Abstract
CCAAT/enhancer-binding protein α (C/EBPα) is one of the key transcription factors that mediate lineage specification and differentiation of multipotent myeloid progenitors into mature granulocytes. Although C/EBPα is known to induce granulopoiesis while suppressing monocyte differentiation, it is unclear how C/EBPα regulates this cell fate choice at the mechanistic level. Here we report that inducers of monocyte differentiation inhibit the alternate cell fate choice, that of granulopoiesis, through inhibition of C/EBPα. This inhibition is mediated by extracellular signal-regulated kinases 1 and/or 2 (ERK1/2), which interact with C/EBPα through an FXFP docking site and phosphorylate serine 21. As a consequence of C/EBPα phosphorylation, induction of granulocyte differentiation by C/EBPα or retinoic acid is inhibited. Our analysis of C/EBPα by fluorescent resonance energy transfer revealed that phosphorylation induces conformational changes in C/EBPα, increasing the distance between the amino termini of C/EBPα dimers. Thus, myeloid development is partly regulated by an ERK1/2-mediated change in the conformation of C/EBPα that favors monocyte differentiation by blocking granulopoiesis.
Myeloid cells are key components of the innate immune system, offering general protection against bacteria and parasites. As with all forms of hematopoiesis, new myeloid cells arise throughout life to replace the old. When myeloid cells are renewed, they must choose between two distinct lineages, monocytes and granulocytes. These cell fate decisions and differentiation events are critically important not only because myeloid cells are necessary for proper immune function but also because their dysregulation leads to myeloid leukemias. Recent work has shown that the transcription factor C/EBPα is fundamental both in normal myeloid differentiation and in the block to differentiation that causes myeloid leukemias (44).
C/EBPα is the prototypical basic-region leucine zipper (bZIP) transcription factor (4). This family is characterized by the presence of a basic region that mediates DNA binding and a leucine zipper that allows dimer formation. In addition to forming homodimers, C/EBPα dimerizes with other members of the C/EBP family (C/EBPβ, -γ, -δ, ɛ, and CHOP) (20) and interacts with other proteins such as TFIIB, TBP, Rb, p300/CBP, p21, and members of the SWI/SNF complex (2, 5, 24, 30, 45). Recently, it has been shown that C/EBPα is regulated by phosphorylation and sumoylation (17, 37, 43). However, it is not clear how these posttranslational modifications affect the functions of C/EBPα in vivo.
A key role of C/EBPα is to regulate differentiation of a select set of cell types. Within the hematopoietic system, C/EBPα is expressed in myeloblast progenitors and granulocytes, but not monocytes. Ectopic expression of C/EBPα in bipotential myeloid cells induces granulopoiesis and blocks monocyte differentiation (34), whereas loss of C/EBPα results in an absence of granulocytes (57). C/EBPα is likewise important for adipose tissue, where it mediates differentiation of preadipocytes into adipocytes and regulates the metabolism of mature adipocytes (4, 36). The requirement of C/EBPα for life, however, is revealed in the liver and lung. Without intervention, C/EBPα−/− mice die within hours of birth of hypoglycemia due to impaired function of hepatocytes (54) or of respiratory failure due to impaired function of type II pneumocytes (8, 22). C/EBPα is also expressed, although its function remains poorly characterized, in the intestine, adrenal gland, skin, mammary gland, placenta, and brain (20).
Several recent studies highlight the crucial antimitotic role of C/EBPα, which inhibits cell growth through a variety of mechanisms. First, C/EBPα induces expression and stability of the cyclin-dependent kinase inhibitor, p21 (48). Second, C/EBPα interacts directly with the cyclin-dependent kinases Cdk2 and Cdk4 and blocks their ability to interact with cyclins, thereby impeding cell cycle progression (14, 52, 53). Finally, C/EBPα directly represses the activity of E2F, a key transcriptional regulator of cell cycle genes (16, 41, 46, 47). Indeed, the ability of C/EBPα to repress E2F may be necessary for induction of differentiation to granulocytes and adipocytes (16, 32). Moreover, there is increasing evidence from myeloid and lung cancers that C/EBPα is a tumor suppressor gene (13). Several papers have reported that the antiproliferative effect of p42C/EBPα is blocked by mutations of C/EBPα in acute myeloid leukemia (AML) (11, 28, 33). While some mutations increase expression of the alternate translation product p30C/EBPα, which behaves functionally as a dominant negative factor for C/EBPα action, other mutations, within the bZIP domain, result in deficient binding to DNA. The differentiation of granulocytic blasts is blocked in these tumors, again underscoring the inseparable association between growth arrest and differentiation, each requiring C/EBPα for proper regulation.
While many studies have uncovered the importance of C/EBPα, both in normal physiology and in disease, little is known about mechanisms that regulate its activity. In this study, we identified serine 21 as a novel site of phosphorylation and showed that this site is directly phosphorylated by extracellular signal-regulated kinases 1 and/or 2 (ERK1/2), which recognize serine 21 of C/EBPα as a substrate through an FXFP docking motif. This phosphorylation induces a conformational change in C/EBPα such that the transactivation domains of two C/EBPα molecules within a dimer move further apart, as demonstrated by fluorescence resonance energy transfer (FRET). Finally, we present evidence that phosphorylation of C/EBPα inhibits granulopoiesis. Our work elucidates a molecular mechanism that inhibits the activity of C/EBPα in the context of myeloid differentiation and may therefore have implications for myeloid leukemias.
MATERIALS AND METHODS
Plasmid vectors.
Wild-type p42C/EBPα is contained within plasmid SER28 (37). S21A, S21D, and F31A mutations were created by site-directed mutagenesis (Stratagene) and are contained in plasmids SER48, SER50, and F31A, respectively. Construction of ΔC/EBPα (in which amino acids 137 to 230 are deleted) was done as previously described (in reference 5, where it is called CR1/2). For glutathione S-transferase (GST) fusion proteins, primers were designed with engineered BamHI and XhoI restriction sites to amplify coding regions for amino acids 1 to 139 of wild-type C/EBPα and S21A, S21D, and F31A mutant C/EBPα proteins from parent plasmids SER28, SER48, SER50, and F31A, respectively. PCR products were cloned into pCR4-TOPO (Invitrogen). Clones were digested with BamHI and XhoI, and isolated DNA was subcloned into pGEX-5X-1 to produce N-terminal GST-C/EBPα expression vectors. Fusions of C/EBPα with cyan fluorescent protein (CFP) or yellow fluorescent protein (YFP) were as follows. Wild-type C/EBPα and S21A and S21D mutant C/EBPα were cloned in frame into the pECFP-C1, pECFP-N1, pEYFP-C1, and pEYFP-N1 expression vectors (Clontech) to create the CFP-C/EBPα, C/EBPα-CFP, YFP-C/EBPα, and C/EBPα-YFP expression vectors, as well as comparable fusion proteins containing C/EBPα mutation S21A or S21D. To create fusion proteins with C/EBPα and the ligand-binding domain of the estrogen receptor (ER), C/EBPα cDNAs were amplified from SER28, SER48, and SER50 by PCR with primers with engineered BamHI sites, which removed the stop codon. After subcloning into vector pGEM-T (Promega), 1.1-kb BamHI fragments were cloned in frame with the ligand-binding domain of pBabe-ERpuro (50, 55), generating pC/EBPα-ER, pC/EBPα(S21A)-ER, and pC/EBPα(S21D)-ER, respectively.
Cell culture.
Mouse 3T3-L1 preadipocytes (12), human embryonic kidney 293T cells, and CV-1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% calf serum. 3T3-L1 cells were induced to differentiate as previously described (15). Where indicated, cells were serum starved in DMEM containing 1% calf serum and 1% bovine serum albumin for 12 h prior to treatment. The human myeloid U937 (American Type Culture Collection; CRL 1593) and erythroleukemic K562 (American Type Culture Collection; CCL 246) cell lines were grown in RPMI 1640 medium (Invitrogen) containing 10% fetal bovine serum (FBS; HyClone) and 2 mM l-glutamine. For deprivation experiments, U937 cells were washed twice in RPMI medium without serum and cultured in phenol red-free RPMI 1640 medium (Invitrogen) supplemented with 10% charcoal-dextran-treated FBS (HyClone or Gemini BioProducts) and 0.5 μg of puromycin (Sigma) per ml. Platelet-derived growth factor (PDGF) was from Intergen, U0126 was from Promega, PD98059 was from Cell Signaling, and β-estradiol was from Sigma.
In vivo phosphorylation and protein purification.
His-tagged C/EBPα was transfected into 293T cells, labeled in vivo, and purified as previously described (37). Briefly, transfected cells were incubated for 3 h in phosphate-free DMEM supplemented with 32Pi and were then lysed in a denaturing urea buffer. His-tagged C/EBPα was purified on nickel resin, concentrated by centrifugation, and separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Gels were zinc stained, and His-tagged C/EBPα was excised. Samples were sent to the Protein Structure Core Facility at the University of Nebraska Medical Center for analyses. In-gel cleavage was performed with trypsin. Peptides were separated on a Vydac C18 column and sequenced with an ABI 477 protein sequencer as described previously (51).
Stable transfection.
For K562 and U937 cells, a total of 107 cells were electroporated in a Gene Pulser (Bio-Rad) with 5 μg of ScaI-linearized plasmid at 220 V and 960 μF in 0.4-cm cuvettes (Molecular BioProducts). Alternatively, U937 cells were nucleofected by the Amaxa system with reagent R and program T-20. Cells were plated on 96-well plates at 105 per well in phenol red-free RPMI medium-10% charcoal-stripped FBS, and selection with 0.5 μg of puromycin per ml was initiated 48 h after transfection.
Retroviral infection.
Ectopic expression of C/EBPα was by retroviral infection as previously described (5). Briefly, 293T cells were transfected with C/EBPα expression vectors and viral packaging vectors. Virus-containing media were collected and applied to 3T3-L1 cells, and infected cells were selected with Geneticin (Invitrogen) or puromycin as indicated.
Protein extracts and separation.
