Abstract
Transcriptional activation by the tumor suppressor p53 is considered to depend on cellular level, although there are few systems where this dependence on cellular level of p53 has been directly addressed. Previously, we reported that transactivation from p53 targets was sensitive to both p53 amount and DNA sequence, with some sequences being responsive to much lower p53 levels than others when examined in yeast model systems or human cells. Because p53 is normally present at low levels and perturbations might lead to small increases, we examined transactivation under limiting p53. Unlike the positive relationship between transactivation and binding affinity from target sequences at high cellular levels of human p53 in yeast, no such relationship was found at low levels. However, transactivation in the yeast system and the torsional flexibility of target sequences were highly correlated, revealing a unique structural relationship between transcriptional function and sequence. Surprisingly, a few sequences supported high transactivation at low p53 levels in yeast or when transfected into human cells. On the basis of kinetic and flexibility analyses the “supertransactivation” property was due to low binding off rates of flexible target sites. Interestingly, a supertransactivation response element can differentiate transcriptional capacities of many breast cancer-associated p53 mutants. Overall, these studies, which are relevant to other transcription factors, address the extent to which transactivation properties of p53 target sequences are determined by their intrinsic physical properties and reveal unique rules of engagement of target sequences at low p53 levels.
Keywords: basal level transcription, DNA twist flexibility, DNA dissociation kinetics
Sequence-specific DNA binding by transcription factors (TFs) to their target sites on DNA is a key step in many cellular functions such as transcription, replication, and recombination. In response to cellular stress p53 acts as a TF by binding to DNA targets, leading to the expression of many genes that participate in a variety of biological processes including cell-cycle arrest, DNA repair, and apoptosis (1). After nearly 30 y, the mechanisms of p53 targeting and transactivation remain unclear although its tumor suppressive abilities are intimately linked to its sequence-specific binding and transactivation (2). Abrogation of p53 sequence-dependent binding is implicated in ∼50% of all known cancers (1), and the DNA-binding domain of p53 contains 95% of the p53 missense mutations identified in human tumors (3). The consensus DNA response element (RE) consists of two decameric half sites with the motif RRRCWWGYYY (R = A, G; W = A, T; Y = C, T) separated by a variable number of base pairs (4). Most (∼95%) validated natural p53 targets deviate from the consensus sequence in at least one position (2, 5), and ∼10% of them are not clearly related to the consensus (2). p53 binds REs cooperatively as a tetramer, composed of a dimer of dimers (6). Large differences in binding affinities were observed between p53 REs belonging to different functional groups (7), where REs belonging to cell-cycle arrest genes appear to bind with higher affinities than most REs related to apoptosis (7). These differences are amplified by posttranslational modifications of p53 residues that directly contact DNA (8). Furthermore, large differences in transactivation levels from various p53 REs have been observed and were linked to variations in the sequence of the individual REs, their internal arrangement, the number of decameric repeats, and location with respect to the start site (9, 10).
We previously showed that the protein–DNA interface varied in p53 DNA-binding domain (p53DBD)/DNA complexes as a function of the DNA base sequence and that there is a correlation between binding affinity of the complexes and the protein–DNA interface geometries (11). The p53DBD/DNA complexes varied in both direct-readout (changes in protein–DNA contacts) and indirect-readout effects (changes in DNA structure). Indirect-readout effects are expected to be dominant in regions that are not contacted by the protein such as the WW doublet in the CWWG center of p53 half sites. The WW position is highly conserved in p53 binding sites (9) even though the WW dinucleotide is not contacted in the p53DBD/DNA complex (11). Recently, we showed that p53DBD binds consensus sequences in this region with different affinities and cooperativity (12). The binding cooperativity can vary over five orders of magnitude and is determined by the structural properties of this region, particularly the torsional flexibility of the CWWG motif. Sequences containing the torsionally flexible CATG motif, which are often identified with binding sites in cell-cycle arrest genes, are bound with high affinity and low cooperativity by p53DBD. Sequences containing the torsionally rigid CTAG and CAAG motifs are bound with lower affinity and high cooperativity. These motifs are abundant in binding sites associated with low-affinity apoptosis-related genes.
Despite the extensive studies addressing sequence-specific transactivation there remains the issue of what determines the differential transactivation of p53 in vivo in response to variations in levels of p53 protein. Moreover, differential p53 interaction with its target sites and the role of DNA physical characteristics in these interactions are not fully resolved. Previously, we developed an in vivo isogenomic yeast-based model system to address human p53 transactivation from different RE sequences in a constant chromatin environment (13). This system, which was created in diploid yeast (13), addresses both the ability of p53 to function from specific target sequences (i.e., on/off) and the extent of transactivation from these sites at variable levels of p53 protein.
Here, we establish that low basal levels of p53 expression result in transactivation of a linked reporter. Further increases in p53 lead to increased transactivation in an RE-specific manner. Using in vitro binding affinities for various p53 constructs, we found a positive relationship between in vivo transactivation and binding affinities, but only at high p53 levels. However, at low p53 levels transactivation was correlated with the DNA torsional flexibility of 29 natural and synthetic target sequences, revealing a unique structural relationship between transcriptional function and p53 RE sequence.
In addition, we discovered two sequences with unusual properties for p53-driven transactivation. They are highly responsive to low basal levels of p53. At high levels, the maximum response was comparable to that for transactivation from other highly responsive REs such at p21-5′ and p53R2. The transcriptional capacity of many p53 mutants associated with breast cancers could be differentiated by transactivation from such an RE. We establish that the highly sensitive response to wild-type (WT) p53 is linked to an increased kinetic stability of p53 on such sequences. Thus, the DNA sequence itself can strongly affect p53 transactivation even under conditions where the number of p53 molecules is small, suggesting that DNA structural properties can be critical factors in p53-dependent gene regulation.
Results
Transactivation at High vs. Low Protein Expression Level.
