Abstract
Tyr142, the C-terminal amino acid of histone variant H2A.X is phosphorylated by WSTF (Williams-Beuren syndrome transcription factor), a component of the WICH complex (WSTF-ISWI chromatin-remodeling complex), under basal conditions in the cell. In response to DNA double-strand breaks (DSBs), H2A.X is instantaneously phosphorylated at Ser139 by the kinases ATM and ATR and is progressively dephosphorylated at Tyr142 by the Eya1 and Eya3 tyrosine phosphatases, resulting in a temporal switch from a postulated diphosphorylated (pSer139, pTyr142) to monophosphorylated (pSer139) H2A.X state. How mediator proteins interpret these two signals remains a question of fundamental interest. We provide structural, biochemical, and cellular evidence that Microcephalin (MCPH1), an early DNA damage response protein, can read both modifications via its tandem BRCA1 C-terminal (BRCT) domains, thereby emerging as a versatile sensor of H2A.X phosphorylation marks. We show that MCPH1 recruitment to sites of DNA damage is linked to both states of H2A.X.
Preserving genomic integrity is vital to the fitness of an organism. Damage to DNA emanating from endogenous and exogenous insults can trigger signaling cascades relayed by posttranslational modifications culminating in the activation of the cell cycle checkpoint and initiation of repair. One of the key initiating events in this process is the phosphorylation at serine 139 (pSer139) of histone H2A.X, a chromatin-bound histone variant (1, 2). Phosphorylated H2A.X (γH2A.X) serves as a platform for the recruitment of downstream mediator proteins as well as chromatin modifying proteins to the affected site (3–5). Recent studies have added a new dimension to the recognition of γH2A.X with the identification of a new phosphorylation site in H2A.X, Tyr142. In response to DNA damage, Tyr142 was found to transition from a phosphorylated (pTyr142) to a nonphosphorylated state in an Eya1/3 phosphatase-dependent manner (6–8). Whereas the dephosphorylation of pTyr142 is gradual, the phosphorylation of Ser139 is prompt, and the overlap in the two processes is thought to give rise to the doubly phosphorylated (pSer139, pTyr142) H2A.X state (di-γH2A.X) following genotoxic insult (6, 7). The existence of di-γH2A.X and proteins that recognize this state remain open questions in the field.
The protein MDC1 (mediator of DNA damage checkpoint 1) has emerged as an interacting partner of γH2A.X. MDC1 was found to directly sense pSer139 and to mobilize the downstream response by virtue of its tandem BRCA1 C-terminal (BRCT) domains (4, 5). An important question therefore is how MDC1, an established γH2A.X binder, responds to the presence of diphosphorylated H2A.X. Interestingly, several groups have independently found that MDC1 does not interact with di-γH2A.X (6, 7, 9). Indeed, efforts to identify binding partners of a di-γH2A.X peptide were redirected at well-established pTyr-binding SH2/PTB domains and led to the second PTB domain of Fe65 as a possible target (7). Is this inability to bind Tyr142-phosphorylated H2A.X reflective of the inherent limitations of tandem BRCT domains, previously identified as pSer or pThr binding domains, to recognize the pTyr state?
Microcephalin (MCPH1), the first gene identified as causative for primary autosomal microcephaly (10), has been linked to various cellular processes including the DNA damage checkpoint, DNA repair by homologous recombination, and DNA transcription (11–15). Previous studies have established that MCPH1 is an early responder in the DNA damage response (DDR) and regulates the recruitment of several downstream mediator proteins, a characteristic shared with MDC1. Similarities between MCPH1 and MDC1 are inescapable. Both proteins include tandem BRCT domains at their C terminus that are necessary for irradiation-induced foci (IRIF) formation in an H2A.X pSer139 phosphorylation-dependent manner (16, 17). MCPH1 and MDC1 can bind a peptide that encompasses phosphorylated Ser139 (16, 18) and in vivo mutation of this residue results in loss of IRIF (16). Because MCPH1 IRIF formation necessitates γH2A.X, presumably because of direct interaction with γH2A.X, we asked whether MCPH1 could also interact with di-γH2A.X.
The discovery that H2A.X Tyr142 can be phosphorylated has raised several interesting points. First, whereas the di-γH2A.X species has been suggested, a deeper characterization of its kinetics and role in DNA damage remains to be attempted. Second, it remains to be determined whether a relationship exists between di-γH2A.X and γH2A.X. Third, it is not known whether MCPH1, a previously described γH2A.X interactor, can also recognize di-γH2A.X. Here, via the generation of an antibody that selectively recognizes a di-γH2A.X peptide (di-pH2A.X), we demonstrate the existence of di-γH2A.X and monitor its dynamics following DNA damage. Furthermore, we show that there exists a direct relationship between pTyr142 and pSer139, whereby the former affects the latter. Most importantly, our work identifies MCPH1 as a BRCT protein that directly interacts with di-pH2A.X and explains the structural basis for this recognition. Hitherto, BRCT domains were thought to function within the realm of pSer or pThr recognition (19–21).