Nuclei were purified from 3T3-L1 preadipocytes or white adipose tissue derived from the epididymal fat pads of C57BL/6 mice by Dounce homogenization in a hypotonic lysis buffer and centrifugation as previously described (37). Purified nuclei were lysed in isoelectric focusing buffer or SDS-PAGE buffer (37). For isoelectric focusing, polyacrylamide-urea minigels were made in accordance with the manufacturer's (Bio-Rad) instructions with a 1:1 mixture of pH 3 to 10 and 5 to 8 ampholytes (Bio-Rad). Protein samples (approximately 20 μg) were loaded and focused for 15 min at 100 V, for 15 min at 200 V, and then for 1 h at 450 V. For SDS-PAGE, nuclear or whole-cell lysates were separated on 11.5% gels. Proteins separated by isoelectric focusing or SDS-PAGE were then transferred onto PVDF-plus membrane for immunoblot analyses.
For K562 and U937 cells, 5 × 106 cells were harvested by centrifugation and washed in 100 μl of phosphate-buffered saline (PBS). Cell pellets were resuspended in PBS, and cells were lysed by addition of an equal volume of 2× Laemmli sample buffer (without bromophenol blue [18]) and boiling for 10 min. Mouse tissues were finely chopped with a razor blade, suspended in a small volume of PBS, and lysed in Laemmli buffer as described above. Protein concentrations were determined with protein assay solution (Bio-Rad). For immunoblot analysis, protein extracts (60 to 80 μg) were electrophoresed on SDS-7.5% discontinuous polyacrylamide gels (18).
Immunoblot analysis.
Immunoblotting was performed essentially as described previously (15), with the following antibodies: a polyclonal C/EBPα antibody raised against a synthetic peptide (amino acids 253 to 265; reference 21), ERK1/2 antibody (Promega), phospho-ERK1/2 antibody (Cell Signaling Technology Inc.), antibody against phosphoserine 21 of C/EBPα (gift from Cell Signaling Technology Inc.), rabbit polyclonal anti-C/EBPα antibody (sc-61; Santa Cruz Biotechnology), goat anti-C/EBPα antibody (sc-9314; Santa Cruz Biotechnology), and mouse anti-β-tubulin monoclonal antibody. Equivalent loading of lanes was determined by Ponceau S staining.
In vitro kinase assay.
GST fusion proteins were grown in 1 liter of BL-21 cells stimulated with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h. Bacteria were then centrifuged at 5,000 rpm (Eppendorf 5415C centrifuge) for 10 min, resuspended in 30 ml of cell lytic B reagent (Sigma), and lysed by sonication. Following centrifugation at 10,000 rpm (Eppendorf 5415C centrifuge) for 10 min, supernatant was incubated with 0.5 ml of glutathione agarose beads (Pharmacia) for 1 h at 4°C. Beads were pelleted and washed four times with lysis buffer. Next, beads were resuspended 1:1 in storage buffer (20 mM HEPES [pH 7.4], 50% glycerol, 150 mM NaCl). For in vitro kinase assays, 5 μg of GST fusion protein was incubated with 25 U of recombinant ERK2 (New England Biolabs) in accordance with the manufacturer's instructions. Reaction products were separated by SDS-PAGE and analyzed by autoradiography.
Immunofluorescent labeling.
Cytospun cells were fixed with Histochoice tissue fixative (Sigma) for 10 min. Preparations were washed twice with PBS, permeabilized in 0.1% Triton X-100 in PBS (5 min), washed in PBS, and blocked in 5% milk-TBST for 1 h at room temperature. Slides were stained with anti-C/EBPα antibody (Santa Cruz) diluted 1:100 in 5% milk-TBST with 10% normal goat serum (Sigma), incubated for 1 h at room temperature, and washed twice in PBS. Following a 1-h incubation at room temperature with anti-rabbit-Alexa 488 conjugate (Molecular Probes) diluted 1:400 in 5% milk-TBST, slides were washed three times in PBS and rinsed in water. Preparations were mounted in ProlongFade (Molecular Probes), and cells were visualized by fluorescence microscopy.
Nitroblue tetrazolium reduction assay.
Reduction of nitroblue tetrazolium by respiratory burst products was assayed with nitroblue tetrazolium tablets (Sigma) in accordance with the manufacturer's protocols. Cells were cytospun and counterstained with safranin.
RNA isolation and Northern blot analysis.
Total RNA was isolated with TriReagent (Molecular Research Center). RNA (20 μg) was denatured in formamide and fractionated on 1% agarose-2.2 M formaldehyde gels (38). RNA was transferred to Biotrans Plus (ICN) membrane in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and blots were hybridized at 40°C in 50% formamide-7.5× Denhardt's solution-5× SSC-50 mM NaPO4 (pH 6.8)-100 μg of salmon sperm DNA per ml-0.5% SDS. DNA fragments were labeled by random priming (7). Blots were washed in 1× SSC-0.5% SDS for 15 min at room temperature, in 1× SSC-0.5% SDS for 15 min at 42°C, and then in 0.1× SSC-1% SDS for 30 min at 65°C. A fragment encompassing bp 289 to 1024 of the human granulocyte colony-stimulating factor (G-CSF) receptor cDNA (10, 42) served as a probe for G-CSF receptor mRNA. The human C/EBPɛ probe was a 0.5-kb PstI fragment of the pJurkat1 clone (1). To ensure uniform levels and integrity of RNA samples loaded in each lane, the blot was stripped and rehybridized with a 1.3-kb PstI fragment of rat glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (9). After autoradiography, relative mRNA levels were quantified with a PhosphorImager and software from Molecular Dynamics.
EMSAs.
Nuclear extracts were prepared, and electrophoretic mobility shift assays (EMSAs) were performed as previously described (3, 35). Briefly, 5 × 107 cells were washed once in PBS, resuspended in an equal volume of ice-cold hypotonic buffer, and incubated on ice for 15 min. Cells were then lysed by five passages through a 26-gauge needle and nuclei were isolated by a 20-s centrifugation at 14,000 rpm (Eppendorf 5415C centrifuge). Nuclear proteins were extracted with a high-salt buffer, and total protein was assayed with the Bio-Rad kit. Complementary oligonucleotides were annealed and labeled at the 5′ ends with [32P]ATP (6,000 Ci/mmol; Amersham, Arlington Heights, Ill.) and T4 polynucleotide kinase (NEB, Beverly, Mass.) and separated from unincorporated nucleotides by passage through a Sephadex G-25 column. EMSAs were performed by incubating 10 μg of nuclear extracts with 50,000 cpm of double-stranded oligonucleotide in a 20-μl reaction mixture. For the supershift assay, 1 μl of polyclonal anti-C/EBPα antibody (sc-61X; Santa Cruz) was added to the binding reaction mixture. Binding reaction products were resolved on a 4% nondenaturing polyacrylamide gel. Oligonucleotides used in EMSAs were derived from the human G-CSF receptor promoter (bp −57 to −38).
FRET measurement of C/EBPα dimer conformation in 3T3-L1 preadipocytes.
Fluorescent C/EBPα fusion proteins were expressed individually or coexpressed with C/EBPα expression vectors tagged with the complementary fluorophore, as indicated. 3T3-L1 cells were transfected with Fugene 6 (Roche Diagnostics), grown for 2 days, and then treated with vehicle (dimethyl sulfoxide) or 10 nM phorbol 12-myristate 13-acetate (TPA; Sigma). Cells were imaged with quantitative fluorescence imaging detection equipment as described previously (39, 56). All images were collected with an Olympus 60× Plan Apochromat objective 20 to 35 min after treatment with TPA. For each cell, three fluorescence channels were collected. The donor channel consisted of CFP excited with 431- to 440-nm light and CFP fluorescence collected at 455 to 485 nm, the acceptor channel consisted of YFP excited with 496- to 505-nm light and YFP fluorescence collected at 520 to 550 nm, and the FRET channel consisted of CFP excited with 431- to 440-nm light and YFP fluorescence collected at 520 to 550 nm.
Calculation of FRET relied on correction for background fluorescence and the relative contributions of CFP and YFP fusion proteins themselves to each of the donor, acceptor, and FRET fluorescence channels for each cell. With corrected fluorescence values, we then examined whether amounts of cyan (donor) fluorescence decreased concomitant with an increase in FRET as energy was transferred from CFP to YFP (39, 56). This was accomplished by determining whether the ratio of corrected fluorescence in the FRET and donor channels (FRET/donor) was increased when CFP- and YFP-tagged C/EBPα proteins were coexpressed compared to the ratio expected from independently expressed CFP- and YFP-tagged C/EBPα proteins. This procedure is described in brief below, but details have been provided previously (39, 56).
Control measurements from cells expressing only CFP-C/EBPα, C/EBPα-CFP, YFP-C/EBPα, or C/EBPα-YFP established the extents to which each CFP or YFP fusion protein independently contributed to the other fluorescence channels. Fluorescence levels in each channel were quantified for each cell following subtraction of the background levels of fluorescence detected within adjacent regions of the same images where the fluorescent fusion proteins were not expressed. Image collection and data analysis were conducted with Metamorph imaging software (Universal Imaging Corporation). Compared to the signal in the donor channel, contributions of the donors, CFP-C/EBPα (−0.02% ± 0.40%; n = 243 cells) and C/EBPα-CFP (0.02% ± 0.43%; n = 296 cells), to the acceptor channel were insignificant, whereas their contributions to the FRET channel were 54.54% ± 1.33% and 54.62% ± 1.21%, respectively. The acceptors, YFP-C/EBPα and C/EBPα-YFP, contributed 0.06% ± 0.37% (n = 201 cells) and 0.00% ± 0.34% (n = 234 cells) to the donor channel and 13.88% ± 0.73% and 14.22% ± 0.75% of that collected in the acceptor channel, respectively. These values are constants reflecting the physical properties of the fluorophores themselves and the collection properties of the detection equipment.
To detect energy transfer from donor CFP to acceptor YFP in cells coexpressing CFP- and YFP-tagged C/EBPα, contributions of YFP-tagged C/EBPα to the FRET and donor channels were subtracted with the constants defined above (39, 56). Similarly, the negligible amount of the donor contribution to the acceptor channel was subtracted. The remaining signal in the donor and FRET channels was the contribution of CFP to the donor and FRET channels, as well as the contributions to the FRET channel of energy transfer from CFP to YFP. If there is energy transfer from CFP to YFP, amounts of fluorescence in the donor channel decrease and amounts of fluorescence in the FRET channel increase. Thus, after correction for the background and bleedthrough, FRET is detected as an increase in FRET/donor fluorescence above the 54.57% contributed by CFP fusion proteins alone.