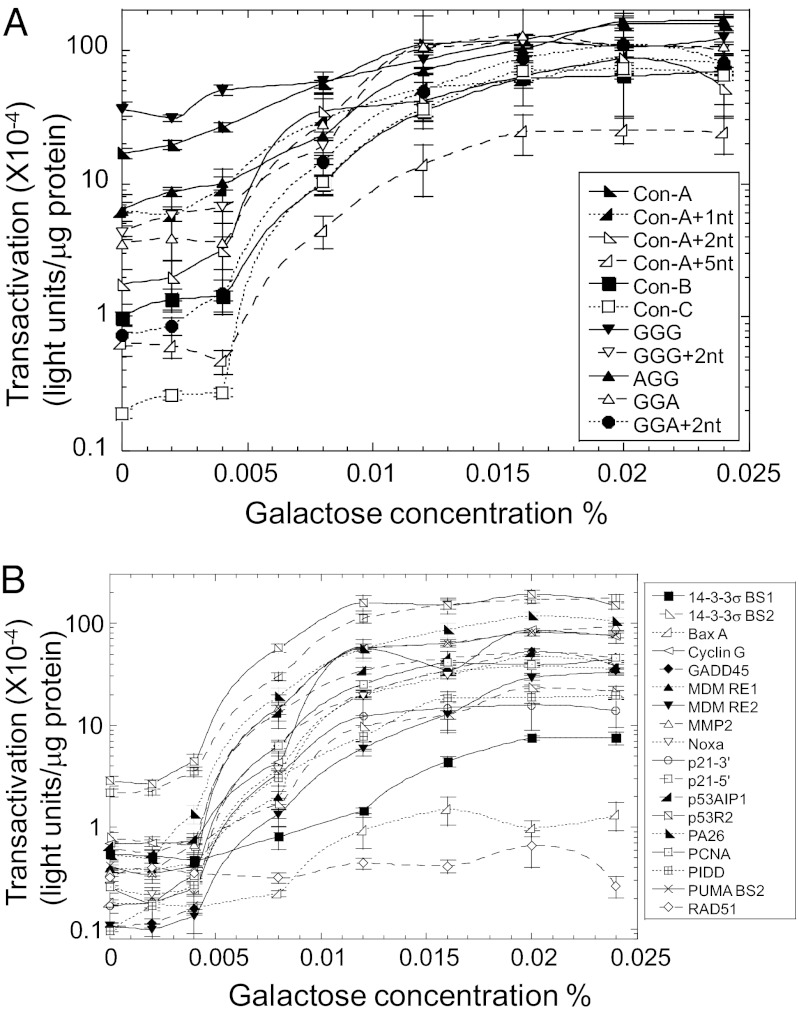
Recently, we developed a luciferase reporter system in diploid yeast that allows p53 transactivation capacities to be addressed with target sequences at variable levels of expression under isogenomic conditions (13). p53 expression is under a rheostatable GAL1 promoter that provides controlled and inducible expression of p53, depending on the carbon source. This assay provides the opportunity to address the relation between p53 induction and transactivation capacity at high and low levels of p53 molecules. In Table S1 we estimate that p53 levels change from ∼300 molecules per cell in the absence of galactose to ∼33,500 molecules at high levels of expression (0.024% galactose). Analysis of the strength of transactivation (relative light units per microgram of protein) from 11 artificial consensus-related targets (Table S2 and Fig. 1A) and 18 natural REs (Table S2 and Fig.1B) under different levels of p53 expression revealed distinct patterns of transactivation. The pattern of change in transactivation level from the natural p53 REs at high protein levels (Fig. 1B) spans three orders of magnitude and is similar to in vitro protein binding patterns (7, 11, 12), with high-affinity binding sites such as p21-5′ and p53R2 providing high levels of transactivation and weak binding sites such as Rad51 remaining at low transactivation levels, regardless of p53 expression (Fig. 1B). Below the 0.004% level of galactose, all natural REs decrease dramatically in their transactivation level, most being below 10−4 light units/μg protein, whereas p21-5′ and p53R2 are slightly above (Fig. 1B). The consensus-like targets are less variable in transactivation at high protein levels and center around 10−2 light units/μg protein (Fig. 1A). However, at low and basal levels of protein the transactivation pattern of the consensus-like sequences changes dramatically from that observed for natural p53 REs with a wide range of transactivation levels (0.2–40 × 10−4 light units/μg protein) from the consensus-like targets.
Fig. 1.
Variation in p53 protein levels reveals distinct transactivation patterns for p53 toward different response elements (REs). The abilities of WT p53 to transactivate different consensus-like (A) and natural (B) REs with increased cellular levels of p53 were measured in 24-h growing cultures of cells incubated with increasing amounts of galactose. For basal levels (galactose 0%) cells were grown on media containing raffinose as a carbon source. Presented are the mean and SEM for 5–30 independent luciferase reporter assay repeats. Sequences are described in Table S2.
Transactivation Capacity Correlates with Binding Affinity at High, but Not Low, Protein Expression Level.
