∶1, 16∶1, 18∶1, 20∶1, and 22∶1 PC liposomes containing different amounts of cholesterol. Interestingly, the addition of cholesterol had no effect on the CD spectra (Fig. 2C) for all acyl chain lengths, and the ellipticity at 222 nm remained unchanged regardless of the addition of 10%, 20%, or 30% cholesterol. The state II tryptophan fluorescence, on the other hand, changed similarly for each set of PC lipids (Fig. 2D): As cholesterol content increased, the fluorescence spectral maxima red-shifted, likely due to greater hydration of the bilayer surface, a known consequence of adding cholesterol to unsaturated lipid bilayers (22).
The addition of cholesterol is known to significantly affect the physical properties of the bilayer. These changes include bilayer thickening and an increase of chain ordering in fluid phase lipids. The observation that pHLIP’s state II helicity at the bilayer surface is affected by acyl chain length but not by cholesterol is surprising and indicates that a physical parameter other than hydrophobic thickness must explain our results (see Discussion).
Acyl Chain Length Affects both pHLIP and P20G Insertion pKas.
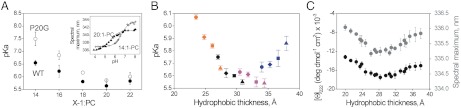
We next explored the influence of lipid composition on pHLIP membrane insertion (the transition from state II to III). We followed the fluorescence changes caused by peptide insertion into bilayers of different lipid acyl chain lengths to determine the insertion pKa. Our results show that the pKa is controlled by the acyl chain length, ranging from 5.6 to pH 7.5 (Fig. 3A). For WT pHLIP, the pKa first decreases from 14∶1-PC to 20∶1-PC and then saturates for 22∶1-PC. A very similar trend was obtained for P20G, with pKa values higher than those of WT. The pKa difference between the two peptides for 14∶1-PC is approximately 1 pH unit, but this difference diminishes with increasing acyl chain length until it is within the experimental error for 22∶1-PC. Because P20G has higher helicity than WT (Fig. 1A), our results suggest helicity may influence the insertion pKa.
Fig. 3.
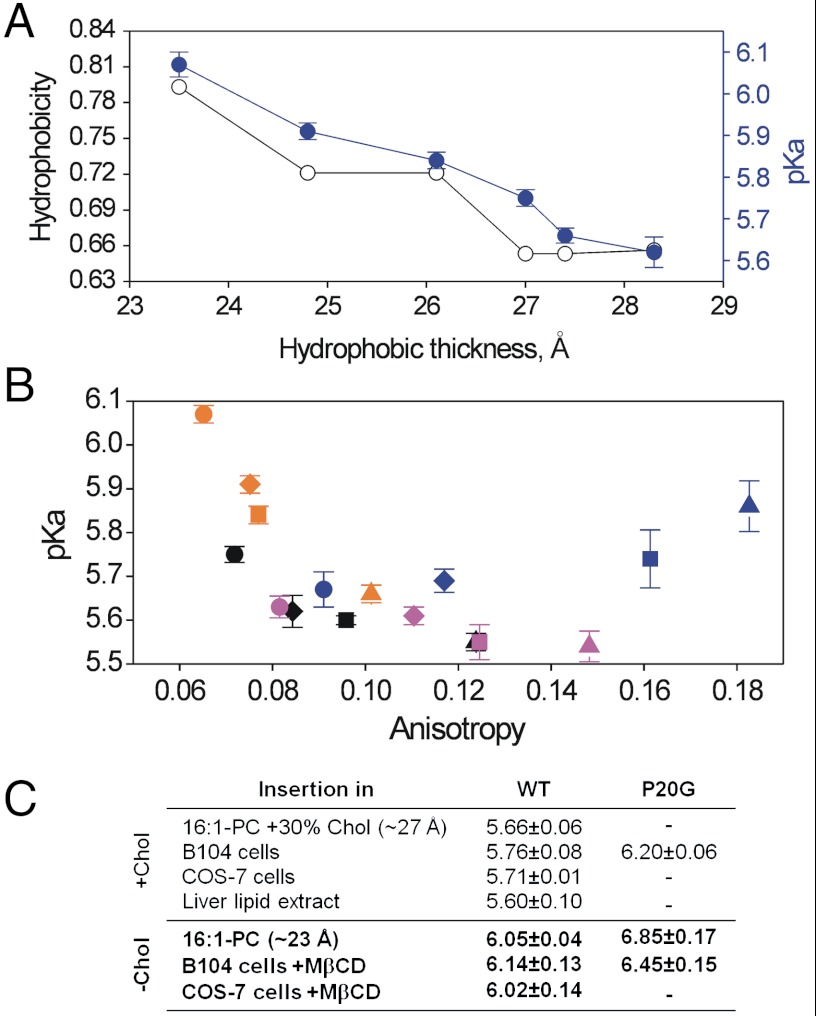
Insertion pKa is affected by both acyl chain length and cholesterol. (A) Membrane insertion pKa for WT (closed symbols) and P20G (open symbols) in liposomes of different acyl chain length. pKa values were determined from fluorescence spectra maxima at different pH values, as the midpoint of the sigmoidal transition (36), as shown in the inset for 20∶1-PC and 14∶1-PC. (B) Correlation between pHLIP insertion pKa and membrane hydrophobic thickness. Liposomes of PC lipids of different chain length (16∶1-PC, orange; 18∶1-PC, black; 20∶1-PC, magenta; 22∶1-PC, blue); containing 0% (circle), 10% (diamond), 20% (square), and 