The amount by which FRET/donor fluorescence increases is dependent on the relative expression of the acceptor and donor fluorophores in the cell (39, 56). As acceptor levels increase, opportunities for interaction with a donor-containing partner increase and the FRET/donor ratio increases. At acceptor/donor levels substantially below those in which all donors are saturated by interactions with acceptors, the slope of FRET/donor versus acceptor/donor graphs represents the extent to which energy transfer, normalized for the relative acceptor and donor amounts within the cell, occurs under different conditions. The extent of FRET is affected by the distance between fluorophores; by the orientation of the fluorophores, which do not transfer energy symmetrically; and by the interaction kinetics of target proteins (39).
In the present studies, FRET for each combination of wild-type C/EBPα and S21A and S21D mutant C/EBPα labeled with CFP and YFP at the amino termini was calculated from 99, 104, and 154 cells measured after vehicle addition and 111, 103, and 132 cells after incubation with TPA. The amounts of FRET from a C-terminal C/EBPα-CFP to an N-terminal YFP-C/EBPα were calculated from 119 wild-type, 116 S21A mutant, and 249 S21D mutant cells incubated with the vehicle and from 120 wild-type, 119 S21A mutant, and 248 S21D mutant cells incubated with TPA. Extents of FRET between CFP and YFP fused at the carboxy termini were calculated from 116 wild-type, 115 S21A mutant, and 153 S21D mutant cells incubated with vehicle and from 118 wild-type, 106 S21A mutant, and 155 S21D mutant cells incubated with TPA.
RESULTS
Phosphorylation of C/EBPα on serine 21.
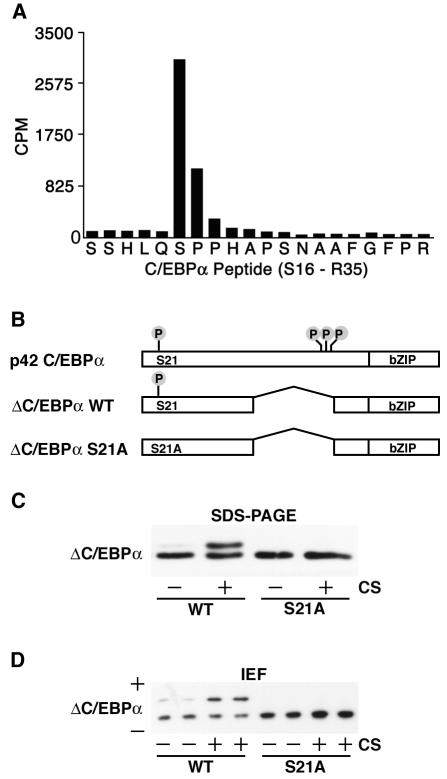
We previously reported that C/EBPα is a phosphoprotein and identified three phosphorylation sites (T222, T226, and S230), two of which (T222 and T226) are phosphorylated by glycogen synthase kinase 3 (15, 23, 37). However, isoelectric focusing of C/EBPα suggested the existence of at least one additional site. To identify this site, we metabolically labeled C/EBPα in vivo, purified the protein, and digested it into peptides by treatment with trypsin. 32P-labeled peptides were sequenced by Edman degradation, and the radioactivity associated with each cycle was quantified. By this approach, we identified the C/EBPα peptide spanning serine 16 through arginine 35 as a phosphopeptide in three independent experiments. The dramatic release of radioactivity with cleavage of serine 21 during cycle 6 (Fig. 1A) identified this site as a phosphoamino acid. Sequence analysis of C/EBPα revealed that serine 21 and its surrounding sequence are highly conserved among mammals (data not shown).
FIG. 1.
Phosphorylation of C/EBPα on serine 21. (A) In vivo labeling of C/EBPα was performed to identify phosphorylated residues. 32P-labeled C/EBPα was purified, cleaved with trypsin, and separated by high-performance liquid chromatography. Fraction 87 was found to contain radioactivity, and sequencing revealed it to be amino acids S16 to R35 of C/EBPα (x axis). The radioactivity associated with each amino acid was quantified (y axis; counts per minute [CPM]). The majority of the radioactivity was found in the sixth cycle, corresponding to serine 21. (B) Schematic representation of C/EBPα proteins engineered for further experiments. Four known phosphoamino acids, S21 (identified herein), T222, T226, and S230 (previously identified [37]) are illustrated. WT, wild type. (C) 3T3-L1 cells were infected with retroviruses carrying the genes for ΔC/EBPα and ΔC/EBPα-S21A. Cells were not treated (−) or treated (+) with 10% calf serum (CS) for 10 min prior to lysis and purification of nuclear proteins. Nuclear lysates were separated by SDS-PAGE and analyzed for C/EBPα by immunoblotting. (D) Samples from panel C were separated by isoelectric focusing (IEF) and analyzed for C/EBPα by immunoblotting. Acidic (+) and basic (−) ends are indicated. Similar results were obtained in at least three independent experiments.
The presence of multiple phosphorylation sites on C/EBPα makes it difficult to monitor the phosphorylation of a particular amino acid by mobility shift upon SDS-PAGE (15, 23, 37). Therefore, to study covalent modification of serine 21, we engineered a C/EBPα protein with an internal deletion of amino acids 137 to 230 (ΔC/EBPα). This protein does not contain phosphoamino acids T222, T226, and S230, and differences in the mobility of ΔC/EBPα are easier to resolve by SDS-PAGE because of its smaller size. To serve as a control for subsequent experiments, we engineered a mutation of the phosphoacceptor residue such that serine 21 is converted to alanine. These expression constructs are illustrated in Fig. 1B.
To confirm that serine 21 is a phosphoamino acid and to investigate the regulation of this site, ΔC/EBPα or ΔC/EBPα-S21A was introduced into 3T3-L1 preadipocytes with a retroviral delivery system. Following selection, serum-deprived cells were left untreated (−) or were treated with 10% calf serum (+) for 10 min. Proteins were separated by SDS-PAGE (Fig. 1C) or isoelectric focusing (Fig. 1D) and analyzed by immunoblot assay for C/EBPα. In the absence of treatment, ΔC/EBPα-S21A and the majority of ΔC/EBPα migrated with a single mobility, as assessed by SDS-PAGE or isoelectric focusing. Treatment of ΔC/EBPα-expressing cells with calf serum resulted in the appearance of a second species with decreased mobility upon SDS-PAGE and increased acidity upon isoelectric focusing, indicative of phosphorylation. In sharp contrast, migration of ΔC/EBPα-S21A was not affected by treatment with calf serum. To confirm that shifts in the mobility of ΔC/EBPα were due to phosphorylation, we treated lysates from calf serum-treated cells with alkaline phosphatase. This treatment resulted in the disappearance of the second, upper band on SDS-PAGE and isoelectric focusing gels (data not shown). These data demonstrate that C/EBPα is phosphorylated on serine 21 in 3T3-L1 cells in response to calf serum.
ERK1/2 phosphorylates C/EBPα on serine 21.
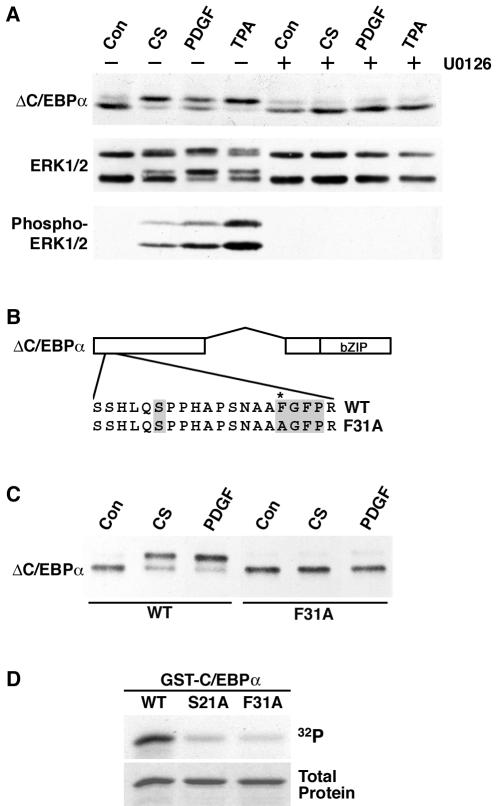
As a first step toward uncovering the mechanism of signaling to C/EBPα, we investigated specific agents that induce serine 21 phosphorylation. We tested a variety of hormones known to be present in calf serum and found that PDGF stimulates serine 21 phosphorylation (Fig. 2A). Further experiments revealed that physiological concentrations of PDGF (50% effective concentration, ∼10 nM) rapidly and transiently stimulate serine 21 phosphorylation, with maximal phosphorylation observed after 20 min (data not shown). Because serine 21 is adjacent to a C-terminal proline, and because PDGF is known to activate ERKs, which are proline-directed kinases, we evaluated whether serine 21 is phosphorylated by ERKs. Treatment of ΔC/EBPα-expressing 3T3-L1 cells with calf serum, PDGF, or the protein kinase C activator TPA stimulates serine 21 phosphorylation, as indicated by a shift in the mobility of ΔC/EBPα (Fig. 2A, top). As expected, these treatments also increased the phosphorylation of ERK1 and ERK2, as assessed by the mobility of ERK1/2 on SDS-PAGE (Fig. 2A, middle) and by immunoblot analysis with antisera against phosphorylated ERK1/2 (Fig. 2A, bottom). Pretreatment of ΔC/EBPα-expressing cells for 30 min with a MEK inhibitor (U0126) not only blocked ERK1/2 activation by calf serum, PDGF, or TPA but also abolished serine 21 phosphorylation (Fig. 2A). In contrast, inhibition of p38 MAP kinase with SB203580 or inhibition of c-Jun N-terminal kinase with SP600125 did not affect the stimulation of serine 21 phosphorylation by these agents (data not shown). These data indicate that activation of ERK1/2 is required for calf serum, PDGF, or TPA to induce phosphorylation of serine 21.
FIG. 2.