Results
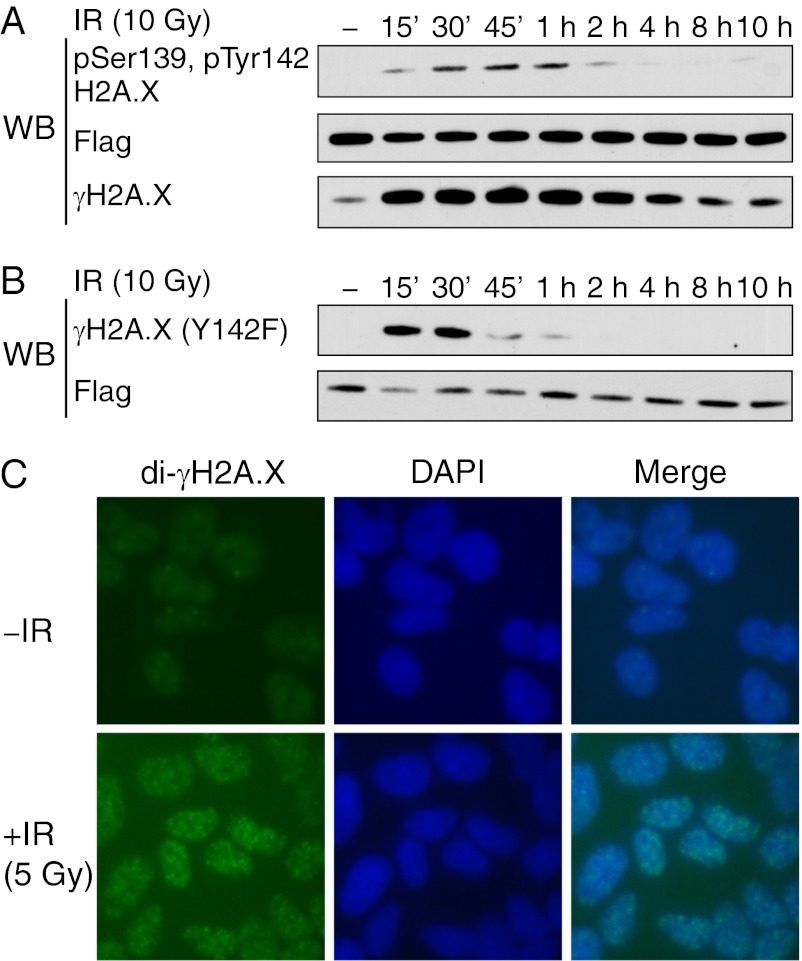
To directly examine the role of di-γH2A.X in the DDR, we developed an antibody specific to di-γH2A.X. This antibody preferentially recognizes di-γH2A.X among the four possible posttranslational states of H2A.X and could be completely blocked with a di-γH2A.X peptide but not with the corresponding nonphosphorylated peptide (Fig. S1). Using this reagent, we tracked the kinetics of di-γH2A.X in H2a.x−/− mouse embryonic fibroblasts (MEFs) reconstituted with wild-type H2A.X and determined that di-γH2A.X levels peak 1 h postirradiation and decline rapidly thereafter (Fig. 1A). Because immunofluorescence allows visualization of γH2A.X as distinct nuclear foci after exposure to ionizing radiation (Fig. S2), we then asked whether di-γH2A.X behaves similarly. We observed that di-γH2A.X, much like γH2A.X, forms IRIF following DNA damage (Fig. 1C). Next, using H2a.x−/− MEFs stably reconstituted with either wild-type or Y142F mutant H2A.X, we monitored the kinetics of γH2A.X in both cell lines (Fig. 1 A and B). Interestingly, the Y142F mutant showed an increase in γH2A.X until 30 min after irradiation, and then was rapidly reduced to undetectable levels by 1 h postirradiation, whereas the wild-type H2A.X reconstituted cells sustained a high level of γH2A.X even at 10 h postirradiation (Fig. 1 A and B). This result implies that Tyr142 phosphorylation in H2A.X is necessary for retaining γH2A.X after DNA damage. A similar defect in the kinetics of γH2A.X has been shown in cells where expression of the Tyr142-specific kinase WSTF (Williams-Beuren syndrome transcription factor) was knocked down (6). The defect in γH2A.X kinetics impairs the formation of MDC1 and phosphorylated ATM foci postirradiation (6). Taken together, these results validate the postulated di-γH2A.X state, confirm the interplay between di-γH2A.X and γH2A.X, and suggest that di-γH2A.X can critically influence the DDR pathway.
Fig. 1.
Diphosphorylated H2A.X is critical in mediating the DNA damage response. (A) H2a.x−/− MEFs were stably reconstituted with Flag–SBP-tagged wild-type (WT) H2A.X. Cells were irradiated for 1 h at 10 Gy and the whole cell lysate was immunoblotted with the indicated antibodies. (B) Same as A but the MEFs were transfected with the Y142F H2A.X mutant. (C) Di-γH2A.X foci were visualized in 293T cells by immunofluorescence staining before and after exposure to ionizing radiation (IR) for 1 h at 5 Gy.