30% (triangle) cholesterol. Hydrophobic thickness refers to the distance between the acyl chain carbon 2 of the two hemilayers, as determined by small-angle X-ray scattering (30). The thickness increase associated with cholesterol was extrapolated from (49). Data points for 14∶1-PC lipids were not included, as explained in Fig. S9. (C) CD and fluorescence values for pHLIP state III. The ellipticity at 222 nm (left axis) and fluorescence spectral maximum (right axis, grey symbols) are plotted for the hydrophobic thickness of the different PC lipids at 0, 10, 20 and 30% cholesterol. Experiments were performed at 28 mM acetic/acetate buffer, pH 4.5.
Insertion pKa and State III Helicity Change Biphasically with Bilayer Thickness.
Bilayer thickness is a parameter we thought should have a significant effect on pHLIP’s insertion pKa. To further examine the relationship between insertion pKa and hydrophobic thickness, we studied the 16∶1- through 22∶1-PC series containing 0, 10, 20, and 30% cholesterol, which gives an overlapping continuum of thicknesses (Fig. 3B). In agreement with Fig. 3A, pHLIP’s insertion pKa first decreases with increasing hydrophobic thickness and then saturates before returning to a pH value of approximately 6. pHLIP state III helicity and fluorescence maxima were also found to follow this biphasic behavior when plotted against hydrophobic thickness (Fig. 3C).
pHLIP Insertion into Cells Mirrors Liposome Results.
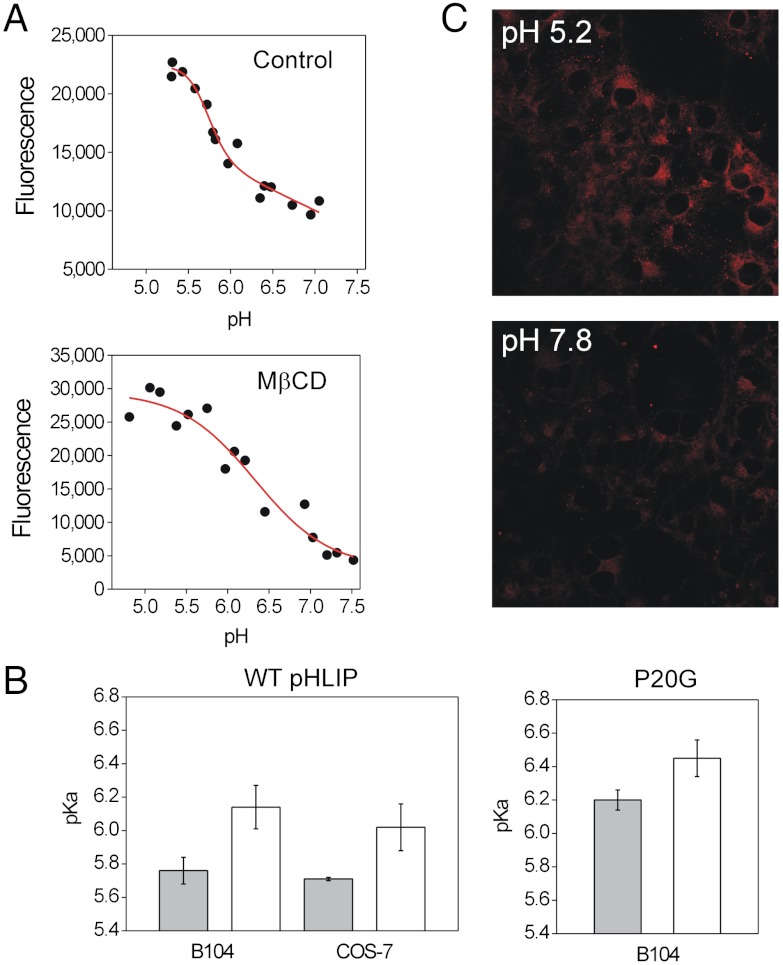
Our results show that the insertion pKa is influenced by the lipid composition of the liposomes. As a way of obtaining a pKa value more representative of the context in vivo, we have developed a plate reader assay to follow the insertion of pHLIP into live cells. In this assay, we covalently linked Alexa-594 to pHLIP. To minimize any influence of the dye on peptide insertion, we linked it to the noninserting, N-terminal peptide end. The resulting Alexa-pHLIP complex was incubated with subconfluent B104 cells at different pHs for short periods of time (to avoid cell death at the acidic pHs), and washed before reading. We observed higher fluorescence intensity in the cells incubated at acidic pH than at neutral pH. These intensities, when plotted, follow a sigmoidal pattern (Fig. 4A) similar to that observed for pHLIP insertion in synthetic liposomes. The midpoint of the transition (which we designate “cell pKa”) was 5.76 ± 0.08. Experiments performed with the P20G variant yielded a higher cell pKa, 6.20 ± 0.06 (Fig. 4B). These values were very close to those obtained for synthetic lipids with di-monounsaturated 16–18 carbon chains and 30% cholesterol, resembling the average composition of an unspecialized plasma membrane (23). To study whether the cell pKa is cell-type dependent, we carried out the plate reader assay using COS-7 cells, obtaining a similar value: 5.71 ± 0.01 (Fig. 4B).
