Abstract
Cancer stem-like cells (CSCs) are a highly tumorigenic cell type present as a minority population in developmentally diverse tumors and cell lines. Using a genetic screen in an inducible model of CSC formation in a breast cell line, we identify microRNAs (miRNAs) that inhibit CSC growth and are down-regulated in CSCs. Aside from the previously identified miR-200 family, these include the miR-15/16 (miR-16, miR-15b) and miR-103/107 (miR-103, miR-107) families as well as miR-145, miR-335, and miR-128b. Interestingly, these miRNAs affect common target genes that encode the Bmi1 and Suz12 components of the polycomb repressor complexes as well as the DNA-binding transcription factors Zeb1, Zeb2, and Klf4. Conversely, expression of the CSC-modulating miRNAs is inhibited by Zeb1 and Zeb2. There is an inverse relationship between the levels of CSC-regulating miRNAs and their respective targets in samples from triple-negative breast cancer patients, providing evidence for the relevance of these interactions in human cancer. In addition, combinatorial overexpression of these miRNAs progressively attenuates the growth of CSCs derived from triple-negative breast cancers. These observations suggest that CSC formation and function are reinforced by an integrated regulatory circuit of miRNAs, transcription factors, and chromatin-modifying activities that can act as a bistable switch to drive cells into either the CSC or the nonstem state within the population of cancer cells.
Keywords: cellular transformation, transcriptional regulation, Zeb 1 repressors
Cancer stem cells (CSCs; also called tumor-initiating cells) are a highly tumorigenic cell type that exist as a minority population within tumors, and have been hypothesized to be play a pivotal role in cancer (1–6). CSCs from developmentally diverse tumors and established cell lines have been isolated by using cell-surface markers. Operationally, CSCs are commonly defined as being highly tumorigenic upon injection in immunodeficient mice and resistant to standard chemotherapeutic drugs (e.g., doxorubicin). Typically, CSCs can grow as spheres (e.g., mammospheres for breast CSCs) under nondifferentiation conditions, but they can differentiate into nonstem cancer cells (NSCCs). Although CSCs have molecular similarities to embryonic and normal adult stem cells (7–11), breast CSCs are not epigenetically stable and do not behave as or arise from classic stem cells (12). Instead, breast CSCs and NSCCs exist in a dynamic equilibrium that is mediated by IL6 (12). This dynamic equilibrium between CSCs and NSCCs resembles the epithelial–mesenchymal transition (EMT) that occurs in normal development and can be simulated in cancer cells.
The properties of CSCs are relevant to cancer treatment, specifically the common observation that tumors relapse even after an effective primary response with chemotherapy. In this CSC hypothesis, chemotherapy kills the vast majority of cancer cells within the tumor, but the CSCs survive, and after treatment, propagate and regenerate the cell types in the tumor, thereby leading to relapse of the disease. In support of the CSC hypothesis, the antidiabetic drug metformin and miR-200 selectively kill CSCs, and they act together with various chemotherapeutic agents to reduce tumor growth and prolong remission (11, 13, 14). As only a small number of CSCs can regenerate a tumor, therapy has to be highly efficient, and in this regard, it would be useful to identify the molecular signature that dictates CSC growth and function.
The miR-200 family of microRNAs (miRNAs) regulates the EMT (15–18) and plays a critical role in CSC formation and growth (9, 11). CSCs have low levels of miR-200 RNAs compared with normal or nontumorigenic cancer cells, and ectopic expression of miR-200 inhibits CSC growth and tumor formation (9, 11). miR-200 RNAs regulate the EMT by directly inhibiting expression of Zeb1 and Zeb2, which are DNA-binding transcriptional repressors of the E-cadherin (Cdh1) gene (15, 17, 19). Conversely, Zeb1 binding to a conserved pair of E-box elements upstream of the promoter inhibits the expression of miR-200 family RNAs, thereby generating a negative feedback loop (16, 18). In addition, miR-200 affects CSC function by directly targeting Bmi1 (9, 20) and Suz12 (11), which, respectively, are components of the PRC1 and PRC2 polycomb complexes that repress transcription. Targets of miR-200 also include proteins involved in secretion (21), cell motility, and anoikis resistance (22), SIRT1 (23), and the Notch pathway (24), and it has been suggested that the stem cell factor Klf4 is also a target (20).
In previous work, we used an inducible model of cellular transformation and CSC formation to identify 22 miRNAs that are differentially expressed in CSCs and NSCCs (12). Here, using a genetic screen, we identify miRNAs that affect CSC growth and function and show that nearly all of these are down-regulated in CSCs. Unexpectedly, the miR-200 targets Zeb2, Suz12, Bmi1, and Klf4 are also directly targeted by other miRNAs that are down-regulated in CSCs and important for CSC function. Conversely, depletion of the Zeb1 and Zeb2 repressors leads to strong induction of the set of miRNAs that are important in CSC growth. In samples from human patients with triple-negative breast cancer, there is a striking inverse relationship between the levels of CSC-regulating miRNAs and their respective targets, providing strong evidence for the relevance of these interactions in human cancer. These observations suggest that CSC function is reinforced by an integrated network of miRNAs, transcription factors, and chromatin-modifying activities.