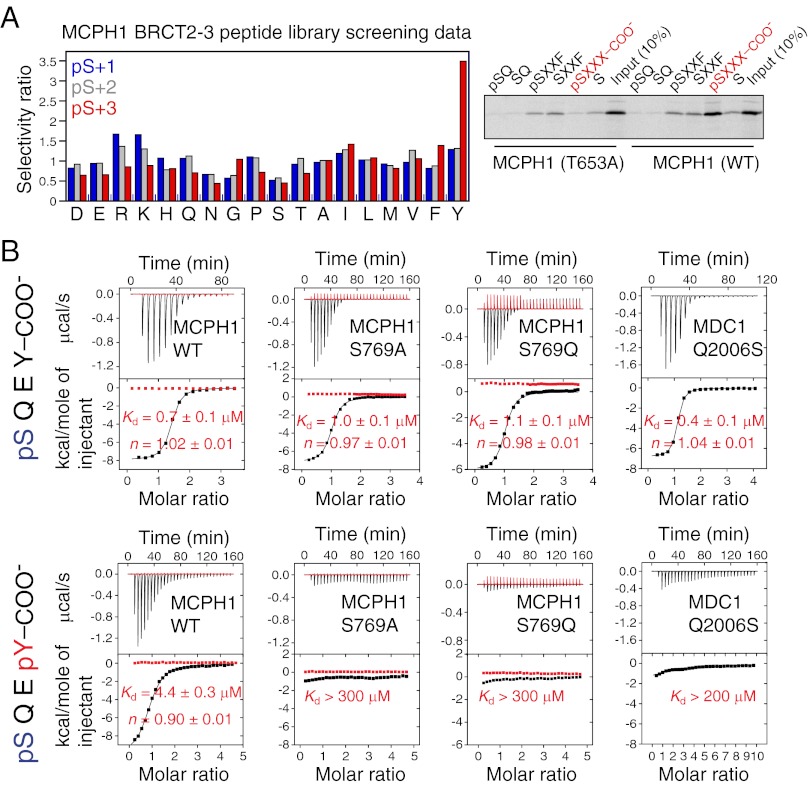
MCPH1 is a DDR protein that is recruited to the sites of DNA damage within minutes in a γH2A.X-dependent fashion and this recruitment necessitates MCPH1 C-terminal tandem BRCT domains (11, 16, 17). Unlike many other repair factors, MCPH1 relocalization to DNA damage sites occurs even in the absence of MDC1 (16). We investigated whether the tandem BRCT domains of MCPH1 directly recognize γH2A.X and di-γH2A.X. Using a phosphoserine-containing peptide library screen, we determined that the BRCT domains optimally bind the motif pSXXY where pS is the phosphoserine, X represents any amino acid, and position pS+3 is a tyrosine (Y), with a strong requirement for a free C-terminal carboxylate group (Fig. 2A). Histone γH2A.X, which exhibits the optimal motif pSQEY, Y being the C-terminal residue of the protein, may represent a preferred binding target. This was validated using pull-down assays that confirmed an interaction between full-length MCPH1 and a γH2A.X peptide that could be disrupted by single point mutations in MCPH1 (see below). Using isothermal titration calorimetry (ITC), we also determined that a Ser139-phosphorylated tetrapeptide corresponding to the C-terminal end of human H2A.X (pH2A.X, residues 139–142) binds the tandem BRCT domains of human MCPH1 with a dissociation constant (Kd) of 0.7 ± 0.1 μM, whereas the peptide devoid of the phosphate group showed no interaction (Fig. 2B and Fig. S3).
Fig. 2.
MCPH1 interacts with mono- and diphosphorylated H2A.X. (A, Left) Amino acid selectivity values for MCPH1 tandem BRCT domains determined using the phosphoserine-oriented degenerate peptide library KKKAYXXXXpSXXX-COO−, where X indicates any amino acid except Cys, and COO− indicates the extreme C terminus. Values ≥1.35 indicate moderate selection and values >2.0 indicate strong selection. (Right) The phosphopeptide library above, with X replaced by specified residues at pS+1 or pS+3 positions, was used in pull-down assays with in vitro translated [35S] Met-labeled MCPH1 tandem BRCT domains (wild-type and T653A mutant). Note the strong preference for the free C-terminal carboxylate group for phosphopeptide binding as highlighted in red. (B) Interaction of the tandem BRCT domains of MCPH1 and MDC1 (wild type and indicated mutants) with pH2A.X and di-pH2A.X tetrapeptides probed by ITC. Shown are the raw data and integrated heat signals (black) plotted against time and molar ratio of peptide to protein. In red are control injections of peptide into buffer solution. A standard one-site binding model was used for curve fitting. Kd and stoichiometry (n) are indicated with the associated SDs determined by nonlinear least-squares analysis.
Whereas MDC1 can directly bind γH2A.X through its tandem BRCT domains, it fails to engage the di-γH2A.X state (6, 7, 9). We found that, unlike MDC1, MCPH1 could bind a diphosphorylated (pSer139, pTyr142) H2A.X tetrapeptide (di-pH2A.X, residues 139–142) with a Kd of 4.4 ± 0.3 μM (Fig. 2B). Furthermore, we observed partial and near complete colocalization of MCPH1 with di-γH2A.X and with γH2A.X, respectively, consistent with their in vitro affinities (see below). Whereas tandem BRCT domains have been previously identified as pSer binding modules (19, 20), MCPH1 represents the singular instance of a tandem BRCT repeat that can interact with both a singly and a doubly phosphorylated motif accompanied by an inherent selectivity for one of the states.