Fig. 4.
Insertion of WT and P20G pHLIP in live cells. (A) Plate reader insertion assay for WT pHLIP covalently labeled with Alexa594 at the N terminus in control and MβCD-treated B104 cells. (B) Cell pKa obtained for WT and P20G in control (grey bars) and MβCD-treated (white bars) B104 and COS-7 cells. (C) Confocal images of the membrane translocation assay with BODIPY 630/650 reversibly linked to the C terminus of pHLIP. Experiments were performed in B104 cells at pH 5.2 and 7.8.
Our liposome results show that cholesterol has a marked effect on pHLIP membrane insertion. To study whether cholesterol has a similar effect in cellular membranes, we used methyl-β-cyclodextrin (MβCD) to deplete cholesterol from the plasma membrane (Fig. S3) (24, 25). Treatment with 5 mM MβCD for 1 h caused a 46% reduction in plasma membrane cholesterol, in agreement with reports elsewhere (25). In parallel with the results in synthetic liposomes, MβCD treatment resulted in an increase in the cell pKa for all peptides and cell types assayed (Fig. 4B). Control experiments were performed to show that neither acidity nor the peptide construct caused significant cell death (Fig. S4). Additional controls showed that the fluorescence of the Alexa-pHLIP construct was constant in the pH range of 5.2–7.0 (Fig. S5).
A critical difference between the cell assay and our previous experiments is the low complexity of the liposomes. To partially reproduce the complexity of a natural membrane in a synthetic setting, we assayed pHLIP insertion in liposomes prepared from a liver lipid extract, obtaining a pKa of 5.60 ± 0.10 (Fig. S6), which is similar to the observed cell pKa of 5.76 ± 0.08. It has been previously reported that pHLIP is able to translocate cargo molecules, including fluorescent dyes, across the cell plasma membrane (12, 26). We performed a cell translocation experiment to confirm that the inserting end (C terminus) of pHLIP crosses the cell plasma membrane in the plate reader conditions. To this end, we attached the fluorescent dye BODIPY-630/650 to the C terminus of pHLIP via a disulfide bond. This bond can be cleaved by the reducing environment of the cytoplasm, resulting in release of the dye upon peptide insertion at low pH (26). When translocation occurs, we expect a greater retention of dye in the cells after washing. Confocal microscopy showed that, indeed, at pH 5.2 intense fluorescence is observed in the cell cytoplasm (Fig. 4B). Conversely, at pH 7.8 a low signal is seen, in agreement with a pH-dependent translocation.
Discussion
P20 Modulates pHLIP Helicity.