ERK1/2 phosphorylates C/EBPα on serine 21. (A) 3T3-L1 cells were infected with a retrovirus carrying the gene for ΔC/EBPα. ΔC/EBPα-expressing cells were pretreated for 15 min in the absence (−) or presence (+) of the MEK inhibitor U0126 (10 μM) prior to treatment with vehicle (Con), 10% calf serum (CS), 25 ng of PDGF per ml, or 100 nM TPA for an additional 10 min. Lysates were separated by SDS-PAGE and subjected to immunoblot analysis for C/EBPα, ERK1/2, or phospho-ERK1/2. ERK1, which migrates with an apparent mobility of 44 kDa, is the upper doublet, and ERK2, which migrates with an apparent mobility of 42 kDa, is the lower. (B) Schematic representation of C/EBPα and a C/EBPα docking site mutant engineered for use in further experiments. The ERK docking site and serine 21 are shaded. The asterisk (*) indicates the phenylalanine that is converted to alanine in the docking site mutant form. WT, wild type. (C) 3T3-L1 cells were infected with retroviruses carrying the genes for wild-type and F31A mutant ΔC/EBPα. Cells were not treated (Con) or treated with 10% calf serum (CS) or 25 ng of PDGF per ml for 10 min. (D) In vitro kinase assays were performed with recombinant ERK2 and GST fusion proteins containing the first 139 amino acids of C/EBPα (WT) or the same region with the indicated point mutations (S21A and F31A). Proteins separated by SDS-PAGE were stained with Coomassie blue (bottom) and subjected to autoradiography (top).
Inspection of the sequence surrounding serine 21 revealed a putative MAP kinase docking site (FXFP; 6) 10 amino acids C terminal to the site of phosphorylation (Fig. 2B). Like serine 21, this binding motif is highly conserved among mammalian C/EBPα proteins (data not shown). We therefore tested the possibility that ERK1/2 interacts with C/EBPα through this site to phosphorylate serine 21. To test this hypothesis, we created a docking site mutant C/EBPα in the context of ΔC/EBPα in which the first phenylalanine in the putative docking site was mutated to an alanine (F31A). 3T3-L1 cells were infected with ΔC/EBPα or ΔC/EBPα-F31A and treated with activators of ERK1/2 prior to immunoblot analysis. As in Fig. 2A, calf serum, PDGF (Fig. 2C), and TPA (data not shown) induced serine 21 phosphorylation in the context of ΔC/EBPα. In contrast, phosphorylation of serine 21 in response to calf serum, PDGF (Fig. 2C), or TPA (data not shown) is dramatically reduced when the ERK docking site is mutated from FGFP to AGFP. These findings suggest that phosphorylation of C/EBPα on S21 by ERK1/2 is dependent on this specific docking sequence.
To ascertain whether ERKs phosphorylate C/EBPα directly, we created a GST-C/EBPα fusion protein (containing amino acids 1 to 139 of C/EBPα) to use as a substrate for in ERK1 in vitro. We also created GST fusion proteins in which the phosphoacceptor residue was mutated to an alanine (S21A) or in which the first phenylalanine of the putative ERK docking site was ablated (F31A). In vitro kinase reactions revealed that wild-type GST-C/EBPα is a substrate for phosphorylation by ERK2 (Fig. 2D), confirming that this kinase directly phosphorylates C/EBPα. However, when either serine 21 or phenylalanine 31 is mutated to alanine, the amount of phosphate incorporated into C/EBPα is greatly reduced. These data indicate that efficient phosphorylation of C/EBPα (amino acids 1 to 139) by ERK2 in vitro requires both the phosphoacceptor residue, serine 21, and an ERK docking motif.
Phosphorylation of serine 21 in vivo.
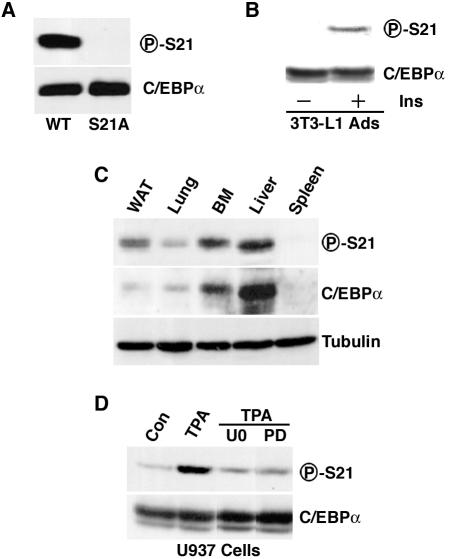
Although our data provide compelling evidence that exogenous C/EBPα is phosphorylated on serine 21 in a number of experimental settings, our experiments did not prove that this modification occurs on endogenous C/EBPα. To address this issue, we initiated development of a phosphospecific antibody that specifically recognizes C/EBPα when phosphorylated on serine 21 (P-S21). To test the specificity of this antibody, CV-1 cells were transiently transfected with expression vectors encoding C/EBPα or C/EBPα-S21A. Lysates were separated by SDS-PAGE and analyzed by immunoblotting with a C/EBPα antibody or the P-S21 antibody. While approximately equivalent amounts of C/EBPα were expressed from these expression vectors, wild-type C/EBPα, but not the S21A mutant form, was recognized by the phosphospecific antibody (Fig. 3A). One of the contexts in which C/EBPα has been most thoroughly studied is the 3T3-L1 adipocyte. Thus, we next investigated whether serine 21 is phosphorylated in this adipocyte model. 3T3-L1 adipocytes were serum deprived and then not stimulated or stimulated with 167 nM insulin for 10 min prior to lysis and separation by SDS-PAGE. Treatment of adipocytes with insulin, a known activator of ERK1/2 in these cells (19), resulted in phosphorylation of C/EBPα on serine 21 (Fig. 3B). As with PDGF, phosphorylation of serine 21 in response to insulin was rapid and transient, with maximal phosphorylation observed 20 min after stimulation (data not shown).
FIG. 3.
Phosphorylation of serine 21 in vivo. (A) CV-1 cells were transfected with an expression vector carrying the gene for either wild-type (WT) or S21A mutant C/EBPα. Lysates were separated by SDS-PAGE and subjected to immunoblot analysis with a phosphospecific antibody designed against serine 21 of C/EBPα (P-S21; top) or total C/EBPα (bottom). (B) After serum deprivation, 3T3-L1 adipocytes (Ads) were not treated (−) or treated (+) with insulin for 10 min. Lysates were separated by SDS-PAGE and subjected to immunoblot analysis for total C/EBPα (bottom) or C/EBPα phosphorylated on serine 21 (top) (C) Lysates from white adipose tissue (WAT), lung, bone marrow (BM), liver, and spleen were separated by SDS-PAGE and subjected to immunoblot analysis with antibody to phosphorylated serine 21 (P-S21) or C/EBPα. (D) U937 cells were washed twice in RPMI medium without serum and suspended in RPMI medium (no serum) at 106/ml. After 2 h at 37°C, cells were treated with 10 μM U0126 (U0) or 50 μM PD98059 (PD) for an additional 2.5 h. TPA was added to a final concentration of 1 μM, and cells were incubated for an additional 30 min. Immunoblot analysis with antisera specific for phosphorylated serine 21 or for C/EBPα was then performed. For all experiments, similar results were obtained in three independent experiments.
We then used the phosphospecific antibody to test whether serine 21 is phosphorylated in tissues. These experiments revealed that C/EBPα is phosphorylated on serine 21 in white adipose tissue, lung, bone marrow, and liver (Fig. 3C). In tissues such as the spleen, where C/EBPα mRNA is not expressed, no C/EBPα protein could be detected, thus demonstrating that both antibodies are specific. Phosphorylation of C/EBPα in bone marrow raised the possibility that modification of serine 21 is regulated within the myeloid lineage. Consistent with this idea, phosphorylation of serine 21 is rapidly induced in U397 myeloid progenitors treated with TPA (Fig. 3D). As expected on the basis of regulation of serine 21 in fibroblasts (Fig. 2A and C), effects of TPA on endogenous C/EBPα phosphorylation are blocked by inhibitors of ERK activation (Fig. 3D). Taken together, these data indicate that C/EBPα is phosphorylated on serine 21 in tissues and cell lines and that phosphorylation of this residue is mediated by ERK1/2.
Phosphorylation of serine 21 alters the conformation of C/EBPα dimers but not binding to DNA.
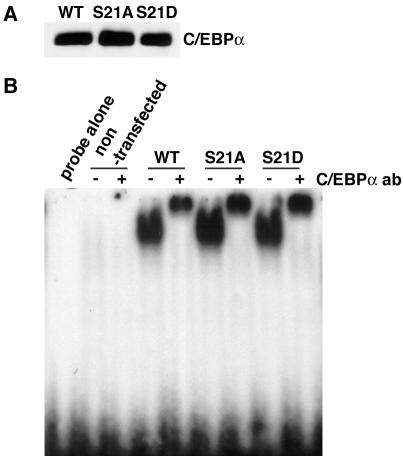
Phosphorylation of transcription factors can influence their activity through many mechanisms, including regulation of binding to DNA. To determine whether serine 21 is involved in the ability of C/EBPα to bind DNA, COS-7 cells were transiently transfected with expression vectors for wild-type C/EBPα, C/EBPα-S21A, or C/EBPα-S21D, and nuclear lysates prepared. Immunoblot analysis revealed that mutant forms were expressed at levels similar to those of wild-type C/EBPα (Fig. 4A). Moreover, EMSA showed that in vitro binding to an oligonucleotide containing the C/EBPα binding site from the human G-CSF receptor promoter was also comparable (Fig. 4B). Thus, serine 21 does not appear to regulate binding to DNA.
FIG. 4.
Mutation of serine 21 does not influence binding to DNA. Expression vectors encoding wild-type (WT) or S21A or S21D mutant C/EBPα were transiently transfected into COS-7 cells, and nuclear lysates were prepared. (A) Immunoblot analysis of wild-type C/EBPα and serine 21 mutant forms. (B) Gel shift analysis with nuclear lysates and a labeled oligonucleotide corresponding to the C/EBPα binding site from the human G-CSF receptor promoter. Supershifting was performed with antibody (ab) to C/EBPα.