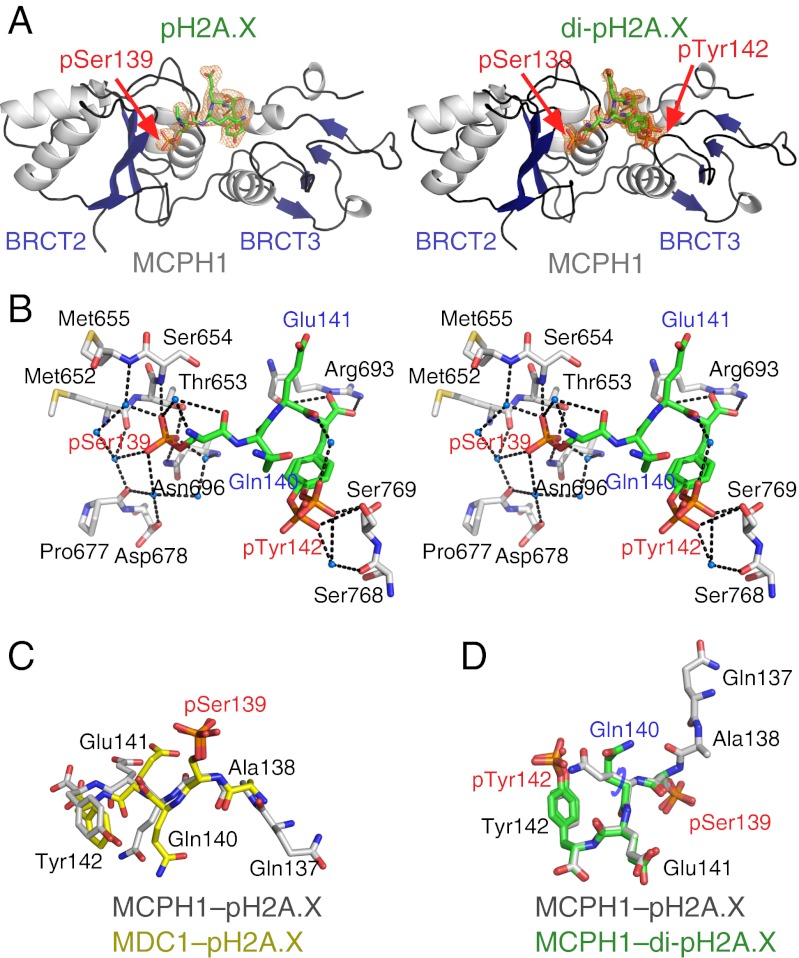
To gain a deeper understanding of these interactions, we determined the structures of MCPH1 tandem BRCT domains (BRCT2–BRCT3) in complex with a pH2A.X decapeptide (MCPH1–γH2A.X) and di-γH2A.X tetrapeptide (MCPH1–di-γH2A.X) to 2.6-Å and 1.5-Å resolution, respectively (Fig. 3 A and B and Table 1 and Figs. S4 and S5). Whereas γH2A.X peptides adopt similar conformations when bound to either MCPH1 or MDC1 (Fig. 3C), a reorientation of di-pH2A.X Gln140 side chain is necessary for binding to MCPH1 (Fig. 3D). In fact, the rigid conformation of Gln140 was held partly responsible for the inability of MDC1 to interact with di-pH2A.X (9).
Fig. 3.
Three dimensional structures of MCPH1 tandem BRCT domains in complex with pH2A.X and di-pH2A.X phosphopeptides. (A) Ribbon diagrams of the structures of MCPH1 C-terminal tandem BRCT domains (BRCT2 and BRCT3; residues 640–835) in complex with (Left) a pH2A.X decapeptide (residues 133–142) and (Right) a di-pH2A.X tetrapeptide (residues 139–142). The 2Fo–Fc omit electron density maps of the peptides are shown contoured at 1σ level (orange mesh). Note the two conformations of pTyr42 in the di-pH2A.X complex. (B) Stereographic representation of key residues of MCPH1 interacting with the di-pH2A.X peptide. Hydrogen bonds and water molecules are depicted as black dashes and blue spheres, respectively. (C) Overlay of a pH2A.X hexapeptide from the MCPH1–pH2A.X structure (gray) with a pentapeptide from the MDC1–pH2A.X structure (yellow). (D) Overlay of a pH2A.X hexapeptide from the MCPH1–pH2A.X structure (gray) and di-pH2A.X tetrapeptide from the MCPH1–di-pH2A.X structure (green). Note the different orientations of Gln140 side chains corresponding to a χ1 angle change of ∼86°.
Table 1.
Data collection and refinement statistics
| MCPH1–pH2A.X | MCPH1–di-pH2A.X | |
| Data collection | ||
| Space group | P1 | P21 |
| Cell dimensions | ||
| a, b, and c, Å | 67.81, 82.23, and 104.42 | 37.69, 46.54, and 55.78 |
| α, β, and γ° | 72.10, 87.87, and 80.94 | 90.00, 97.07, and 90.00 |
| Resolution (Å) | 50.0–2.63 (2.74–2.63)* | 50.0–1.50 (1.55–1.50) |
| Rmerge | 0.064 (0.471) | 0.062 (0.437) |
| I/σI | 13.57 (2.0) | 25.7 (2.3) |
| Completeness (%) | 90.2 (97.8) | 96.7 (80.1) |
| Redundancy | 2.6 (2.6) | 3.0 (2.6) |
| Refinement | ||
| Resolution (Å) | 50.0–2.63 (2.89–2.63) | 23.79–1.50 (1.65–1.50) |
| No. reflections | 56,418 (14,911) | 29,928 (7,036) |
| Rwork/Rfree | 20.1/25.2 | 12.9/17.1 |
| No. atoms | 14,108 | 2,063 |
| Protein | 13,294 | 1,720 |
| Ligand/ion | 422 | 54 |
| Water | 392 | 289 |
| B factors | ||
| Protein | 66.4 | 20.9 |
| Ligand/ion | 56.3 | 24.9 |
| Water | 47.4 | 34.5 |
| R.m.s. deviations | ||
| Bond lengths, Å | 0.009 | 0.005 |
| Bond angles, ° | 1.05 | 1.071 |
| Ramachandran plot | ||
| Favored regions, % | 98.0 | 99.0 |
| Allowed regions, % | 1.8 | 1.0 |
Each dataset was collected on a single crystal.
*Values in parentheses are for highest-resolution shell.