In this work, we explore the physical variables influencing peptide attachment at the membrane interface and transmembrane helix formation. We studied the contribution of peptide secondary structure by replacing P20 with glycine. In aqueous solution (state I) the P20G mutation did not induce α-helix formation. Conversely, in the presence of all lipids studied at pH 7.8 (state II) the P20G mutation induced a marked α-helix increase as reported by CD. This result agrees with the observation that glycine has higher alpha helical propensity than proline in a hydrophobic environment (27). The helical increase could arise from: (i) the formation of a short helical segment in state II, (ii) helical structure transiently distributed over the entire peptide sequence, or (iii) a subpopulation of peptide that is entirely helical transiently. We favor the first option because helix formation is a cooperative process (28) and helix propensity will vary along the sequence. The P20G mutation induces a smaller helical increase for the TM state compared to the attached state, probably because state III is highly structured in pHLIP. We do not know where in the sequence the helical increase occurs, but it is tempting to suggest that this increase takes place near the site of the mutation. This observation could reasonably imply that the TM helix of pHLIP is distorted at position 20, and from the magnitude of the ellipticity change, the helical disruption involves several residues. Such a disruption would not be unprecedented, because significant local distortion around proline residues has been reported for other TM peptides (29).
Acyl Chain Length But Not Cholesterol Modulates State II Helicity.
Our results show that, surprisingly, the structure of pHLIP at the bilayer surface (state II) is influenced by variations of the lipid acyl chain length (Fig. 2). The reason that this result is unexpected is that the area per molecule in the bilayer is relatively constant as the acyl chain lengthens (30). The increases in helical content and tryptophan burial for longer acyl chains support the idea of a deeper location of the peptide at the water/lipid interface. This result suggests that state II may populate an ensemble of interconverting conformations with longer lipid acyl chains shifting the equilibrium towards the more helical state/s. It would be very interesting to understand the physical basis underlying this process. However, the interpretation becomes complicated because changing the acyl chain length alters several bilayer properties including, but not limited to, thickness and fluidity.
Examining the results obtained in cholesterol-containing liposomes is informative, because cholesterol affects both membrane thickness and fluidity (6) in the same direction as increasing acyl length (Fig. 5). However, while longer acyl chains promoted helical structure in state II, cholesterol did not (Fig. 2), indicating that a physical parameter other than thickness and fluidity must explain our results. We suggest that the bilayer elastic bending modulus (kc) may influence α-helix formation in state II. The elastic bending modulus is the energetic term for the elastic, out-of-plane bending fluctuations to which membranes are subject (31). While kc increases with acyl chain length (32), it does not change with the addition of cholesterol to di-monounsaturated lipids (33, 34), correlating with the state II α-helix changes we observe. The kc parameter has been reported to modulate the coupled membrane attachment and insertion of the bacterial β-barrel porin OmpA (35). For pHLIP, both processes are independent, since after membrane attachment, insertion cannot proceed until key acidic groups in pHLIP are protonated (36). We therefore propose that kc influences pHLIP’s state II but not state III. This influence suggests that in the case of OmpA, kc might have a more important effect on membrane attachment than on folding-insertion.
Fig. 5.
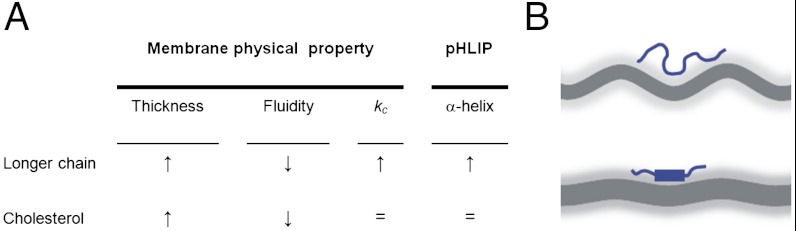
(A) Changes in bilayer thickness, fluidity and elastic bending modulus (kc) caused by acyl chain length and cholesterol (6, 31–34). (B) Scheme summarizing the proposed effect of kc on pHLIP helicity. Upper. Lipids with shorter acyl chains have important elastic, out of plane bending fluctuations, being kc the energy scale for the fluctuations. These fluctuations favor peptide structural disorder. Lower. Lipids with longer acyl chains have reduced fluctuations, and pHLIP is more helical. The bilayer core is represented in dark grey, and the bilayer-water interface as a diffuse light grey layer. The magnitude of the membrane fluctuation has been modified for illustrative purposes.
P20 Affects the Insertion pKa.
TM helices are stabilized by backbone hydrogen bonds, because the low dielectric constant of the bilayer core makes hydrogen bonds more favorable than in water (37). We have found that the P20G mutation has a significant influence on the membrane insertion pKa (Fig. 3A), which is higher for P20G than for WT pHLIP in all PC lipids except 22∶1-PC. The ellipticity increase at 222 nm from state II to III is greater for WT than for P20G (Fig. 1). The higher helicity of P20G in state III suggests that the transition to state III is influenced by the number of backbone hydrogen bonds occurring in the final TM state, more than by the net increase in hydrogen bonds from state II to III. Accordingly, in the case of 22∶1-PC, while the secondary structure of state II for each peptide is different (Fig. 2B), the α-helix content in state III is similar (Fig. S2), in agreement with the finding of comparable pKa values.