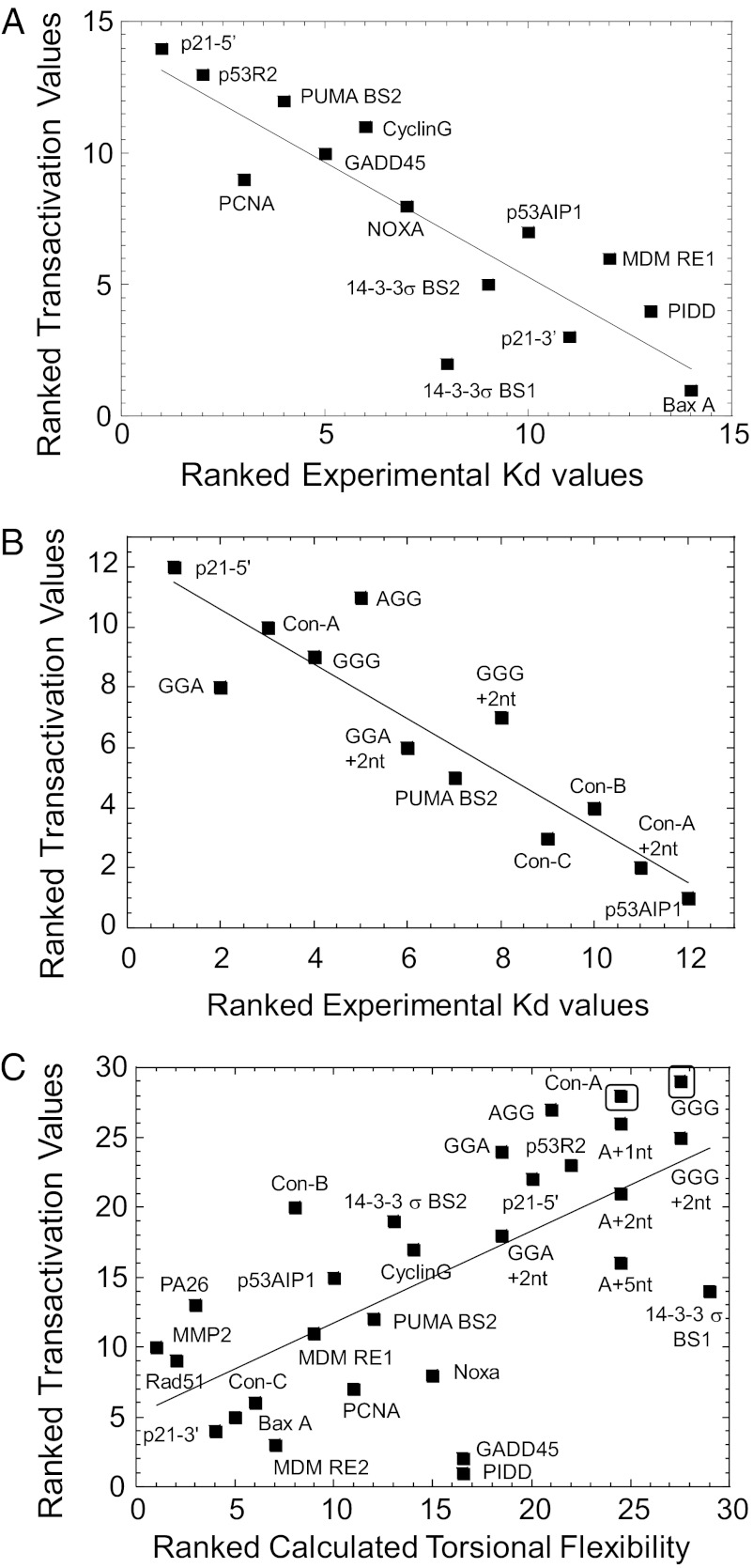
Our observation that at high p53 protein levels the transactivation correlates with in vitro binding (7, 11, 12) led to a statistical evaluation between these variables, using the Spearman rank correlation coefficient. Only 14 sequences were in common with the in vitro studies of Weinberg et al. (7). As shown in Fig. 2A, there is a strong (ρ = 0.87) and highly significant correlation (t test for the significance of the coefficient = −6.19, P value = 0.002) between transactivation level at 0.024% galactose (high induction of p53) and binding affinity measured for a p53 construct containing the DBD and the tetramerization domain (p53CT) (7). At low cellular levels of p53 (0.002% galactose) a weak (ρ = 0.41) and not significant correlation (t test = −1.58, P value = 0.134) is observed between these 14 sequences (Fig. S1A). A similar strong (ρ = 0.91) and significant (t test = −6.90, P value = 0.003) correlation at 0.024% galactose is observed when a p53 construct containing only the DBD was used in the binding affinity measurements (Fig. 2B), using 12 p53 REs for which we measured binding affinity (Table S3, Fig. S2, and ref. 12) as well as transactivation levels. These correlations demonstrate that the relationship between transactivation and p53 binding is independent of whether p53 binding characteristics are measured with or without the tetramerization domain, as well as being independent of the method of measurement [electrophoretic mobility shift assay (EMSA), Fig. 2B, vs. fluorescence anisotropy, Fig. 2A].
Fig. 2.
Transactivation from p53 REs correlates to different DNA-related characteristics at high vs. low p53 expression levels. (A) Transactivation at high p53 expression level is correlated to DNA-binding affinity of p53CT to p53 REs (7). There are 14 common sequences between this study and that of Weinberg et al. (7). (B) Transactivation at high p53 expression level is correlated to DNA-binding affinity of p53DBD to p53 REs. The 12 common data points are from published data (11, 12) or this study (Fig. S2 and Table S3). (C) Transactivation at low p53 expression level is correlated with DNA torsional flexibility of p53 REs. Calculated torsional flexibility is based on the values of Olson et al. (14). Torsional flexibility calculations were carried out on both decameric parts of all p53 sequences examined, excluding spacer sequences; hence the ranking of identical sites differing only in the spacer sequence is identical. The squares around “GGG” and “Con-A” mark them as supertransactivation sequences. Spearman’s rank correlation was used to test for the strength of the correlation. ρ = 0.87, 0.91, and 0.66 in A–C and P value = 0.002, 0.003, and 0.0006 in A–C, respectively. Sequences are described in Table S2.
Contrary to the weak and lack of significant correlation of transactivation at low p53 levels and KD, there is a strong (ρ = 0.66) and highly significant correlation (t test = 4.55, P value = 0.0006) between the torsional flexibility of p53 REs and transactivation (Fig. 2C). We recently showed that torsional (twist) flexibilities of p53 REs are important in p53 interactions with its REs (12). Moreover, we showed that the torsional flexibility of unmeasured sequences can be estimated using the dispersion of twist angle values from their average values, as determined from crystal structures of protein–DNA complexes (14). This approach is valid because the sequence-dependent torsional deformability described by Olson et al. (14) correlates with our experimental values (12). Using this methodology we assessed the relationship between transactivation at low cellular levels of p53 and calculated torsional flexibility (14, 15) of the entire sequence for each RE, using rank-order correlation calculations (Fig. 2C) for all binding sites studied here (29 sequences). Fig. 2C shows that the relationship between the calculated torsional flexibility (14, 15) of p53 sites and transactivation level is strong and highly significant, as detailed above. These torsional flexibility calculations were carried out along both decameric parts of all p53 sequences examined, regardless of whether they had a spacer between half sites or not. (Comparable level of significance for calculations along the entire sequence of each RE: ρ = 0.62, t test = 4.09, P value = 0.0012.) At high p53 expression level the correlation between transactivation level and torsional flexibility of p53 decamers is less significant (ρ = 0.37, t test = 2.07, P value = 0.05, Fig. S1B; for the entire full site the values are ρ = 0.3, t test = 1.71, P value = 0.1).