In the MCPH1–γH2A.X and MCPH1–di-pH2A.X structures, the phosphate of pSer139 is recognized via direct hydrogen bonding to MCPH1 Thr653, Ser654, and Asn696 (Fig. 3B and Fig. S5). Additionally, the higher resolution MCPH1–di-pH2A.X structure shows a network of water-mediated interactions involving the phosphate of pSer139 and the backbone carbonyl groups of Met652, Met655, and Pro677 as well as the backbone amide atoms of Asn696 (Fig. 3B). Thr653 resides in a position structurally identical to Thr1898 in MDC1 and Ser1655 in BRCA1 and binds the phosphate analogously (Fig. S6). Mutation of Thr653 to Ala dramatically reduced binding of the MCPH1 BRCT domains to the pSXXX C-terminal peptide library (Fig. 2A), whereas mutation of MCPH1 Asn696 to an aspartate resulted in greater than 500-fold reduction in affinity (Fig. S3). Noticeably, in MDC1 (4) and BRCA1 (22–25), and in other structures of tandem BRCT domain–phosphopeptide complexes, such as human TopBP1 (21) or Schizosaccharomyces pombe Crb2 (26) and Brc1 (27), the residue corresponding to MCPH1 Asn696 is a lysine (Figs. S6 and S7A). Whereas the vast majority of phosphopeptide binding modules characterized to date carry at least one lysine or arginine in their phosphate binding site (28), the structures of MCPH1 complexes demonstrate that presence of a lysine or an arginine is not an absolute requirement for phosphate recognition.
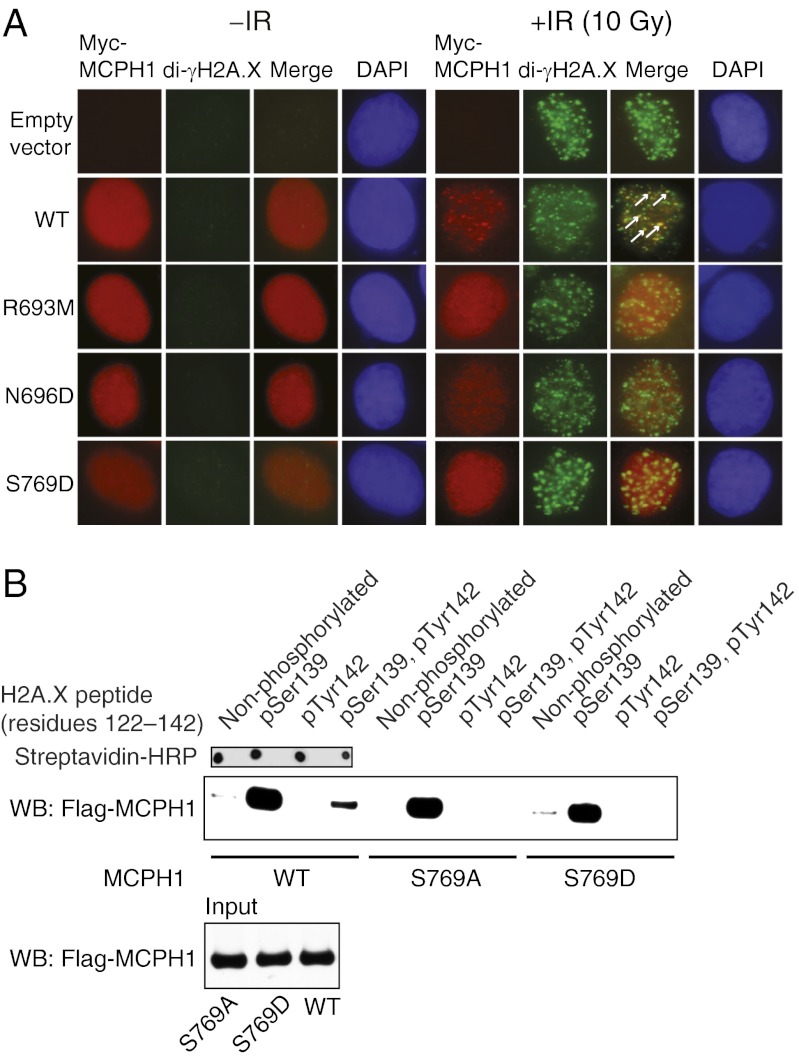
The two structures of MCPH1 complexes show that the carboxylate group of H2A.X Tyr142 is anchored to MCPH1 via evolutionarily conserved Arg693. Amidation of this carboxylate group in pH2A.X resulted in a more than 100-fold decrease in affinity (Fig. S3). Whereas the guanidinium group of Arg693 forms two hydrogen bonds to the C-terminal carboxylate, its main chain carbonyl supplements this interaction by hydrogen bonding to Tyr142 amide proton (Fig. 2B and Fig. S6). Mutation of Arg693 to methionine profoundly affected binding between pH2A.X and MCPH1 (Kd > 270 μM) (Fig. S3). Because the tandem BRCT domains of MCPH1 are important for damage-induced foci formation, we asked whether the R693M and N696D mutants affect this process. We found that both the R693M and N696D MCPH1 mutants show a marked reduction in their ability to form IRIF, reflecting their inability to engage γH2A.X or di-γH2A.X (Fig. 4A and Fig. S2).
Fig. 4.
Probing the MCPH1–di-γH2A.X interaction in cells. (A) U2OS cells were transfected with Myc-tagged wild-type (WT) or mutant MCPH1 and cells were coimmunostained with anti-Myc and anti-γH2A.X antibodies before and after exposure to 10 Gy of ionizing radiation (IR) for 1 h. Note that MCPH1 foci are almost completely colocalized with γH2A.X foci (Fig. S2), whereas they are partially colocalized with di-γH2A.X foci, some of which are highlighted with white arrows. (B) 293T cells were transfected with Flag-tagged wild-type (WT) or indicated MCPH1 mutants, and nuclear extracts from the irradiated cells were incubated with a synthetic biotinylated H2A.X peptide (unmodified or phosphorylated as indicated) affixed to streptavidin-coated magnetic beads. Resulting pull-down products were analyzed by Western blot (WB) using an anti-Flag antibody.