TM Segment Hydrophobicity Affects Insertion pKa in Thinner Bilayers.
Bilayer thickness has been reported to influence the structure and function (38–40), oligomerization (41), and dynamics (42) of prokaryotic and eukaryotic membrane proteins, most notably ion channels and membrane transporters (6). A general feature of the lipid influence on the activity profile of these membrane proteins is that the changes are biphasic, an observation often attributed to hydrophobic mismatch between protein and membrane domains (6). Our results show there is an intermediate range of bilayer thicknesses for which pHLIP’s pKa value is minimal, suggesting similar mismatch behavior. We propose a model in which pHLIP insertion pKa might be affected by different physical properties in bilayers of short to intermediate hydrophobic core thickness and bilayers of intermediate to long core thickness. For short to intermediate core thickness, we observe the insertion pKa correlates with the per residue average hydrophobicity of pHLIP peptide segments predicted to span each thickness (Fig. 6A). These hydrophobicity predictions were obtained using the Wimley-White octanol scale (20) as described in Fig. S7. Although these predictions make a number of assumptions (such as the formation of a straight helix) and ignore certain complexities of the system (e.g., side chain conformation), it is pleasing to see that there is a good correlation. This correlation supports the idea that incorporation of new residues into an expanding TM domain serves as the main determinant of insertion favorability as lipid bilayer thickness increases. In short, if the new residues added to the expanding hydrophobic core are hydrophilic, the average ΔG for the spanning segment becomes more positive and the insertion less favorable, resulting in a lower insertion pKa (i.e., it takes more acid to insert the peptide).
Fig. 6.
Contribution of membrane thickness and fluidity to transmembrane helix formation. (A) The per residue hydrophobicity (in kcal/mol units) was calculated from the octanol scale (20), as described in SI Text. The thickness increase associated with cholesterol was extrapolated from (49). (B) A comparison of insertion pKa values obtained in cells and liposomes with or without cholesterol. We note the degree of similarity between the results obtained in cells and the results for 16∶1-PC lipids. For liposome experiments, the core thickness values, in Angstroms, are provided. (C) Correlation between pHLIP insertion pKa and anisotropy of DPH embedded in PC liposomes of different chain length (16∶1-PC, orange; 18∶1-PC, black; 20∶1-PC, magenta; 22∶1-PC, blue); containing 0% (circle), 10% (diamond), 20% (square) and 30% (triangle) cholesterol. Data points for 14∶1 PC lipids were not included as discussed in Fig. S9.
The plateau of pHLIP state III CD and tryptophan fluorescence occurs concurrently with saturation of the insertion pKa at intermediate core thicknesses (Fig. 3). This event occurs when the arginine at position 11 is predicted to start incorporating into the frame of the putative spanning segment (Fig. S7). This finding points to the possibility that inhibition of arginine snorkeling (43) may be a key feature that generates mismatch strain in our peptide for intermediate and longer lipids, constituting an upper thickness limit for unstrained insertion, but further experiments will be needed to test this idea. In combination with the cell pKa results described below, we propose that the pKa changes observed for intermediate to long lipids may be attributed to changes in membrane fluidity, perhaps relating to how the membrane accommodates this strained arginine.
Membrane Fluidity Governs Insertion pKa for Thicker Lipids.
Our cell pKa experiments yield results comparable to those from the liposome pKa assays. It has been reported previously by our lab that MβCD-mediated cholesterol removal from eukaryotic plasma membranes has little or no influence on the average bilayer thickness when membrane proteins are present (24). This observation provides us an opportunity for separating the role of thickness and fluidity in TM helix formation (with the qualification that there may be nanodomains of different thickness where pHLIP may insert preferentially). This previous work allows us to omit the possibility that thickness alteration alone can account for the cell pKa changes we observe, prompting us to seek an alternative explanation.