Con-A and GGG Are Supertransactivation Sequences.
Among the sequences studied here, transactivation at the “Con-A” and “GGG” REs (Table S2) was nearly 10-fold higher than at p21-5′ at basal levels of p53 expression or when cells were incubated at low levels of galactose (up to 0.004% galactose, Fig. 1 A and B). We refer to them as “supertransactivation” REs. Transactivation of Con-A was high even under the repressed (glucose) levels of p53 (Fig. S3A). Changing the CWWG core from CATG (Con-A) to CTAG (Con-B) or CAAG (Con-C) abolished the high transactivation level at basal levels of p53 (Fig. 1A). Furthermore, supertransactivation was dramatically reduced when one or more bases were inserted between half-site decamers (Fig. 1A). There was limited transactivation at high levels of p53 protein from 3/4-site and 1/2-site derivatives of Con-A; however, none of the noncanonical versions of Con-A supported a significant response at low levels of p53 (Fig. S3B). Thus, p53 capability for transactivation from the supertransactivation Con-A RE is dependent on the availability of a full-site RE, without spacers between half sites and with a CATG center. Neither of these supertransactivation REs was found among the >200,000 p53-like REs (16) in the human genome; however, the probability of any 20-base sequence in the genome is low.
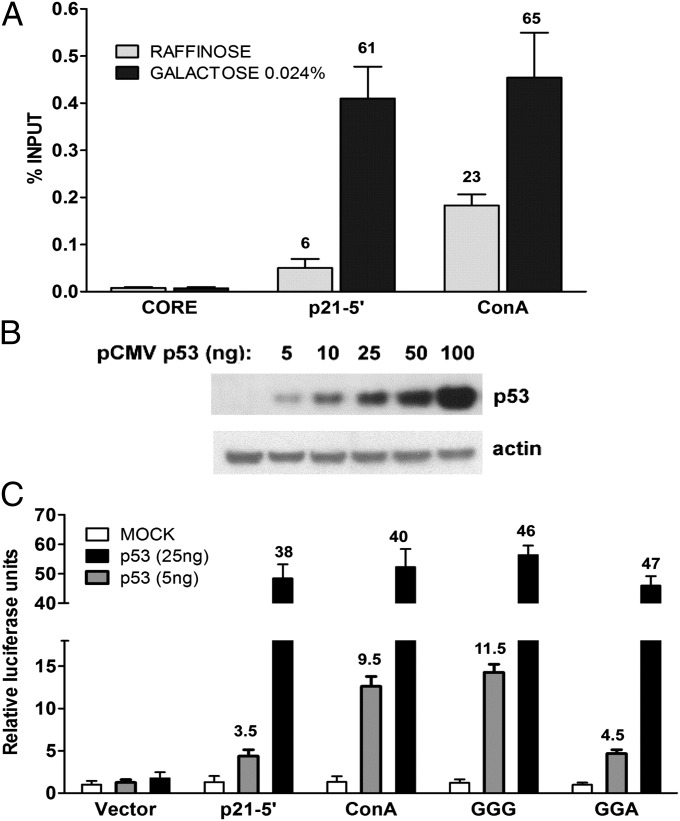
To determine whether the differences at basal p53 levels between supertransactivation sequences and other RE sequences are due to increased binding of p53 to these specific REs, we investigated in vivo occupancy, using quantitative chromatin immunoprecipitation-PCR (ChIP-qPCR) in yeast (Fig. 3A). As shown in Fig. 3A, no significant difference in p53 binding to p21-5′ or Con-A REs was observed at high protein levels (0.024% galactose, solid bars). However, at basal p53 levels, when cells were grown in raffinose (shaded bars), there was an approximate fourfold difference in p53 binding to the Con-A sequence compared with the p21-5′ RE, consistent with the transactivation results. No p53 binding was found in cells not induced for p53 (grown in raffinose) and containing only the empty CORE cassette used to clone the p53 REs (Fig. 3A). Differential occupancy by p53 was also determined using standard ChIP-PCR approaches (Fig. S4A).
Fig. 3.
Validation of supertransactivation response at low levels of p53 in yeast and mammalian cells. (A) Supertransactivation of Con-A RE correlates with high p53 binding at low protein levels. Occupancies of p53 at p21-5′ and Con-A REs were evaluated in yeast by ChIP-qPCR at low (raffinose) and high (0.024% galactose) levels of cellular p53 protein after 24 h growth. Cells lacking p53 (denoted “CORE”) were used as controls. Numbers above the bars are fold enrichment over the control values. Presented are the average and SD from two biological repeats. (B) Supertransactivation responses in human cells. p53 null SaOS2 cells were transfected with increasing amounts of pCMV-p53 WT vector. The p53 protein levels were checked by immunoblotting with p53-specific antibody 48 h posttransfection. A representative gel is shown. Actin served as a loading control. (C) Transactivation in human SaOS2 cells. Luciferase reporter constructs containing p21-5′, Con-A, GGG, and GGA REs were cotransfected along with 5 ng (low expression) or 25 ng (high expression) of the CMV-p53 WT vector. Transactivation was assessed with the luciferase assay 48 h later. Relative luciferase activity was compared with a mock transfection containing the promoterless pGL3 plasmid and is represented by the average and SDs of three independent experiments, each containing three replicates.
p53 Requirements for DNA-Dependent Supertransactivation in Yeast and Human Cells.