The MCPH1–di-pH2A.X structure reveals that tandem BRCT domains can recognize the dual phosphorylation status. This is achieved through recognition of pSer139 by the first BRCT domain in the tandem repeat (BRCT2) and recognition of pTyr142 by the second BRCT domain (BRCT3) (Fig. 3 A and B). The phosphate of pTyr142 is detected by a direct hydrogen bond to the hydroxyl group of Ser769 as well as water-mediated hydrogen bonding to the hydroxyl group of Ser768 (Fig. 3B). The MCPH1 S769A mutation, that cannot stabilize pTyr by hydrogen bonding, selectively disrupts the MCPH1–di-pH2A.X interaction (Kd > 200 μM) while leaving the interaction with pH2A.X virtually intact (Kd = 1.0 ± 0.1 μM) (Fig. 2B). We confirmed that the single S769A or S769D mutations when introduced into full-length MCPH1 prevent binding to di-pH2A.X but not pH2A.X (Fig. 4B). Therefore, a single amino acid mutation is sufficient to cause MCPH1 to behave like MDC1 by interacting only with monophosphorylated H2A.X. We also observed that the S769D mutant has reduced ability to form MCPH1 IRIF (Fig. 4A), suggesting that the MCPH1–di-γH2A.X interaction may play a role in MCPH1 recruitment to DNA damage sites.
Like MCPH1, MDC1 directly recognizes the carboxylate group of Tyr142 in the pH2A.X peptide. Why does MDC1 not engage the peptide when Tyr142 is phosphorylated? A recent study quantified the interaction between MDC1 and di-pH2A.X and found a dramatic >300-fold decrease in affinity compared with pH2A.X (9). To explain the loss in affinity, the authors modeled two different conformations of pTyr142. The loss of affinity was partly attributed to a potential clash between either the tyrosine phosphate and Pro2009 in MDC1 specificity loop, or alternately, pTyr142 and the di-pH2A.X Gln140 side chain. However, we observed that in the MCPH1–di-pH2A.X complex this barrier is overcome by an altered conformation of di-pH2A.X that permits accommodation of the phosphotyrosine. This conformational change now results in the side chain of Gln140 pointing away from pTyr142 (Fig. 3D). Furthermore, the proline residue (Pro770), also present in the specificity loop of MCPH1 (Fig. S7A), does not hamper binding to di-pH2A.X. Because in MDC1 a glutamine (Gln2008) resides at a position equivalent to MCPH1 Ser769 that forms a hydrogen bond with the phosphate of pTyr142 in di-pH2A.X (Fig. S7A), we asked whether the Q2008S mutation in MDC1 would allow it to bind di-pH2A.X. This mutation, however, is insufficient to change its binding proclivity with respect to di-pH2A.X. MDC1 Q2008S retains tight binding to pH2A.X (Kd = 0.4 ± 0.1 μM) but, like the wild-type protein, does not bind di-pH2A.X (Kd > 200 μM) (Fig. 2B). In contrast to MDC1, MCPH1 presents a more favorable electropositive surface in BRCT3 for the phosphotyrosine, possibly providing a permissive environment for binding (Fig. S7B). This could explain why a single amino acid mutation is insufficient to convert MDC1 to an MCPH1-like state.
Discussion
DNA damage initiates a complex cascade of chromatin modification, signaling, and repair events that are just beginning to be understood. Although there exist dedicated proteins attending to each of these processes, other proteins have a broader coordinating role in these pathways. The protein Microcephalin or MCPH1 is a case in point. MCPH1 is implicated in chromatin remodeling by virtue of its interaction with the SWI/SNF complex (15), in signaling programs via its association with Chk1 among other proteins (11, 29–32), and aids the DNA repair process by interacting with BRCA2 (33) and Condensin II (34, 35). Because of its involvement in multiple processes that converge to maintain genomic integrity, MCPH1 has been termed a “guardian of the genome” (13). Indeed, ablation of MCPH1 resulted in a menagerie of chromosomal defects that includes changes in both the nature and number of chromosomes, highlighting its importance in regulating genomic stability (12, 13). Furthermore, diminished DNA copy numbers as well as reduced MCPH1 mRNA and protein expression levels have been noted in several breast and ovarian cell lines and in ovarian cancer tissues, suggesting that MCPH1 is a putative tumor suppressor (12). Whereas considerable progress has been made regarding the biology of MCPH1, a central unanswered question is: how is this key regulator of genomic stability recruited to sites of DNA damage?
To probe MCPH1 recruitment to sites of DNA damage, its IRIF were visualized in cells devoid of various sensor and mediator proteins. MCPH1 foci were found to depend on γH2A.X but were independent of ATM, MDC1, or NBS1 (16). Interestingly, whereas the down-regulation of MCPH1 resulted in several proximal as well as distal DDR proteins to be absent from the sites of DNA damage including ATM, ATR, and Rad51, the formation of γH2A.X foci remained unaffected (12). This suggests that MCPH1 recruitment is γH2A.X-dependent and that MCPH1 does not regulate γH2A.X foci formation.