To see if membrane fluidity may yield an alternative correlation with our observed insertion pKa changes, we measured the fluorescence anisotropy of diphenylhexatriene (DPH) embedded in our different synthetic lipid compositions. Anisotropy measures the rotational membrane diffusion of the DPH probe (44) and is correlated with the overall membrane fluidity at the center of the bilayer. The anisotropy values were plotted against pHLIP insertion pKa (Fig. 6C) to show a relationship similar to that observed for hydrophobic thickness (Fig. 3B), indicating that a dependence on fluidity is also a valid interpretation, especially for intermediate to thicker lipids. To see if this correlation with fluidity persists in cells, we measured the DPH anisotropy in cell plasma membranes with or without MβCD extraction. We found the control membranes were less fluid (anisotropy = 0.238 ± 0.005) than the extracted membranes (anisotropy = 0.197 ± 0.009), in agreement with membrane fluidity being a potential governing factor. While TM helices respond to membrane thickness changes in different ways, by tilting for instance (45), adapting to fluidity changes appears to be less straightforward. Although we make no direct guess at how state III peptide may be changing with very thick bilayers, we do offer pHLIP as a mechanistic example of how mismatch strain can generate structural constraints on a TM peptide, modulating its function. Large scale membrane curvature (compared with the short range fluctuations from the bending modulus (Fig. 5) is a parameter that potentially can also influence pHLIP membrane interaction, so it is important to ask if the vesicles in these experiments have constant size. We used dynamic light scattering experiments to determine vesicle size under our experimental conditions. We found that in all cases vesicle size was close to 50 nm, in agreement with previous reports (46), and that neither acidity nor pHLIP binding impact the size of the vesicles (Fig. S8).
Implications for Therapeutic Use of pHLIP.
Tumor tissue is characterized by an acidic extracellular pH, in the range of 6.5 to 6.9 (47). These values are higher than the cell pKa of pHLIP (approximately 5.7), potentially reducing pHLIP efficacy in tumor imaging and therapy despite our documented success of effective use (13). While there may be other properties that need to be understood, the use of pHLIP mutants with higher cell pKas, like P20G, might improve peptide targeting.
Interestingly, it has been reported that the cell membranes of several tumor types (lung, cervix and neural cancers, lymphoma, and leukemia) are more fluid than the membranes of normal cells (14–19). Increased fluidity has also been correlated with poorer prognosis and increased metastasis (17–19). Our cell experiments show that pHLIP insertion is favored in MβCD-treated cells (pKa increase of 0.3–0.4 pH units), which resulted in more fluid membranes (DPH anisotropy was 0.24 in control cells and 0.20 in MβCD-treated cells). Such a fluidity difference is similar to the DPH anisotropy change observed amongst normal lymphocytes (0.22) and lymphocytes from patients with chronic lymphocytic leukemia (0.18) (48), suggesting that, in addition to acidity, increased membrane fluidity might foster targeting of pHLIP to tumors. Our results suggest extracellular pH may not be the only parameter controlling targeting to tumors and that pHLIP-based cancer diagnosis and treatment may also be influenced by the physical properties of the tumor cell membrane. Because significant variations of membrane fluidity are expected for different tumor types, fluidity measurements might help guide pHLIP use for better diagnosis, and pHLIP designs might be tailored to optimize targeting.
Materials and Methods
Peptide synthesis, liposome preparation, analytical ultracentrifugation, fluorescence, and CD spectroscopy protocol are described elsewhere (36). A detailed description of the experimental methods is available in the SI Text.
Supplementary Material
ACKNOWLEDGMENTS.
We thank the following for the helpful experimental advice: Miriam Alonso, Joseph S. Wolenski, Joseph Watson, and Daiane Santana Alves, and to Maureen Gilmore-Hebert and David F. Stern for providing cell lines. This work was supported by National Institutes of Health grants R01-GM073857-04, and R01-CA133890-02.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212665109/-/DCSupplemental.
References
- 1.Johnson RM, Harrison SD, Maclean D. Therapeutic applications of cell-penetrating peptides. Methods Mol Biol. 2011;683:535–551. doi: 10.1007/978-1-60761-919-2_38. [DOI] [PubMed] [Google Scholar]