We analyzed transactivation at supertransactivation sequences by p53 structural mutants that are compromised for oligomerization and tetramerization. Although p53 tetramers are the functional unit essential for high-level transactivation, p53 can transactivate from half-site REs in yeast and human cells (13), and p53 dimers have been shown to bind consensus half-site sequences in vitro (17). The L344P mutation or deletion of the tetramerization domain (ΔTet, lacking residues 325–357) yields p53 monomers, whereas L344A results in a dimeric protein (summarized in ref. 13, see references in Fig. S3). As shown in Fig. S3C, these mutant proteins were not able to drive transactivation from the Con-A sequence at low or even high levels of p53. Thus, p53 capability for transactivation from the supertransactivation Con-A RE is also dependent on p53 being able to form tetramers.
We also determined the impact of p53 protein levels on transactivation from the supertransactivation sequences in human cells. Osteosarcoma SaOS2 cells, which are p53 null, were transfected with increasing amounts of p53 expression vector along with a constant amount of luciferase reporter constructs containing the p53 REs. The p53 protein levels were confirmed by Western blot analysis (Fig. 3B). We determined p53 transactivation from the supertransactivation sequences Con-A and GGG as well as from GGA and p21-5′ at low (5 ng) and high p53 levels (25 ng; Fig. 3C). These levels were chosen on the basis of transactivation from p21-5′ and Con-A REs with increasing amounts of transfected p53 expression vector (Fig. S4B). As shown in Fig. 3C, low-level p53 expression resulted in an approximate threefold greater transactivation from Con-A and GGG REs compared with that from the p21-5′ RE and the GGA sequence. At high p53 levels there were no significant differences between any of the REs (Fig. 3C).
Among the thousands of cancer-associated p53 mutations identified (International Agency for Research on Cancer, version R15, November 2010, ref. 18) many have very low transactivation capabilities (19), which might be more responsive with the supertransactivation Con-A RE. We examined 50 missense p53 mutants associated with breast cancer for their ability to transactivate from the Con-A RE, using a yeast ADE2 color assay (Fig. S5 A and B and ref. 13) and the luciferase reporter assay (Fig. S5C). The 50 mutants were previously analyzed for “functional fingerprints” in terms of capability for transactivation from 11 human target REs, where mutants could alter the spectrum of targets transactivated as well as levels (20). Among these, 29 mutants lacked transactivation capability and the functional fingerprint of 21 mutants was altered. Using the ADE2 plate assay and a high galactose level (0.128%), we found that the lack of any transactivation capability by these mutants was best predicted by Con-A, compared with any of the other 11 human p53-RE targets used (Fig. S5A). Among the altered function mutants all but P151H and R174K eliminated the basal-level response characteristic of WT p53 (Fig. S5B); however, they all retained good induction from Con-A at higher p53 levels. The transactivation capability of several mutants was confirmed with the luciferase reporter assay. Several mutants exhibited substantial transactivation from the p21-5′ and Con-A REs at elevated p53 levels; however, only Con-A supported high levels of transactivation at basal levels of some of the p53 mutants (Fig. S5C). Thus, transactivation from the Con-A sequence can be used as a diagnostic tool to address transcriptional capabilities of p53 mutants.
Binding Kinetics and the Molecular Basis of Supertransactivation.
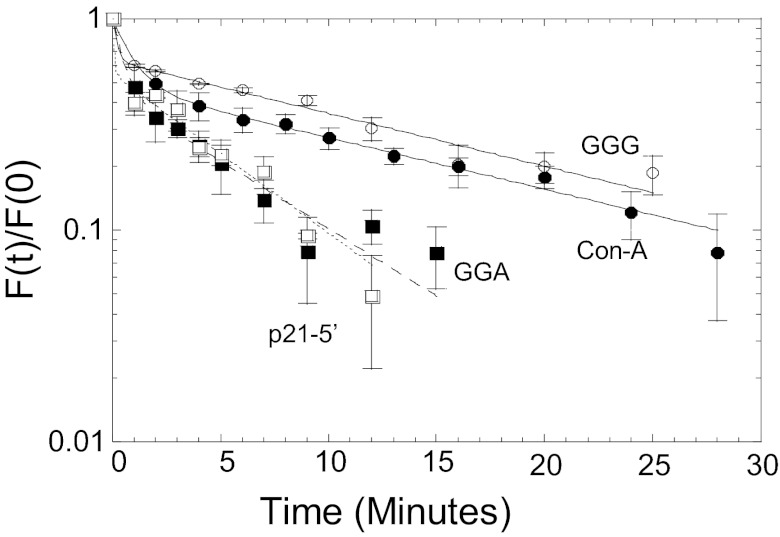
What is the molecular origin of the supertransactivation phenomenon? The two supertransactivation sequences do not reach the high-affinity levels of well-established targets such as p21-5′ and GGA (Table S3, Fig. S2, and refs. 7, 8, and 11), as noted above. The Con-A and GGG sequences exhibit a high degree of torsional flexibility (12), more than other p53 REs; however, the difference in torsional flexibility between these sequences and the GGA target may not account fully for the supertransactivation phenomenon. We, therefore, determined the binding stability (i.e., the kinetic off rate) of WT p53DBD toward four DNA-binding sites: the supertransactivation targets, Con-A and GGG, and high-affinity targets p21-5′ and GGA (Fig. 4 and Fig. S6A). Analysis of the tetramer band in Fig. S6A (Table S4) shows that there is a biphasic dissociation kinetic behavior of p53 tetramers from all four sequences. The half-life of fraction “A” was always fast (a few seconds) and variable between experiments (with a curve-fitting error of the same magnitude or larger), whereas the half-life of fraction “B” was slower and reproducible between experiments and, hence, had a low curve-fitting error. This biphasic behavior means that there are two linked macroscopic dissociation processes and that the kinetics of dissociation are determined by a mechanism involving binding intermediates. On the basis of the half-lives of complexes undergoing process B dissociation (Fig. 4 and Table S4), the supertransactivation sequences form more kinetically stable complexes with p53DBD, with off rates that are twice as large as those with the other sequences.