Histone variant H2A.X is considered to occupy a sentinel position in the DDR, serving as a critical intermediary in relaying information pertaining to double-strand breaks (DSBs) to the outside environment. To understand how MCPH1 is mobilized to DSBs, we began investigating the interaction of the C-terminal end of MCPH1, previously deemed important for this anchoring process, with the different H2A.X states. Because chronologically pTyr142-containing H2A.X is present in the absence of DNA damage, we explored and found that MCPH1, like MDC1, does not recognize this state of H2A.X. At the present time, readers of pTyr142 H2A.X remain undiscovered. Next, we found that with regards to γH2A.X, MCPH1 and MDC1 can both recognize this state with similar affinities. In the absence of a clear affinity advantage of either MDC1 or MCPH1 for γH2A.X, their respective levels may determine the amounts recruited. This contention is supported by an earlier study reporting that overexpression of MCPH1 could result in a reduction of MDC1 foci (16). Therefore, overexpressed MCPH1 may outcompete and directly displace γH2A.X-bound MDC1. It thus appears that tipping the concentration balance in favor of MCPH1 or MDC1 could significantly alter the composition at γH2A.X nucleosomes. Also, the ability of MDC1 to dimerize in response to DNA damage may considerably change its capacity to bind γH2A.X with avidity effects coming into play (36, 37). This may explain how MDC1 independently engages γH2A.X in the presence of MCPH1. Furthermore, both MCPH1 and MDC1 may be present in high molecular weight multicomponent complexes, which could alter the affinities measured using purified recombinant domains. Thus, whereas the BRCT domains of MCPH1 and MDC1 are direct readers of γH2A.X, this ability may be harnessed as part of larger DNA damage sensing multiprotein assemblies to gain access to sites of DSBs. Such is likely the case for MCPH1, an established component of the BRCA2/Rad51 (33), SWI/SNF (15), and Condensin II (34, 35) complexes that need to be directed to damaged sites. Future work is required to decipher the amounts of MCPH1 parsed into each of these and other complexes and the contribution of MCPH1–γH2A.X interaction toward their targeting.
Whereas considerable strides have been made toward understanding how Ser139-phosphorylated H2A.X controls downstream mediators and effectors, much less is known about H2A.X phosphorylated at both Ser139 and Tyr142. In this study, we have first confirmed the existence of previously hypothesized di-γH2A.X in vivo, a form of H2A.X that exists in the very early phases of DDR. Next, we have shown a clear and unambiguous difference in the ability of two early mediator proteins to engage modified H2A.X. Whereas MDC1 can bind the canonical γH2A.X state, it shows no affinity toward di-γH2A.X. In contrast, MCPH1 shows the requisite plasticity to sense both the pSer139- and pSer139, pTyr142-modified H2A.X. Our work therefore uncovers the latent ability of a subset of tandem BRCT domains to recognize the pSXXpY motif with important implications for future biology as it provides a distinct recognition strategy for selective recruitment of MCPH1 in early phases of the DDR. The biological significance of the ability of MCPH1 to read both γH2A.X and di-γH2A.X is yet to be explored. On the basis of the kinetics of γH2A.X and di-γH2A.X, we speculate that the ability of MCPH1 to bind di-γH2A.X recruits MCPH1 to the damaged sites as one of the earliest responses to DNA damage. As the repair process proceeds, pTyr42 is dephosphorylated, which would allow even stronger interaction between γH2A.X and MCPH1. This increased affinity could be necessary for the recruitment of other repair factors. If pTyr42 is not dephosphorylated, factors involved in apoptosis might eventually displace MCPH1 to initiate apoptosis, which has previously been suggested to be one of the roles of di-γH2A.X (7, 8). Specifically, our finding forms the basis for selectively inhibiting the MCPH1–H2A.X pathway over the MDC1–H2A.X pathway using di-pH2A.X mimetics. Lastly, our study broadens the scope of tandem BRCT domain recognition sequences, previously restricted to pSer or pThr, to include phosphotyrosine and multiple phosphorylation marks. This discovery speculates that BRCT domains may play a larger role in biological functions than previously anticipated.
Materials and Methods
Detailed methods for protein production, purification, crystallization, structure determination, ITC measurements, antibody generation, pull-down assays, and immunofluorescence are described in SI Materials and Methods. Briefly, MCPH1 and MDC1 were expressed in Escherichia coli as N-terminal His6-tag fusion constructs and purified by metal affinity and, after cleavage of the His6-tag, by size exclusion chromatography. Three dimensional structures of the MCPH1–pH2A.X and MCPH1–di-pH2A.X complexes were determined by molecular replacement from diffraction data recorded at beamline 19-ID of the Advanced Photon Source (APS) at Argonne National Laboratory and beamline X29 of the National Synchrotron Light Source (NLS) at Brookhaven National Laboratory, respectively. Data collection and refinement statistics are presented in Table 1.
Supplementary Material
Acknowledgments
We are grateful to Y. Kim at Advanced Photon Source (APS) for assistance with X-ray data collection, Z. Dauter for insightful comments regarding structure refinement, and A. Nussenzweig for providing the H2a.x−/− mouse embryonic fibroblasts. The MCPH1 construct used for peptide library screening was a kind gift of R. Chahwan and S. P. Jackson, University of Cambridge, Cambridge, United Kingdom. This work was supported by National Institutes of Health (NIH) Grants CA132878 (to G.M.), CA097134, DK018477, DK39949, and NS034934 (to M.G.R.), CA116167 (to F.J.C.), and CA112967 and ES015339 (to M.B.Y.). M.G.R. is a Howard Hughes Medical Institute investigator. We acknowledge the use of beamline 19-ID at APS and beamline X29 at National Synchrotron Light Source (NLS). APS is operated by University of Chicago’s Argonne National Laboratory, for the US Department of Energy under Contract DE-AC02-06CH11357. Funding for NLS comes from the offices of Biological and Environmental Research and Basic Energy Sciences of the US Department of Energy and from NIH Grant P41RR012408.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates for the structures of MCPH1–pH2A.X and MCPH1–di-pH2A.X complexes have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3SZM and 3U3Z).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212366109/-/DCSupplemental.