The bZIP domain of C/EBPα is predicted, on the basis of X-ray crystallography of other bZIP proteins, to form an alpha-helical coiled coil (26, 40). In contrast, the highly flexible nature of the amino terminus of C/EBPα hinders structural analysis of transactivation domains. One technique for indirectly assessing structural aspects of C/EBPα, including the transactivation domains, is FRET combined with fluorescence microscopy. Transfer of energy from a donor fluorophore (i.e., CFP) to an acceptor fluorophore (i.e., YFP) is detectable if the fluorophores are less than 80 angstroms apart. Since FRET decreases to the sixth power as the distance between fluorophores increases (29, 49), FRET nanoscopy can be used to measure distances between two fluorophore-tagged proteins that are in close proximity. Because of the association of C/EBPα molecules within a dimer, it is therefore possible to measure FRET when a CFP-C/EBPα molecule resonates with a YFP-C/EBPα molecule. Here we used FRET nanoscopy to probe basic structural features of C/EBPα dimers in living cells and to determine how the conformation of dimers is regulated by phosphorylation of serine 21.
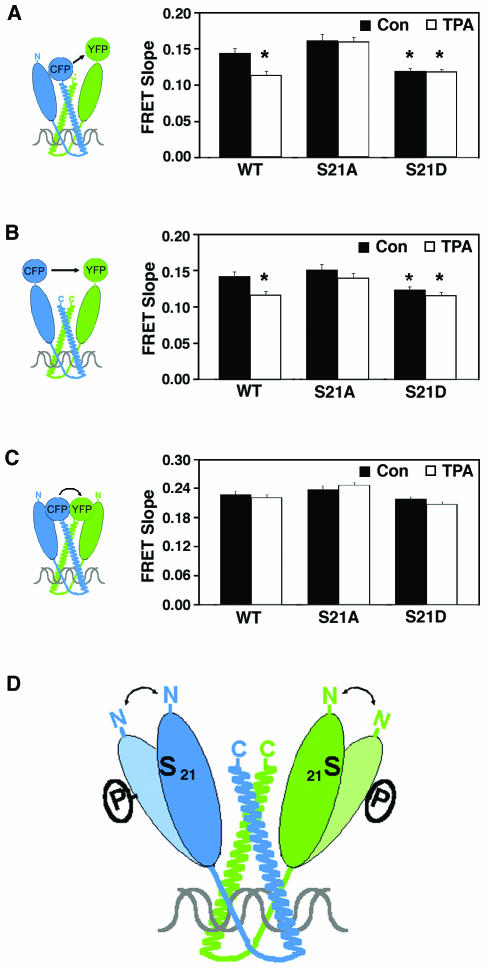
For our experiments, we constructed expression vectors encoding fusion proteins in which CFP or YFP was fused to either wild-type C/EBPα or the C/EBPα mutant forms C/EBPα-S21A and C/EBPα-S21D. In addition, fluorophores were fused to either the amino (N) terminus or the carboxy (C) terminus of C/EBPα, as illustrated in Fig. 5. Various pairwise combinations of C/EBPα-fluorophore fusion proteins were transfected into undifferentiated 3T3-L1 cells. The level of endogenous C/EBPα in undifferentiated 3T3-L1 cells is exceedingly low, such that it would not compete for dimerization between the fluorophore-labeled proteins. Two days after transfection, cells were treated with 10 nM TPA or vehicle for 20 min, and images were collected. Energy transfer from CFP to YFP was measured as described previously (37). No FRET was observed if CFP and YFP were not fused to C/EBPα (37). For FRET determination, each image was corrected for the amounts of background fluorescence and for the contribution of each fluorophore to the donor, acceptor, and FRET channels. Calculations are described in detail in Materials and Methods. Transfection variation resulted in different levels of CFP- and YFP-C/EBPα expressed in each cell. This enabled us to accurately calculate the amount of FRET according to the previously described (37) relationship between the amount of FRET and the relative amounts of the acceptor (YFP) and donor (CFP) fluorophores (see Materials and Methods). Overall, expression of the fluorophore-tagged factors in undifferentiated 3T3-L1 cells was approximately equivalent to the amount of endogenous C/EBPα expressed in 3T3-L1 cells differentiated into adipocytes (37). In addition, the N- or C-terminal fluorescent tags did not influence the stability or DNA binding of C/EBPα, as assessed by immunoblot and gel shift analyses, respectively (data not shown). Finally, fluorophore-tagged C/EBPα, like endogenous C/EBPα, concentrates in the nucleus at pericentromeric AT-rich DNA (39). Whether C/EBPα dimers had one N- and one C-terminal fluorophore (Fig. 5A), two N-terminal fluorophores (Fig. 5B), or two C-terminal fluorophores (Fig. 5C), the fluorophores are close enough to transfer energy. These data are consistent with the idea that C/EBPα bends such that the N and C termini are in close proximity, as illustrated in Fig. 5.
FIG. 5.
Phosphorylation of serine 21 alters the conformation of C/EBPα dimers. Expression vectors encoding wild-type (WT) or S21A or S21D mutant C/EBPα fused at the N or C terminus with CFP or YFP were constructed. Various pairwise combinations of C/EBPα-fluorophore fusion proteins were transfected into 3T3-L1 cells. Differences in FRET relative to wild-type C/EBPα in untreated cells are indicated by an asterisk (P < 0.05). (A) CFP at the C terminus and YFP at the N terminus of wild-type or S21A or S21D mutant C/EBPα. Con, control. (B) CFP and YFP at the N terminus of wild-type or S21A or S21D mutant C/EBPα. (C) CFP and YFP at the C terminus of wild-type or S21A or S21D mutant C/EBPα. Two days after transfection, cells were treated with 10 nM TPA or vehicle for 20 min and images were collected. Analysis of FRET between CFP and YFP is described in Materials and Methods. Effect of TPA on energy transfer between fluorophores (FRET slope) is represented graphically for each C/EBPα dimer combination. (D) Model illustrating that C/EBPα bends such that the N and C termini are in close proximity and that when phosphorylated at serine 21, the transactivation domain of C/EBPα moves away from the N and C termini of the other molecule in the dimer.
Here we found that treatment of cells with TPA for 20 min caused a statistically significant decrease in FRET when C/EBPα dimers were tagged at the N and C termini (Fig. 5), consistent with the idea that TPA triggers a dynamic conformational change in C/EBPα and increased distance between fluorophores. This effect was not observed when cells expressing fluorophore-tagged C/EBPα-S21A were treated with TPA (Fig. 5A). The finding that FRET associated with C/EBPα-S21A was not affected by TPA and was similar to untreated, wild-type C/EBPα suggests that phosphorylation of S21 underlies the conformational change. In further support of this conclusion, FRET associated with expression of C/EBPα-S21D was also not affected by TPA treatment. However, in this case, the extent of FRET was significantly less than that measured from untreated cells expressing wild-type C/EBPα. Indeed, whether cells expressing C/EBPα-S21D were treated with TPA or not, the extent of FRET was very similar to that observed from wild-type C/EBPα treated with TPA. Taken together, these observations suggest that TPA-induced phosphorylation of C/EBPα causes an increase in the distance between the N and C termini of the molecules in a dimer. Furthermore, our data suggest that the C/EBPα-S21A mutant mimics the dephosphorylated form of C/EBPα, whereas the C/EBPα-S21D mutant mimics the phosphorylated form.
Virtually identical results were obtained when the same experiment was performed with C/EBPα fusion proteins in which fluorophores were placed on the N terminus (Fig. 5B). These findings suggest that phosphorylation of serine 21 causes a conformational change in C/EBPα that not only moves the N terminus away from the C-terminal end of the heterodimeric partner (Fig. 5A) but which also moves the N termini apart. In sharp contrast, when both fluorophores were attached to the bZIP domain at the C terminus of C/EBPα, no differences in FRET were observed under any condition (Fig. 5C). These observations suggest that interactions between the bZIP domains are similar irrespective of the charge at amino acid 21. This finding is important because it indicates that phosphorylation of C/EBPα has no effect on the stability of dimer formation; rather, it is specific to the conformation of C/EBPα dimers.
Phosphorylation of C/EBPα on serine 21 inhibits granulopoiesis.
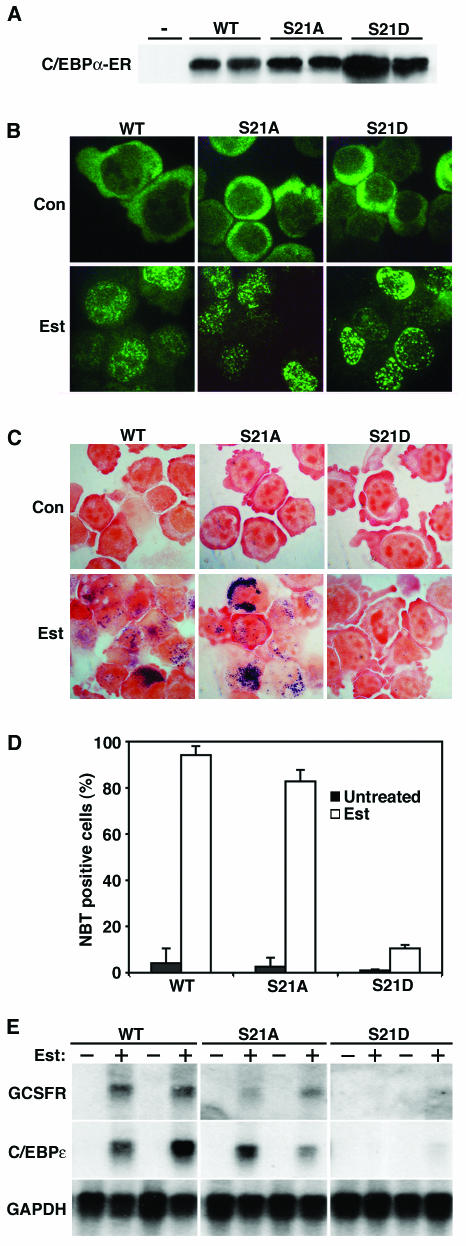
Given that phosphorylation of serine 21 alters the conformation of C/EBPα dimers, we hypothesized that this modification might alter the function of C/EBPα. We and others (28, 34, 55) previously showed that ectopic expression of C/EBPα directs granulocytic differentiation. To determine whether phosphorylation of serine 21 regulates the ability of C/EBPα to mediate this process, we expressed either wild-type C/EBPα or phosphorylation mutant forms in K562 cells. These leukemia-derived cells were chosen because they do not express endogenous C/EBPα and they undergo rapid granulocytic differentiation in response to ectopic C/EBPα (H. S. Radomska, P. Zhang, and D. G. Tenen, unpublished data). For these experiments, the ligand-binding domain of ER was fused to C/EBPα to create a protein that translocates to the nucleus (where it is active) upon addition of β-estradiol. Several independent K562 clonal lines were created for each C/EBPα-ER expression vector, and these showed similar levels of ectopic protein expression (Fig. 6A). Control experiments revealed that while C/EBPα-ER proteins are almost exclusively cytosolic in the basal state, they translocate to the cell nucleus within 1 h of β-estradiol treatment (Fig. 6B and data not shown) (27, 55). As expected, β-estradiol treatment of cells expressing C/EBPα-ER induced granulocyte development, as assessed by detection of respiratory burst activity with nitroblue tetrazolium (NBT; Fig. 6C). Quantitation of this assay revealed that approximately 90% of C/EBPα-ER-expressing cells became NBT positive upon β-estradiol treatment (Fig. 6D). This C/EBPα-induced granulopoiesis was accompanied by an increase in the expression of granulocyte-specific genes (e.g., those for the G-CSF receptor and C/EBPɛ; Fig. 6E). When clones expressing C/EBPα-ER-S21A were treated with β-estradiol, they likewise underwent granulopoietic development, with approximately 85% of the cells staining positively for NBT (Fig. 6C and D), suggesting that the dephosphorylated form of C/EBPα is sufficient to mediate granulopoiesis. In marked contrast, when serine 21 was mutated to aspartate (S21D) to mimic the phosphorylated state of C/EBPα, this mutant form was unable to induce granulopoiesis (Fig. 6C and D). In this case, only 10% of the cells were positive upon NBT staining and the granulocyte markers G-CSF receptor and C/EBPɛ were undetectable (Fig. 6E). These data suggest that phosphorylation of C/EBPα on serine 21 drastically reduces the granulocyte-inducing activity of C/EBPα.