- 2.Andreev OA, Engelman DM, Reshetnyak YK. pH-sensitive membrane peptides (pHLIPs) as a novel class of delivery agents. Mol Membr Biol. 2010;27:341–352. doi: 10.3109/09687688.2010.509285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charalambous K, et al. Engineering de novo membrane-mediated protein-protein communication networks. J Am Chem Soc. 2012;134:5746–5749. doi: 10.1021/ja300523q. [DOI] [PubMed] [Google Scholar]
- 5.Khandelia H, Ipsen JH, Mouritsen OG. The impact of peptides on lipid membranes. Biochim Biophys Acta. 2008;1778:1528–1536. doi: 10.1016/j.bbamem.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Andersen OS, Koeppe RE. Bilayer thickness and membrane protein function: An energetic perspective. Annu Rev Biophys Biomol Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 7.Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunt JF, Rath P, Rothschild KJ, Engelman DM. Spontaneous, pH-dependent membrane insertion of a transbilayer alpha-helix. Biochemistry. 1997;36:15177–15192. doi: 10.1021/bi970147b. [DOI] [PubMed] [Google Scholar]
- 9.Reshetnyak YK, Segala M, Andreev OA, Engelman DM. A monomeric membrane peptide that lives in three worlds: In solution, attached to, and inserted across lipid bilayers. Biophys J. 2007;93:2363–2372. doi: 10.1529/biophysj.107.109967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An M, Wijesinghe D, Andreev OA, Reshetnyak YK, Engelman DM. pH-(low)-insertion-peptide (pHLIP) translocation of membrane impermeable phalloidin toxin inhibits cancer cell proliferation. Proc Natl Acad Sci USA. 2010;107:20246–20250. doi: 10.1073/pnas.1014403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vavere AL, et al. A novel technology for the imaging of acidic prostate tumors by positron emission tomography. Cancer Res. 2009;69:4510–4516. doi: 10.1158/0008-5472.CAN-08-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreev OA, et al. Mechanism and uses of a membrane peptide that targets tumors and other acidic tissues in vivo. Proc Natl Acad Sci USA. 2007;104:7893–7898. doi: 10.1073/pnas.0702439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reshetnyak YK, et al. Measuring tumor aggressiveness and targeting metastatic lesions with fluorescent pHLIP. Mol Imaging Biol. 2011;13:1146–1156. doi: 10.1007/s11307-010-0457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sok M, Sentjurc M, Schara M. Membrane fluidity characteristics of human lung cancer. Cancer Lett. 1999;139:215–220. doi: 10.1016/s0304-3835(99)00044-0. [DOI] [PubMed] [Google Scholar]
- 15.Preetha A, Banerjee R, Huilgol N. Effect of temperature on surface properties of cervical tissue homogenate and organic phase monolayers. Colloids Surf B. 2007;60:12–18. doi: 10.1016/j.colsurfb.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 16.Taraboletti G, et al. Membrane fluidity affects tumor-cell motility, invasion and lung-colonizing potential. Int J Cancer. 1989;44:707–713. doi: 10.1002/ijc.2910440426. [DOI] [PubMed] [Google Scholar]
- 17.Sok M, Sentjurc M, Schara M, Stare J, Rott T. Cell membrane fluidity and prognosis of lung cancer. Ann Thorac Surg. 2002;73:1567–1571. doi: 10.1016/s0003-4975(02)03458-6. [DOI] [PubMed] [Google Scholar]
- 18.Baritaki S, et al. Reversal of tumor resistance to apoptotic stimuli by alteration of membrane fluidity: Therapeutic implications. Adv Cancer Res. 2007;98:149–190. doi: 10.1016/S0065-230X(06)98005-1. [DOI] [PubMed] [Google Scholar]
- 19.Hendrich AB, Michalak K. Lipids as a target for drugs modulating multidrug resistance of cancer cells. Curr Drug Targets. 2003;4:23–30. doi: 10.2174/1389450033347172. [DOI] [PubMed] [Google Scholar]
- 20.White SH, Wimley WC. Membrane protein folding and stability;Physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 21.Wallace BA. Protein characterization by synchrotron radiation circular dichroism spectroscopy. Q Rev Biophys. 2009;42:317–370. doi: 10.1017/S003358351000003X. [DOI] [PubMed] [Google Scholar]
- 22.Gallova J, Uhrikova D, Kucerka N, Teixeira J, Balgavy P. Hydrophobic thickness, lipid surface area and polar region hydration in monounsaturated diacylphosphatidylcholine bilayers: SANS study of effects of cholesterol and beta-sitosterol in unilamellar vesicles. Biochim Biophys Acta. 2008;1778:2627–2632. doi: 10.1016/j.bbamem.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Sampaio JL, et al. Membrane lipidome of an epithelial cell line. Proc Natl Acad Sci USA. 2011;108:1903–1907. doi: 10.1073/pnas.1019267108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitra K, Ubarretxena-Belandia I, Taguchi T, Warren G, Engelman DM. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc Natl Acad Sci USA. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kilsdonk EP, et al. Cellular cholesterol efflux mediated by cyclodextrins. J Biol Chem. 1995;270:17250–17256. doi: 10.1074/jbc.270.29.17250. [DOI] [PubMed] [Google Scholar]