Fig. 4.
Dissociation kinetics of WT p53DBD tetramers from different binding sites. Shown is a plot of the fraction of molecules bound to p53DBD at time (t) divided by the fraction of molecules bound at time 0 as a function of time. The lines are from the best fit to a double-exponential curve. Solid circles, Con-A; open circles, GGG; Solid squares, GGA; open squares, p21-5′. The experimental points are the average and SEM of four to nine independent experiments conducted with each DNA target. The half-life of tetramer complexes formed on the GGG and Con-A sites is 12 ± 2 min, and half-lives for the GGA and p21-5′ sites are 5.1 ± 1 min and 5.2 ± 0.8 min, respectively (Table S4).
These observations point to kinetic stability of p53DBD tetramers on Con-A and GGG being responsible for the supertransactivation behavior. We then asked whether the same trend between kinetic stability and supertransactivation is observed for p53DBD dimers, using DNA targets with just one specific decameric half site (HS), flanked by nonspecific sequences (Table S4 and Fig. S6B). Table S4 shows that p53DBD forms stable dimers on the supertransactivation sequence Con-A HS and the nonsupertransactivation sequence GGA HS, but not on the supertransactivation sequence GGG HS.
Discussion
The level of a transcription activator is expected to influence the extent of target gene expression. Despite that expectation, there are few examples in which it has been possible to directly examine this issue. Here, we used a rheostatable p53 expression system along with various physical analyses to address specifically three aspects of the relationship between transactivation and p53 target sequence: high vs. low p53 level, impact of cancer-associated mutations, and unique supertransactivation sequences.
Transactivation at Low p53 Levels.
At high p53 levels, the positive relationship between transactivation and DNA-binding affinity that we observed suggests that under conditions of constant chromatin environment, binding affinity is a major determinant of transactivation. However, this relationship breaks down at low p53 levels and suggests that under conditions of limited stress the factors responsible for p53-dependent transactivation are different from those at highly induced p53 levels. Under conditions of low p53 levels we found that transactivation levels are largely determined by the torsional flexibility of the DNA sequences. We attribute this difference to the functional form of p53 being a tetramer. p53 dimerizes during cotranslation in polysomes (21) and under 50 nM protein is normally present as a dimer in solution (22, 23). Thus, the functional tetramer form is induced by DNA binding (12, 23, 24). Our estimate (Table S1) is that 0.024% galactose corresponds to 33,500 p53 molecules. Assuming a yeast cell radius of 2–4 μm, the estimated number of molecules corresponds to ∼40 nM of p53 within the cell at high protein expression levels (levels in human cells appear to be sometimes higher; Table S1). When the protein level is low, the chance that two dimeric p53 molecules will arrive together at the RE to bind as a tetramer is low. Therefore, at low protein levels the binding of the tetrameric form can be accomplished only when the first dimer species is bound stably enough on the DNA, such that it is able to stay bound until the next dimer molecules arrives at the RE. Torsional flexibility of the RE can contribute to the kinetic stability of p53 dimers on the RE, because we have previously shown (12) that torsional flexibility of p53 REs facilitates the reorientation of two p53 monomers within each dimer to stabilize intradimer interactions with concomitant decrease in binding cooperativity. Hence, at low levels of expression of p53 and on torsionally flexible DNA targets, the complex of p53 dimers on DNA can transiently accumulate as intermediates. Therefore, the transactivation level at low protein concentrations is correlated with the torsional flexibility of the target site and not with the binding affinity of the p53/DNA complex. Consistent with our previous study (12), all target sites on the right side of Fig. 2B (above “ranked” position 17) have the very torsionally flexible CATG center, whereas those on the left (ranked ≤17) have other DNA motifs at the center.
Supertransactivation.
Many studies have addressed the function of p53 toward a variety of REs that have different capabilities in supporting transactivation. Using our system that examines p53 function over a broad range of protein expression, we discovered that two sequences were highly effective at supporting high levels of transactivation even under very low expression conditions, which has the potential for greatly increasing the functionality of p53.
The supertransactivation character of Con-A and GGG REs at low protein expression requires a full-site consensus binding element and depends on the ability of p53 to form a tetramer, the same structural requirements necessary for high response at high protein levels. Transactivation with few molecules suggests unique properties of these supertransactivation REs. Other REs often require at least 10 times more p53 to achieve even low levels of transactivation, and several weak REs required ∼100 times more p53 protein over basal levels to reach transactivation levels comparable to those obtained from Con-A or GGG REs at basal p53 expression. As reported for other REs with high transactivation responses, a CATG core motif and absence of spacers between half sites are required for a strong response by the supertransactivation REs. However, these requirements are not enough for the supertransactivation phenomenon, as discussed below.
We found that torsional flexibility is a prerequisite for high transactivation at low protein levels. However, this amount of torsional flexibility is not sufficient, because the GGA and AGG targets also have above average torsional flexibilities (12). The supertransactivation phenomenon additionally requires kinetic stability of the tetrameric complex, i.e., low off rate of p53 from full-site RE, which is observed only for the GGG and Con-A targets. A low off rate might be due to DNA breathing (base pair opening), which would be lower in GGG and Con-A relative to other REs, because they contain either no A·T base pairs (GGG) or only one (Con-A), outside the central CATG region. If so, it may be possible to identify additional supertransactivation sequences using the yeast approaches described here.