References
- 1.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 2.Scully R. A histone code for DNA repair. Nat Rev Mol Cell Biol. 2010;11:164. doi: 10.1038/nrm2855. [DOI] [PubMed] [Google Scholar]
- 3.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 4.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 5.Lou Z, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 6.Xiao A, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook PJ, et al. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnan N, et al. Dephosphorylation of the C-terminal tyrosyl residue of the DNA damage-related histone H2A.X is mediated by the protein phosphatase eyes absent. J Biol Chem. 2009;284:16066–16070. doi: 10.1074/jbc.C900032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell SJ, Edwards RA, Glover JN. Comparison of the structures and peptide binding specificities of the BRCT domains of MDC1 and BRCA1. Structure. 2010;18:167–176. doi: 10.1016/j.str.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Jackson AP, et al. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet. 2002;71:136–142. doi: 10.1086/341283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin SY, Rai R, Li K, Xu ZX, Elledge SJ. BRIT1/MCPH1 is a DNA damage responsive protein that regulates the Brca1-Chk1 pathway, implicating checkpoint dysfunction in microcephaly. Proc Natl Acad Sci USA. 2005;102:15105–15109. doi: 10.1073/pnas.0507722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rai R, et al. BRIT1 regulates early DNA damage response, chromosomal integrity, and cancer. Cancer Cell. 2006;10:145–157. doi: 10.1016/j.ccr.2006.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaplet M, Rai R, Jackson-Bernitsas D, Li K, Lin SY. BRIT1/MCPH1: A guardian of genome and an enemy of tumors. Cell Cycle. 2006;5:2579–2583. doi: 10.4161/cc.5.22.3471. [DOI] [PubMed] [Google Scholar]
- 14.Bartek J. Microcephalin guards against small brains, genetic instability, and cancer. Cancer Cell. 2006;10:91–93. doi: 10.1016/j.ccr.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 15.Peng G, et al. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat Cell Biol. 2009;11:865–872. doi: 10.1038/ncb1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wood JL, Singh N, Mer G, Chen J. MCPH1 functions in an H2AX-dependent but MDC1-independent pathway in response to DNA damage. J Biol Chem. 2007;282:35416–35423. doi: 10.1074/jbc.M705245200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeffers LJ, Coull BJ, Stack SJ, Morrison CG. Distinct BRCT domains in Mcph1/Brit1 mediate ionizing radiation-induced focus formation and centrosomal localization. Oncogene. 2008;27:139–144. doi: 10.1038/sj.onc.1210595. [DOI] [PubMed] [Google Scholar]
- 18.Shao Z, et al. Specific recognition of phosphorylated tail of H2AX by the tandem BRCT domains of MCPH1 revealed by complex structure. J Struct Biol. 2012;177:459–468. doi: 10.1016/j.jsb.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 19.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 20.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 21.Leung CC, Gong Z, Chen J, Glover JN. Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control. J Biol Chem. 2011;286:4292–4301. doi: 10.1074/jbc.M110.189555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Botuyan MV, et al. Structural basis of BACH1 phosphopeptide recognition by BRCA1 tandem BRCT domains. Structure. 2004;12:1137–1146. doi: 10.1016/j.str.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiozaki EN, Gu L, Yan N, Shi Y. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: Implications for signaling. Mol Cell. 2004;14:405–412. doi: 10.1016/s1097-2765(04)00238-2. [DOI] [PubMed] [Google Scholar]
- 24.Williams RS, Lee MS, Hau DD, Glover JN. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat Struct Mol Biol. 2004;11:519–525. doi: 10.1038/nsmb776. [DOI] [PubMed] [Google Scholar]
- 25.Clapperton JA, et al. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol. 2004;11:512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- 26.Kilkenny ML, et al. Structural and functional analysis of the Crb2-BRCT2 domain reveals distinct roles in checkpoint signaling and DNA damage repair. Genes Dev. 2008;22:2034–2047. doi: 10.1101/gad.472808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams JS, et al. gammaH2A binds Brc1 to maintain genome integrity during S-phase. EMBO J. 2010;29:1136–1148. doi: 10.1038/emboj.2009.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joughin BA, Tidor B, Yaffe MB. A computational method for the analysis and prediction of protein:phosphopeptide-binding sites. Protein Sci. 2005;14:131–139. doi: 10.1110/ps.04964705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X, Lee J, Stern DF. Microcephalin is a DNA damage response protein involved in regulation of CHK1 and BRCA1. J Biol Chem. 2004;279:34091–34094. doi: 10.1074/jbc.C400139200. [DOI] [PubMed] [Google Scholar]
- 30.Tibelius A, et al. Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1. J Cell Biol. 2009;185:1149–1157. doi: 10.1083/jcb.200810159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Passemard S, et al. VIP blockade leads to microcephaly in mice via disruption of Mcph1-Chk1 signaling. J Clin Invest. 2011;121:3071–3087. doi: 10.1172/JCI43824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gruber R, et al. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat Cell Biol. 2011;13:1325–1334. doi: 10.1038/ncb2342. [DOI] [PubMed] [Google Scholar]
- 33.Wu X, et al. Microcephalin regulates BRCA2 and Rad51-associated DNA double-strand break repair. Cancer Res. 2009;69:5531–5536. doi: 10.1158/0008-5472.CAN-08-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood JL, Liang Y, Li K, Chen J. Microcephalin/MCPH1 associates with the Condensin II complex to function in homologous recombination repair. J Biol Chem. 2008;283:29586–29592. doi: 10.1074/jbc.M804080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamashita D, et al. MCPH1 regulates chromosome condensation and shaping as a composite modulator of condensin II. J Cell Biol. 2011;194:841–854. doi: 10.1083/jcb.201106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, et al. Structural mechanism of the phosphorylation-dependent dimerization of the MDC1 forkhead-associated domain. Nucleic Acids Res. 2012;40:3898–3912. doi: 10.1093/nar/gkr1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jungmichel S, et al. The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator. Nucleic Acids Res. 2012;40:3913–3928. doi: 10.1093/nar/gkr1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.