- 26.Thevenin D, An M, Engelman DM. pHLIP-mediated translocation of membrane-impermeable molecules into cells. Chem Biol. 2009;16:754–762. doi: 10.1016/j.chembiol.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li SC, Deber CM. A measure of helical propensity for amino acids in membrane environments. Nat Struct Biol. 1994;1:368–373. doi: 10.1038/nsb0694-368. [DOI] [PubMed] [Google Scholar]
- 28.Scholtz JM, Baldwin RL. The mechanism of alpha-helix formation by peptides. Annu Rev Biophys Biomol Struct. 1992;21:95–118. doi: 10.1146/annurev.bb.21.060192.000523. [DOI] [PubMed] [Google Scholar]
- 29.Thomas R, Vostrikov VV, Greathouse DV, Koeppe RE., II Influence of proline upon the folding and geometry of the WALP19 transmembrane peptide. Biochemistry. 2009;48:11883–11891. doi: 10.1021/bi9016395. [DOI] [PubMed] [Google Scholar]
- 30.Lewis BA, Engelman DM. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J Mol Biol. 1983;166:211–217. doi: 10.1016/s0022-2836(83)80007-2. [DOI] [PubMed] [Google Scholar]
- 31.Marsh D, Shanmugavadivu B, Kleinschmidt JH. Membrane elastic fluctuations and the insertion and tilt of beta-barrel proteins. Biophys J. 2006;91:227–232. doi: 10.1529/biophysj.105.079004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Z, Rand RP. The influence of cholesterol on phospholipid membrane curvature and bending elasticity. Biophys J. 1997;73:267–276. doi: 10.1016/S0006-3495(97)78067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan J, Tristram-Nagle S, Nagle JF. Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation. Phys Rev E. 2009;80:021931. doi: 10.1103/PhysRevE.80.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleinschmidt JH, Tamm LK. Secondary and tertiary structure formation of the beta-barrel membrane protein OmpA is synchronized and depends on membrane thickness. J Mol Biol. 2002;324:319–330. doi: 10.1016/s0022-2836(02)01071-9. [DOI] [PubMed] [Google Scholar]
- 36.Barrera FN, et al. Roles of carboxyl groups in the transmembrane insertion of peptides. J Mol Biol. 2011;413:359–371. doi: 10.1016/j.jmb.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowie JU. Membrane protein folding: How important are hydrogen bonds? Curr Opin Struct Biol. 2011;21:42–49. doi: 10.1016/j.sbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 39.Cornelius F. Modulation of Na,K-ATPase and Na-ATPase activity by phospholipids and cholesterol. I. Steady-state kinetics. Biochemistry. 2001;40:8842–8851. doi: 10.1021/bi010541g. [DOI] [PubMed] [Google Scholar]
- 40.de Planque MR, et al. Sensitivity of single membrane-spanning alpha-helical peptides to hydrophobic mismatch with a lipid bilayer: Effects on backbone structure, orientation, and extent of membrane incorporation. Biochemistry. 2001;40:5000–5010. doi: 10.1021/bi000804r. [DOI] [PubMed] [Google Scholar]
- 41.Anbazhagan V, Schneider D. The membrane environment modulates self-association of the human GpA TM domain—Implications for membrane protein folding and transmembrane signaling. Biochim Biophys Acta. 2010;1798:1899–1907. doi: 10.1016/j.bbamem.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 42.Xu Q, et al. Membrane hydrocarbon thickness modulates the dynamics of a membrane transport protein. Biophys J. 2008;95:2849–2858. doi: 10.1529/biophysj.108.133629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Killian JA, von Heijne G. How proteins adapt to a membrane-water interface. Trends Biochem Sci. 2000;25:429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- 44.Lentz BR. Use of fluorescent probes to monitor molecular order and motions within liposome bilayers. Chem Phys Lipids. 1993;64:99–116. doi: 10.1016/0009-3084(93)90060-g. [DOI] [PubMed] [Google Scholar]
- 45.Holt A, Killian JA. Orientation and dynamics of transmembrane peptides: The power of simple models. Eur Biophys J. 2010;39:609–621. doi: 10.1007/s00249-009-0567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuzaki K, et al. Optical characterization of liposomes by right angle light scattering and turbidity measurement. Biochim Biophys Acta. 2000;1467:219–226. doi: 10.1016/s0005-2736(00)00223-6. [DOI] [PubMed] [Google Scholar]
- 47.Robey IF, et al. Bicarbonate increases tumor pH and inhibits spontaneous metastases. Cancer Res. 2009;69:2260–2268. doi: 10.1158/0008-5472.CAN-07-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liebes LF, Pelle E, Zucker-Franklin D, Silber R. Comparison of lipid composition and 1,6-diphenyl-1,3,5-hexatriene fluorescence polarization measurements of hairy cells with monocytes and lymphocytes from normal subjects and patients with chronic lymphocytic leukemia. Cancer Res. 1981;41:4050–4056. [PubMed] [Google Scholar]
- 49.Kucerka N, Pencer J, Nieh MP, Katsaras J. Influence of cholesterol on the bilayer properties of monounsaturated phosphatidylcholine unilamellar vesicles. Eur Phys J E Soft Matter. 2007;23:247–254. doi: 10.1140/epje/i2007-10202-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.