The Con-A RE was originally derived from a mutagenesis approach of three tandem repeats of the upstream p53-binding element of the human RGC sequence (CCAGGCAAGT)3 done to determine the DNA-binding sequence specificity of wild-type p53 (25). The GGG RE is a self-complementary sequence derived from the Con-A RE (11). The observation that neither of the supertransactivation REs is found in the genome (the closest sequence found has 17 of the 20 nt without a mismatch) may indicate that such sequences would lack the p53 stress response associated with the other targets. Previously, we observed with a bead-binding assay that p53 binding is ∼2-fold greater for Con-A RE than p21-5′ RE (26), using basal levels of p53 from untreated nuclear U2OS cell extracts. Consistent with this, we show here that at low levels, transactivation from p53 is 10-fold better in vivo to the Con-A RE than to p21-5′ RE (Fig. 1), which is generally considered one of the most responsive natural p53 REs at basal p53 expression levels (27).
Binding Kinetics and Implications for the Mechanism of RE Binding of p53.
Kinetic analysis of four p53 targets revealed that p53 tetramers dissociated with a biphasic behavior, indicating that dissociation proceeds through a mechanism involving binding intermediates. Petty et al. (28) suggested an induced-fit binding mechanism of p53 to its REs. This mechanism entails two binding (or dissociation) steps: a rigid body interaction followed by a conformational change. Our results show a considerably greater biphasic character to p53 off rate than for the WT p53 complex studied by Petty et al. (28). This may be due to the p53 construct used here as well as to the conditions of the binding interactions. The biphasic behavior may be due to quaternary changes in p53 conformation (11, 29) and/or changes in the L1-loop conformation observed in several p53 structures upon DNA binding (11, 28, 30, 31). However, they may also be due in part to other kinds of binding intermediates that do not involve an induced-fit mechanism, for example a dimer intermediate on the way to forming or abolishing the functional tetrameric form of p53. From Table S4 it is clear that different intermediates occur when tetramers dissociate from p53 full sites, compared with dissociation of dimeric complexes from p53 half sites, on the basis of differences in the biphasic characteristics of the dissociation process; however, the nature of the various species is unknown at present.
p53 Cancer-Associated Mutants and Implications of Supertransactivation.
Many p53 mutants retain transcription capability and can even result in a “change of spectrum” of the transactivated REs (32, 33). Importantly, transactivation from the Con-A sequence appears to distinguish transcriptional capacity between many p53 mutations associated with breast cancers and WT p53 (20). The supertransactivation Con-A sequence appears diagnostic of residual transactivation function of mutant p53 proteins. Among the 50 cancer-associated p53 mutants examined, transactivation by mutant p53 from Con-A predicts transactivation from at least one additional RE (Fig. S5A). However, in the absence of transactivation from Con-A, even at high p53 expression, there is also no transactivation from any of an additional 11 natural REs studied (Fig. S5A). The supertransactivation property of Con-A might be useful for determining which p53 mutants could be remedied.
The present results at low p53 levels lead us to raise a general question of whether such sequences exist for other transcription factors. Our findings establish the possibility of screening for supertransactivation simply by presenting potential sequences with an associated reporter at low levels of the relevant transcription factor, such as NKX2.5, in yeast (34) or mammalian cells. Supertransactivation response sequences may have general diagnostic and therapeutic utilities for p53 as well as for other transcription factors.
Materials and Methods
A detailed description of materials and methods is available in SI Materials and Methods.
Protein and DNA.
Human p53 core domain (residues 94–293, referred to as p53DBD) was a kind gift from Zippora Shakked (Weizmann Institute of Science, Rehovot, Israel). All DNA sequences for EMSA were synthesized by Sigma Genosys and purified by a reverse-phase cartridge.
Binding Experiments.
Binding affinity and dissociation kinetics measurements were conducted as previously described (12, 35, 36). In kinetics experiments radiolabeled hairpin duplexes (0.4 nM) were incubated with 270 nM or 160 nM p53DBD (for half-site and full-site sequences, respectively) in the same binding buffer used in KD measurements before adding unlabeled competitor DNA of the same sequence (1.58 μM).
Twist Flexibility Calculations.
The dispersion of averaged structural parameters from their values in protein–DNA complexes (14), measured by the SD, was taken as a measure of the flexibilities of individual DNA base pair steps. The flexibility calculation was carried out on the entire group of p53 REs (with or without spacers), using a sliding window of dinucleotides.
Statistical Analysis.
In testing the strength of relationships between variables, we calculated Spearman’s rank correlation coefficient (denoted by ρ) as a nonparametric measure of correlation (37). We assumed that the relationship between the variables being compared is monotonic. Reported P values are two-tailed.
Transactivation Reporter and ChIP Assays in Yeast and Human Cells.
The reporter assays are described in ref. 13. Details of the qualitative ADE2 reporter assay and the quantitative luciferase reporter assay are presented in Fig. S5. For the chromatin immunoprecipitation assays p53 DO-1 (Santa Cruz Biotechnology) and H3 ab1791 (Abcam) antibodies were used. Human SaOS2 osteosarcoma cells (HTB-85; American Type Culture Collection) were routinely maintained following standard conditions and procedures for culturing mammalian cells.
Supplementary Material
Acknowledgments
We thank Zippi Shakked for purified WT p53DBD protein and for helpful discussions, Alberto Inga during development of the yeast system and Con-A discussions, and Carl Anderson, Leping Li, and Michael Fried for comments on the manuscript. Support for this work was provided by Department of Defense Breast Cancer Research Program Predoctoral Trainee Award BC051212 (to J.J.J.); by National Institute on Environmental Health Sciences intramural research funds, Project Z01-ES065079 (to M.A.R., D.M., and J.J.J.); and by Israel Science Foundation Grant 104405, Israel Cancer Association Grant 20112012, and Israel Cancer Research Fund Grant 800005 (to T.E.H., J.S., K.R., and I.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205971109/-/DCSupplemental.