FIG. 6.
C/EBPα-S21D is unable to induce granulopoiesis in K562 cells. (A) K562 cells were stably transfected with C/EBPα-ER fusion proteins encoding either wild-type (WT) C/EBPα or a C/EBPα mutant form in which serine 21 was replaced with alanine (S21A) or aspartate (S21D). Lysates from the parental K562 line (−) and two independent stable lines for each C/EBPα-ER fusion protein (wild-type and S21Aand S21D mutant forms, as indicated) were separated by SDS-PAGE and subjected to immunoblot analysis with an antibody against C/EBPα. (B) C/EBPα-ER-expressing stable lines from panel A were left untreated (control [Con]) or treated with 1 μM β-estradiol (Est) for 24 h. Cells were fixed and visualized by immunofluorescence with an antibody to C/EBPα. (C) C/EBPα-ER-expressing stable lines from panel A were left untreated (Con) or treated with 1 μM β-estradiol (Est) for 3 days, at which point cells were analyzed for NBT reduction. (D) Cells were scored for reduction of NBT, and quantified data are presented. (E) C/EBPα-ER-expressing stable lines from panel A were left untreated (−) or treated with 1 μM β-estradiol (+) for 3 days. RNA was collected and analyzed by Northern blotting with probes for the G-CSF receptor (GCSFR), C/EBPɛ, and GAPDH.
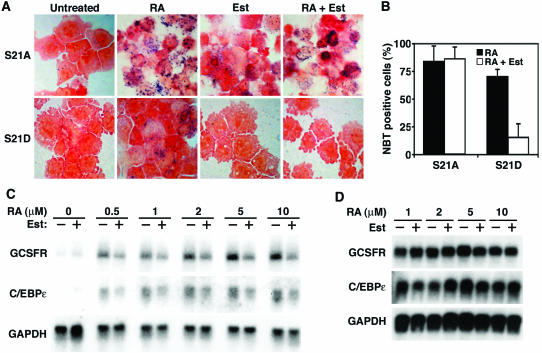
We next addressed the question of whether phosphorylation of C/EBPα is sufficient to block granulocyte differentiation. For these experiments, we used U937 cells, a bipotential myeloid line that differentiates into granulocytes upon treatment with retinoic acid (RA) (25). Endogenous C/EBPα is readily detected in U937 cells and increases upon induction of granulopoiesis. As expected, RA induced granulocyte differentiation in U937 cells expressing either C/EBPα-ER-S21A or C/EBPα-ER-S21D in the absence of β-estradiol (Fig. 7A and B). Whereas approximately 85% of the U937 cells expressing “cytoplasmic” C/EBPα-ER-S21A become NBT positive in response to RA for 48 h, only 70% of the U937 cells expressing cytoplasmic C/EBPα-ER-S21D are positive for NBT with this treatment. It is likely that this slight inhibition of granulocyte differentiation is due to leaky translocation of C/EBPα-ER-S21D from the cytosol to the nucleus, despite the absence of exogenous β-estradiol. Activation of C/EBPα-ER-S21A with β-estradiol in U937 cells induced granulocyte differentiation in the absence of RA, indicating that the dephosphorylated form of C/EBPα stimulates granulopoiesis in both U937 and K562 cells (Fig. 6 and 7A). However, when serine 21 was mutated to aspartate (S21D) to mimic the phosphorylated state of C/EBPα, this mutant form was again unable to induce granulocyte differentiation, as seen in the NBT-stained cells in Fig. 7A. While treatment of C/EBPα-ER-S21A clones with β-estradiol and RA caused granulopoiesis, activation of C/EBPα-ER-S21D with β-estradiol inhibited granulocyte differentiation in response to RA. Counting of NBT-positive cells revealed that approximately 85% of the C/EBPα-ER-S21A-expressing cells were granulocytic in response to β-estradiol and RA, whereas only 15% of the C/EBPα-ER-S21D-expressing cells were granulocytic under these conditions (Fig. 7B). Thus, in the presence of nuclear C/EBPα-ER-S21D, RA is unable to efficiently induce respiratory activity, as shown by NBT reduction assay (Fig. 7A). Moreover, the expression of granulocyte markers, such as the G-CSF receptor and C/EBPɛ, was inhibited approximately twofold (Fig. 7C) in cells that express nuclear C/EBPα-ER-S21D. In contrast, activation of C/EBPα-ER-S21A with estrogen did not inhibit induction of the G-CSF receptor and C/EBPɛ by RA (Fig. 7D). These data suggest that phosphorylation of serine 21 partially represses RA-induced granulocytic differentiation of U937 cells.
FIG. 7.
RA-induced granulopoiesis in U937 cells is partially inhibited by C/EBPα-S21D. (A) U937 cells were stably transfected with expression vectors for C/EBPα-ER fusion proteins in which serine 21 of C/EBPα was replaced with alanine or aspartate. C/EBPα-ER-S21A (S21A) and C/EBPα-ER-S21D (S21D) expressing stable lines were left untreated or treated with 10 μM RA, 1 μM β-estradiol (Est), or 10 μM RA-1 μM β-estradiol (RA + Est). Two days later, cells were analyzed for NBT reduction. (B) Quantification of NBT staining from experiment described in panel A. (C) U937 cells expressing C/EBPα-ER-S21D were treated with RA at the indicated concentrations in the absence (−) or presence (+) of 1 μM β-estradiol. Two days later, RNA was collected and analyzed by Northern blotting with probes for the G-CSF receptor (GCSFR), C/EBPɛ, and GAPDH. (D) U937 cells expressing C/EBPα-ER-S21A were treated with RA at the indicated concentrations in the absence (−) or presence (+) of 1 μM β-estradiol (Est). Two days later, RNA was collected and analyzed by Northern blotting with probes for the G-CSF receptor, C/EBPɛ, and GAPDH.
DISCUSSION
C/EBPα has important roles in the differentiation and metabolism of several cell types, including granulocytes. This transcription factor is expressed in bipotential myeloid cells concomitantly with their commitment to the myeloid lineage. Without it, no mature granulocytes form, whereas its conditional expression induces granulopoiesis and inhibits monocyte differentiation. Given that C/EBPα is expressed prior to granulocyte differentiation, it is likely that extracellular factors influence the timing and cell fate decision between monocyte and granulocyte differentiation in part by regulating C/EBPα activity. Signaling pathways that directly regulate C/EBPα activity, however, are poorly characterized. Here we show that phosphorylation of C/EBPα at serine 21 regulates the activity of C/EBPα in its role as an inducer of granulocyte differentiation. Specifically, only the dephosphorylated form of C/EBPα is able to induce granulopoiesis. Indeed, C/EBPα-S21D partially inhibits RA-induced granulopoiesis, suggesting that the phosphorylated form of C/EBPα may actively repress granulocyte differentiation. Consistent with this function, when U937 cells are induced to undergo monocyte differentiation by treatment with TPA, C/EBPα becomes phosphorylated at serine 21 (Fig. 3D) and therefore inactive in its role as an inducer of granulopoiesis. These findings imply that part of the mechanism whereby TPA induces monocyte differentiation of U937 cells is suppression of the alternate cell fate pathway, that of granulocyte differentiation. This study therefore provides a link between the agents that induce differentiation and the mechanism whereby the posttranslational modification of a transcription factor regulates cell fate determination.
While phosphorylation has long been hypothesized to alter the tertiary structure of proteins, this idea has only been proven in a small number of cases. For transcription factors in the bZIP family, structural information has been largely limited to the DNA-binding domain because this region is rigid enough for analysis by X-ray crystallography (26, 40). Herein, we used FRET nanoscopy as a tool with which to investigate the structural parameters of interacting molecules within living cells. Specifically, we probed elements of C/EBPα structure and determined how the conformation of C/EBPα dimers is regulated by phosphorylation. Although it is conceivable that differences in FRET are influenced by differential binding to coactivators or corepressors, or effects on bending of DNA, the interpretation we favor is that the N-terminal (transactivation) domain of C/EBPα changes conformation upon phosphorylation of C/EBPα. Our data are consistent with the idea that phosphorylation of S21 causes the transactivation domains of C/EBPα to move further apart, as illustrated in Fig. 5D. Distances between fluorophores can be calculated from distance sensitivity in the efficiency of energy transfer between fluorophores (39). On the basis of these calculations, the N terminus is calculated to move 3.1 and 4.2 angstroms further from the C terminus upon treatment with TPA or mutation of serine 21 to aspartate, respectively (Fig. 5A). Similarly, effects of TPA and S21D correspond to a 2.6- to 3.5-angstrom change in the position of N-terminal fluorophores, respectively (Fig. 5B). However, FRET would also change if phosphorylation caused rotation of a domain within C/EBPα such that donor and acceptor fluorophores attained a less favorable orientation for FRET. We cannot distinguish the degree to which changes within the C/EBPα domain distance or orientation contribute to the observed differences in FRET. Regardless, the FRET measurements show that the N-terminal domain of C/EBPα, containing serine 21, specifically adopts a different conformation within the C/EBPα dimer upon phosphorylation at serine 21. Although phosphorylation-induced conformational changes are widely assumed to occur, this is, to our knowledge, the first evidence of their detection in a living cell.
In our analysis to determine whether phosphorylation of serine 21 alters the activity of C/EBPα, we performed a variety of experiments in which we compared the activity of C/EBPα to that of those the C/EBPα-S21A and C/EBPα-S21D mutant forms. On the basis of FRET analysis (Fig. 5A and B), it is likely that the S21A and S21D mutant forms mimic the dephosphorylated and phosphorylated states of C/EBPα, respectively. Immunoblot analysis reveals that, whether introduced into cells by transfection or retroviral infection, all three forms of C/EBPα are expressed at similar levels (data not shown). Likewise, mutation of serine 21 has no effect on the ability of C/EBPα to bind DNA, as revealed by gel shift analysis (Fig. 4). In transient-transfection assays, wild-type C/EBPα and the S21A and S21D mutant forms are similar in the ability to transactivate leptin-, PPARγ-, or G-CSF receptor-derived promoters or to repress c-myc- or E2F-dependent reporter genes (data not shown). Similarly, chimeric transcription factors with the N-terminal transactivation domain of wild-type C/EBPα or the S21A or S21D mutant form fused to the DNA-binding domain of Gal4 similarly induced the expression of a reporter gene (data not shown). Finally, we found that wild-type C/EBPα, C/EBPα-S21A, and C/EBPα-S21D are equipotent in the ability to induce the morphological characteristics associated with adipogenesis in both 3T3-L1 cells and C/EBPα−/− mouse embryonic fibroblasts. Thus, our studies raised the possibility that phosphorylation of serine 21 regulates the function of C/EBPα in specific contexts. We favor the idea that this phosphorylation event regulates a subset of binding partners with which C/EBPα interacts. For instance, serine 21 might regulate C/EBPα activity in a tissue-restricted manner through interaction with transcription factors, coactivators, and/or corepressors expressed in certain lineages, such as myeloid cells, but not in others, such as adipocytes. Further experiments are under way to identify putative binding partners whose interaction with C/EBPα is regulated by phosphorylation of serine 21.
Our studies in which ectopic expression of C/EBPα-S21D is shown to partially block RA-induced differentiation of U937 cells (Fig. 7) suggest that phosphorylated C/EBPα acts as a dominant negative protein. Incidentally, it should be noted that C/EBPα-S21D did not block monocyte differentiation (data not shown). Thus, C/EBPα-S21D is not a general suppressor of differentiation but rather specifically inhibits differentiation into granulocytes. We favor the model in which phosphorylated C/EBPα blocks granulopoiesis because it binds to C/EBPα binding sites, thereby blocking the binding of nonphosphorylated C/EBPα, but is inactive in the ability to promote granulopoiesis, perhaps because the phosphorylated form is unable to interact with specific binding partners that are necessary for granulocyte differentiation. Further studies are under way to substantiate this model.
Numerous studies suggest that dysregulation of C/EBPα is central in the development of myeloid leukemias, such as AML and chronic myeloid leukemia (CML) (11, 28, 33). For instance, AML is frequently caused by a translocation that gives rise to a fusion protein of RUNX1 and CBF2T1 (also known as AML-ETO). Patients with AML caused by this translocation have reduced levels of C/EBPα mRNA, and it is thought that absence of C/EBPα in these patients is an underlying cause of the leukemia that develops (27). Analogously, progression of CML to the stage of blast crisis is triggered by a translocation resulting in the BCR-ABL fusion protein. Recently, it has been shown that expression of BCR-ABL results in loss of C/EBPα protein by inhibiting the translation of C/EBPα mRNA (31). A direct causal connection between myeloid leukemia and C/EBPα was uncovered when it was found that approximately 17% of AML patients with a normal karyotype have mutations within the C/EBPA gene. These mutations result in the translation of a mutant C/EBPα protein with dominant negative properties that inhibits the activity of endogenous C/EBPα. Μutations identified in other AML patients in the bZIP domain of C/EBPα interfere with DNA binding. Thus, multiple mechanisms that result in inhibition of C/EBPα protein function contribute to myeloid leukemias (44).
Here we show that ERK1/2 activity inhibits C/EBPα function through phosphorylation of serine 21. About 30% of AML patients carry activating mutations in the FLT3 receptor tyrosine kinase gene, which results in constitutive activation of the downstream ERK1/2 pathway. Indeed, in the course of the present work, we found that in this subtype of AML, C/EBPα is hyperphosphorylated on serine 21, which contributes to the differentiation block of the leukemic blasts (H. S. Radomska et al., unpublished data). Our studies therefore raise the possibility that inhibition of ERK1/2 in myeloid progenitors, which would allow dephosphorylation and thereby activation of C/EBPα, may help promote differentiation from a proliferative myeloid blast to a terminally differentiated granulocyte. Thus, a mechanistic understanding of C/EBPα function may help direct future research aimed at developing therapies for the treatment of myeloid leukemias.
Acknowledgments
We are grateful to Lorean Serra, Lisa M. Johansen, and Erica K. Evans for efforts on this project. We also thank John Blenis and his laboratory members for identification of the ERK docking site and for helpful suggestions.
This research was supported by grants to O.A.M. from the American Diabetes Association and from the National Institutes of Health (DK51563), to F.S. from the National Institutes of Health (DK54345) and the UCSF Diabetes Center Pilot Project Program, to D.G.T. from the National Institutes of Health (CA72009), and to H.S.R. from the National Institutes of Health (DK62064).
REFERENCES
- 1.Antonson, P., B. Stellan, R. Yamanaka, and K. G. Xanthopoulos. 1996. A novel human CCAAT/enhancer binding protein gene, C/EBPepsilon, is expressed in cells of lymphoid and myeloid lineages and is localized on chromosome 14q11.2 close to the T-cell receptor locus. Genomics 35:30-38. [DOI] [PubMed] [Google Scholar]
- 2.Chen, P. L., D. J. Riley, Y. Chen, and W. H. Lee. 1996. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 10:2794-2804. [DOI] [PubMed] [Google Scholar]
- 3.D'Alo, F., L. M. Johansen, E. A. Nelson, H. S. Radomska, E. K. Evans, P. Zhang, C. Nerlov, and D. G. Tenen. 2003. The amino terminal and E2F interaction domains are critical for C/EBPα-mediated induction of granulopoietic development of hematopoietic cells. Blood 102:1363-1371. [DOI] [PubMed]
- 4.Darlington, G. J., S. E. Ross, and O. A. MacDougald. 1998. The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 273:30057-30060. [DOI] [PubMed] [Google Scholar]
- 5.Erickson, R. L., N. Hemati, S. E. Ross, and O. A. MacDougald. 2001. p300 coactivates the adipogenic transcription factor CCAAT/enhancer-binding protein alpha. J. Biol. Chem. 276:16348-16355. [DOI] [PubMed] [Google Scholar]
- 6.Fantz, D. A., D. Jacobs, D. Glossip, and K. Kornfeld. 2001. Docking sites on substrate proteins direct extracellular signal-regulated kinase to phosphorylate specific residues. J. Biol. Chem. 276:27256-27265. [DOI] [PubMed] [Google Scholar]
- 7.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 8.Flodby, P., C. Barlow, H. Kylefjord, L. Ahrlund-Richter, and K. G. Xanthopoulos. 1996. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein α. J. Biol. Chem. 271:24753-24760. [DOI] [PubMed] [Google Scholar]
- 9.Fort, P., L. Marty, M. Piecharczyk, S. El Sabouty, C. Dani, P. Jeanteur, and J. M. Blanchard. 1985. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 11:1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukunaga, R., Y. Seto, S. Mizushima, and S. Nagata. 1990. Three different mRNAs encoding human granulocyte colony-stimulating factor receptor. Proc. Natl. Acad. Sci. USA 87:8702-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gombart, A. F., W.-K. Hofmann, S. Kawano, S. Takeuchi, U. Krug, S. H. Kwok, R. J. Larsen, H. Asou, C. W. Miller, D. Hoelzer, and H. P. Koeffler. 2002. Mutations in the gene encoding the transcription factor CCAAT/enhancer binding protein alpha in myelodysplastic syndromes and acute myeloid leukemias. Blood 99:1332-1340. [DOI] [PubMed] [Google Scholar]
- 12.Green, H., and M. Meuth. 1974. An established pre-adipose cell line and its differentiation in culture. Cell 3:127-133. [DOI] [PubMed] [Google Scholar]
- 13.Halmos, B., C. S. Huettner, O. Kocher, K. Ferenczi, D. D. Karp, and D. G. Tenen. 2002. Downregulation and antiproliferative role of C/EBPα in lung cancer. Cancer Res. 62:528-534. [PubMed] [Google Scholar]
- 14.Harris, T. E., J. H. Albrecht, M. Nakanishi, and G. J. Darlington. 2001. CCAAT/enhancer binding protein-alpha cooperates with p21 to inhibit cyclin-dependent kinase-2 activity and induces growth arrest independent of DNA binding. J. Biol. Chem. 276:29200-29209. [DOI] [PubMed] [Google Scholar]
- 15.Hemati, N., S. E. Ross, R. L. Erickson, G. E. Groblewski, and O. A. MacDougald. 1997. Signaling pathways through which insulin regulates CCAAT/enhancer binding protein α (C/EBPα) phosphorylation and gene expression in 3T3-L1 adipocytes: correlation with GLUT4 gene expression. J. Biol. Chem. 41:25913-25919. [DOI] [PubMed] [Google Scholar]