References
- 1.Lane D, Levine A. p53 Research: The past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9:724–737. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- 3.Olivier M, et al. The IARC TP53 database: New online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 4.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 5.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 6.Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu Rev Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 7.Weinberg RL, Veprintsev DB, Bycroft M, Fersht AR. Comparative binding of p53 to its promoter and DNA recognition elements. J Mol Biol. 2005;348:589–596. doi: 10.1016/j.jmb.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Arbely E, et al. Acetylation of lysine 120 of p53 endows DNA-binding specificity at effective physiological salt concentration. Proc Natl Acad Sci USA. 2011;108:8251–8256. doi: 10.1073/pnas.1105028108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian H, Wang T, Naumovski L, Lopez CD, Brachmann RK. Groups of p53 target genes involved in specific p53 downstream effects cluster into different classes of DNA binding sites. Oncogene. 2002;21:7901–7911. doi: 10.1038/sj.onc.1205974. [DOI] [PubMed] [Google Scholar]
- 10.Inga A, Storici F, Darden TA, Resnick MA. Differential transactivation by the p53 transcription factor is highly dependent on p53 level and promoter target sequence. Mol Cell Biol. 2002;22:8612–8625. doi: 10.1128/MCB.22.24.8612-8625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitayner M, et al. Structural basis of DNA recognition by p53 tetramers. Mol Cell. 2006;22:741–753. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 12.Beno I, Rosenthal K, Levitine M, Shaulov L, Haran TE. Sequence-dependent cooperative binding of p53 to DNA targets and its relationship to the structural properties of the DNA targets. Nucleic Acids Res. 2011;39:1919–1932. doi: 10.1093/nar/gkq1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan JJ, et al. Noncanonical DNA motifs as transactivation targets by wild type and mutant p53. PLoS Genet. 2008;4:e1000104. doi: 10.1371/journal.pgen.1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc Natl Acad Sci USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balasubramanian S, Xu F, Olson WK. DNA sequence-directed organization of chromatin: Structure-based computational analysis of nucleosome-binding sequences. Biophys J. 2009;96:2245–2260. doi: 10.1016/j.bpj.2008.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris CR, et al. p53 responsive elements in human retrotransposons. Oncogene. 2009;28:3857–3865. doi: 10.1038/onc.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLure KG, Lee PWK. How p53 binds DNA as a tetramer. EMBO J. 1998;17:3342–3350. doi: 10.1093/emboj/17.12.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petitjean A, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 19.Kato S, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci USA. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordan JJ, et al. Altered-function p53 missense mutations identified in breast cancers can have subtle effects on transactivation. Mol Cancer Res. 2010;8:701–716. doi: 10.1158/1541-7786.MCR-09-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholls CD, McLure KG, Shields MA, Lee PW. Biogenesis of p53 involves cotranslational dimerization of monomers and posttranslational dimerization of dimers. Implications on the dominant negative effect. J Biol Chem. 2002;277:12937–12945. doi: 10.1074/jbc.M108815200. [DOI] [PubMed] [Google Scholar]
- 22.Brandt T, Petrovich M, Joerger AC, Veprintsev DB. Conservation of DNA-binding specificity and oligomerisation properties within the p53 family. BMC Genomics. 2009;10:628–642. doi: 10.1186/1471-2164-10-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajagopalan S, Huang F, Fersht AR. Single-molecule characterization of oligomerization kinetics and equilibria of the tumor suppressor p53. Nucleic Acids Res. 2011;39:2294–2303. doi: 10.1093/nar/gkq800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinberg RL, Veprintsev DB, Fersht AR. Cooperative binding of tetrameric p53 to DNA. J Mol Biol. 2004;341:1145–1159. doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
- 25.Halazonetis TD, Davis LJ, Kandil AN. Wild-type p53 adopts a ‘mutant’-like conformation when bound to DNA. EMBO J. 1993;12:1021–1028. doi: 10.1002/j.1460-2075.1993.tb05743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noureddine MA, et al. Probing the functional impact of sequence variation on p53-DNA interactions using a novel microsphere assay for protein-DNA binding with human cell extracts. PLoS Genet. 2009;5:e1000462. doi: 10.1371/journal.pgen.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 28.Petty TJ, et al. An induced fit mechanism regulates p53 DNA binding kinetics to confer sequence specificity. EMBO J. 2011;30:2167–2176. doi: 10.1038/emboj.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitayner M, et al. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat Struct Mol Biol. 2010;17:423–429. doi: 10.1038/nsmb.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malecka KA, Ho WC, Marmorstein R. Crystal structure of a p53 core tetramer bound to DNA. Oncogene. 2009;28:325–333. doi: 10.1038/onc.2008.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho WC, Fitzgerald MX, Marmorstein R. Structure of the p53 core domain dimer bound to DNA. J Biol Chem. 2006;281:20494–20502. doi: 10.1074/jbc.M603634200. [DOI] [PubMed] [Google Scholar]
- 32.Resnick MA, Inga A. Functional mutants of the sequence-specific transcription factor p53 and implications for master genes of diversity. Proc Natl Acad Sci USA. 2003;100:9934–9939. doi: 10.1073/pnas.1633803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menendez D, Inga A, Resnick MA. The biological impact of the human master regulator p53 can be altered by mutations that change the spectrum and expression of its target genes. Mol Cell Biol. 2006;26:2297–2308. doi: 10.1128/MCB.26.6.2297-2308.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inga A, Reamon-Buettner SM, Borlak J, Resnick MA. Functional dissection of sequence-specific NKX2-5 DNA binding domain mutations associated with human heart septation defects using a yeast-based system. Hum Mol Genet. 2005;14:1965–1975. doi: 10.1093/hmg/ddi202. [DOI] [PubMed] [Google Scholar]
- 35.Faiger H, Ivanchenko M, Cohen I, Haran TE. TBP flanking sequences: Asymmetry of binding, long-range effects and consensus sequences. Nucleic Acids Res. 2006;34:104–119. doi: 10.1093/nar/gkj414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faiger H, Ivanchenko M, Haran TE. Nearest-neighbor non-additivity versus long-range non-additivity in TATA-box structure and its implications for TBP-binding mechanism. Nucleic Acids Res. 2007;35:4409–4419. doi: 10.1093/nar/gkm451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehman EL, D’Abrera HJM. Non Parametric: Statistical Methods Based on Ranks. Rev Ed. Englewood Cliffs, NJ: Prentice-Hall; 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.