- 16.Johansen, L. M., A. Iwama, T. A. Lodie, K. Sasaki, D. W. Felsher, T. R. Golub, and D. G. Tenen. 2001. c-Myc is a critical target for C/EBPα in granulopoiesis. Mol. Cell. Biol. 21:3789-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, J., C. A. Cantwell, P. F. Johnson, C. M. Pfarr, and S. C. Williams. 2002. Transcriptional activity of CCAAT/enhancer-binding proteins is controlled by a conserved inhibitory domain that is a target for sumoylation. J. Biol. Chem. 277:38037-38044. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Lazar, D. F., R. J. Wiese, M. J. Brady, C. C. Mastick, S. B. Waters, K. Yamauchi, J. E. Pessin, P. Cuatrecasas, and A. R. Saltiel. 1995. Mitogen-activated protein kinase kinase inhibition does not block the stimulation of glucose utilization by insulin. J. Biol. Chem. 270:20801-20807. [DOI] [PubMed] [Google Scholar]
- 20.Lekstrom-Himes, J., and K. G. Xanthopoulos. 1998. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 273:28545-28548. [DOI] [PubMed] [Google Scholar]
- 21.Lin, F.-T., O. A. MacDougald, A. M. Diehl, and M. D. Lane. 1993. A 30 kilodalton alternative translation product of the CCAAT/enhancer binding protein α message: transcriptional activator lacking antimitotic activity. Proc. Natl. Acad. Sci. USA 90:9606-9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linhart, H. G., K. Ishimura-Oka, F. DeMayo, T. Kibe, D. Repka, B. Poindexter, R. J. Bick, and G. J. Darlington. 2001. C/EBPα is required for differentiation of white, but not brown, adipose tissue. Proc. Natl. Acad. Sci. USA 98:12532-12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacDougald, O. A., P. Cornelius, R. Liu, and M. D. Lane. 1995. Insulin regulates transcription of the CCAAT/enhancer binding protein (C/EBP) α, β, and δ genes in fully-differentiated 3T3-L1 adipocytes. J. Biol. Chem. 270:647-654. [DOI] [PubMed] [Google Scholar]
- 24.Nerlov, C., and E. B. Ziff. 1995. CCAAT/enhancer binding protein-α amino acid motifs with dual TBP and TFIIB binding ability co-operate to activate transcription in both yeast and mammalian cells. EMBO J. 14:4318-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oberg, F., J. Botling, and K. Nilsson. 1993. Functional antagonism between vitamin-D3 and retinoic acid in the regulation of CD14 and CD23 expression during monocytic differentiation of U937 cells. J. Immunol. 150:3487-3495. [PubMed] [Google Scholar]
- 26.O'Shea, E. K., J. D. Klemm, P. S. Kim, and T. Alber. 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded parallel coiled coil. Science 254:539-544. [DOI] [PubMed] [Google Scholar]
- 27.Pabst, T., B. U. Mueller, H. Harikawa, C. Schoch, T. Haferlach, G. Behre, W. Hiddemann, D.-E. Hang, and D. G. Tenen. 2001. AML1-ETO downregulates the granulocytic differentiation factor C/EBPα in t(8;21) myeloid leukemia. Nat. Med. 7:444-451. [DOI] [PubMed] [Google Scholar]
- 28.Pabst, T., B. U. Mueller, P. Zhang, H. S. Radomska, S. Narravula, S. Schnittger, G. Behre, W. Hiddemann, and D. G. Tenen. 2001. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPα), in acute myeloid leukemia. Nat. Genet. 27:263-270. [DOI] [PubMed] [Google Scholar]
- 29.Patterson, G. H., D. W. Piston, and B. G. Barisas. 2000. Forster distances between green fluorescent protein pairs. Ann. Biochem. 284:438-440. [DOI] [PubMed] [Google Scholar]
- 30.Pedersen, T. A., E. Kowenz-Leutz, A. Leutz, and C. Nerlov. 2001. Cooperation between C/EBPα TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 15:3208-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perrotti, D., V. Cesi, R. Trotta, C. Guerzoni, G. Santilli, K. Campbell, A. Iervolino, F. Condorelli, C. Gambacorti-Passerini, M. A. Caligiuri, and B. Calabretta. 2002. BCR-ABL suppresses C/EBPα expression through inhibitory action of hnRNP E2. Nat. Genet. 30:48-58.11753385 [Google Scholar]
- 32.Porse, B. T., T. A. Pedersen, X. Xu, B. Lindberg, U. M. Wewer, L. Friis-Hansen, and C. Nerlov. 2001. E2f repression by C/EBPα is required for adipogenesis and granulopoiesis in vivo. Cell 107:247-258. [DOI] [PubMed] [Google Scholar]
- 33.Preudhomme, C., C. Sagot, N. Boissel, J.-M. Cayuela, I. Tigaud, S. de Botton, X. Thomas, E. Raffoux, C. Lamandin, S. Castaigne, P. Fenaux, and H. Dombret. 2002. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA). Blood 100:2717-2723. [DOI] [PubMed] [Google Scholar]
- 34.Radomska, H. S., C. S. Huettner, P. Zhang, T. Cheng, D. T. Scadden, and D. G. Tenen. 1998. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol. Cell. Biol. 18:4301-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radomska, H. S., C. P. Shen, T. Kadesch, and L. A. Eckhardt. 1994. Constitutively expressed Oct-2 prevents immunoglobulin gene silencing in myeloma × T cell hybrids. Immunity 1:623-634. [DOI] [PubMed] [Google Scholar]
- 36.Rosen, E. D., and B. M. Spiegelman. 2000. Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol. 16:145-171. [DOI] [PubMed] [Google Scholar]
- 37.Ross, S. E., R. L. Erickson, N. Hemati, and O. A. MacDougald. 1999. Glycogen synthase kinase 3 is an insulin-regulated C/EBPα kinase. Mol. Cell. Biol. 19:8433-8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Schaufele, F., X. Wang, X. Liu, and R. N. Day. 2003. Conformation of CCAAT/enhancer binding protein α dimers varies with intranuclear location in living cells. J. Biol. Chem. 278:10578-10587. [DOI] [PubMed] [Google Scholar]
- 40.Schumacher, M. A., R. H. Goodman, and R. G. Brennan. 2000. The structure of a CREB bZIP somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced DNA binding. J. Biol. Chem. 275:35242-35247. [DOI] [PubMed] [Google Scholar]
- 41.Slomiany, B. A., K. L. D'Arigo, M. M. Kelly, and D. T. Kurtz. 2000. C/EBPα inhibits cell growth via direct repression of E2F-DP-mediated transcription. Mol. Cell. Biol. 20:5986-5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, L. T., S. Hohaus, D. A. Gonzalez, S. E. Dziennis, and D. G. Tenen. 1996. PU.1 (Spi-1) and C/EBP regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood 88:1234-1247. [PubMed] [Google Scholar]
- 43.Subramanian, L., M. D. Benson, and J. A. Iniguez-Lluhi. 2003. A synergy control motif within the attenuator domain of C/EBPα inhibits transcriptional synergy through its PIASy-enhanced modification by SUMO-1 or SUMO-3. J. Biol. Chem. 278:9134-9141. [DOI] [PubMed]
- 44.Tenen, D. G. 2003. Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. 3:89-101. [DOI] [PubMed] [Google Scholar]
- 45.Timchenko, N. A., E. H. Thurl, M. Wilde, T. Bilyeu, B. L. Burgess-Beusse, M. J. Finegold, and G. J. Darlington. 1997. CCAAT/enhancer binding protein alpha regulates p21 protein and hepatocyte proliferation in newborn mice. Mol. Cell. Biol. 17:7353-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Timchenko, N. A., M. Wilde, and G. J. Darlington. 1999. C/EBPα regulates formation of S-phase-specific E2F-p107 complexes in livers of newborn mice. Mol. Cell. Biol. 19:2936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Timchenko, N. A., M. Wilde, P. Iakova, J. H. Albrecht, and G. J. Darlington. 1999. E2F/p107 and E2F/p130 complexes are regulated by C/EBPα in 3T3-L1 adipocytes. Nucleic Acids Res. 27:3621-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Timchenko, N. A., M. Wilde, M. Nakanishi, J. R. Smith, and G. J. Darlington. 1996. CCAAT/enhancer-binding protein α (C/EBPα) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SCI-1) protein. Genes Dev. 10:804-815. [DOI] [PubMed] [Google Scholar]
- 49.Tsien, T. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 50.Umek, R. M., A. D. Friedman, and S. L. McKnight. 1991. CCAAT-enhancer binding protein: a component of a differentiation switch. Science 251:288-292. [DOI] [PubMed] [Google Scholar]
- 51.Volle, D. J., J. A. Fulton, O. V. Chaika, K. McDermott, H. Luang, L. A. Steinke, and R. E. Lewis. 1999. Phosphorylation of the kinase suppressor of Ras by associated kinases. Biochemistry 38:5130-5137. [DOI] [PubMed] [Google Scholar]
- 52.Wang, H., T. Goode, P. Iakova, J. H. Albrecht, and N. A. Timchenko. 2002. C/EBPα triggers proteosome-dependent degradation of cdk4 during growth arrest. EMBO J. 21:930-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, H., P. Iakova, M. Wilde, A. Welm, T. Goode, W. J. Roessler, and N. A. Timchenko. 2001. C/EBPα arrests cell proliferation through direct inhibition of cdk2 and cdk4. Mol. Cell 8:817-828. [DOI] [PubMed] [Google Scholar]
- 54.Wang, N. D., M. J. Finegold, A. Bradley, C. N. Ou, S. V. Abdelsayed, M. D. Wilde, L. R. Taylor, D. R. Wilson, and G. J. Darlington. 1995. Impaired energy homeostasis in C/EBPα knockout mice. Science 269:1108-1112. [DOI] [PubMed] [Google Scholar]
- 55.Wang, X., E. Scott, C. L. Sawyers, and A. D. Friedman. 1999. C/EBPα bypasses granulocyte colony-stimulating factor signals to rapidly induce PU.1 gene expression, stimulate granulocyte differentiation, and limit proliferation in 32D cl3 myeloblasts. Blood 94:560-571. [PubMed] [Google Scholar]
- 56.Weatherman, R. V., C. Y. Chang, N. J. Clegg, D. C. Carroll, R. N. Day, J. D. Baxter, D. P. McDonnell, T. S. Scanlan, and F. Schaufele. 2002. Ligand-selective interactions of ER detected in living cells by fluorescence resonance energy transfer. Mol. Endocrinol. 16:487-496. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, D.-E., P. Zhang, N.-D. Wang, C. J. Hetherington, G. J. Darlington, and D. G. Tenen. 1997. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proc. Natl. Acad. Sci. USA 94:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]