Results
Identification of miRNAs That Inhibit CSC Growth and Mammosphere Formation.
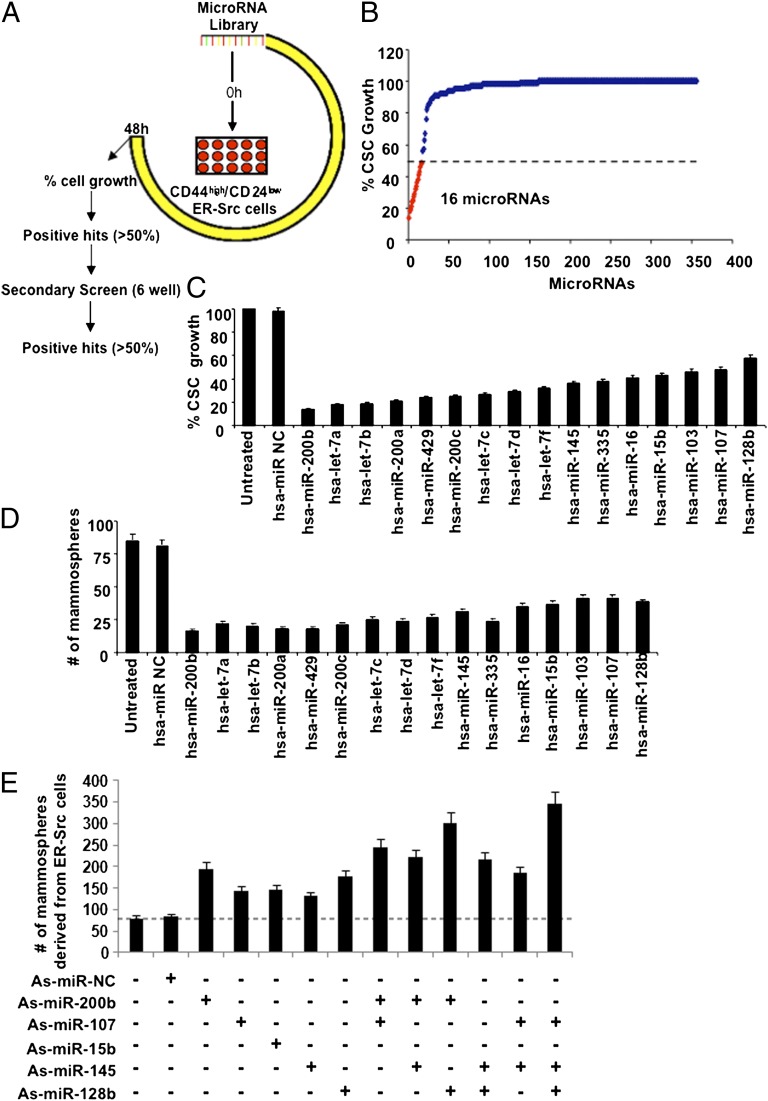
Treatment of nontransformed breast epithelial cells (MCF-10A) carrying an inducible Src oncogene (ER-Src) with tamoxifen induces cellular transformation (10, 25). A subpopulation of these transformed cells are CSCs, as defined by expression of the CD44 marker, mammosphere formation, and the ability to cause tumors in immunodeficient mice (10, 12). To identify miRNAs that regulate CSC growth, we performed a genetic screen in which CSCs isolated from transformed ER-Src cells were transfected with a library of 355 miRNAs (Fig. 1A). Positive hits were defined as miRNAs that inhibit cell growth by more than 50% (Fig. 1B), and these were further validated in secondary screens in six-well plates (Fig. 1C).
Fig. 1.
A microRNA (miRNA)-based genetic screen identifies breast CSC-specific inhibitors. (A) Schematic representation of the miRNA library screen experiment in CD44low/CD24high ER-Src cells. (B) Number of miRNAs inhibiting the growth of CD44low/CD24high cells according to the primary screen. (C) CSC growth following transfection with the respective miRNAs (secondary screen). (D) Number of mammospheres per 1,000 ER-Src cells generated by CSCs following transfection with the respective miRNAs. (E) Number of mammospheres per 1,000 ER-Src cells, 48 h post transfection with different combinations of antisense miRNAs.
From these screens, we identified 16 miRNAs that suppress CSC growth, including members of the miR-200 (miR-200b, miR-200a, miR-429, miR-200c), let-7 (let-7a, let-7b, let-7c, let-7d, let-7f), miR-15/16 (miR-16, miR-15b), and miR-103/107 (miR-103, miR-107) families as well as miR-145, miR-335, and miR-128b (Fig. 1 B and C). Furthermore, individual overexpression of these miRNAs inhibits the formation of mammospheres by at least 50% (Fig. 1D). Conversely, as observed previously for miR-200 family members (11), inhibition of representative miRNAs (via antisense RNA) results in increased efficiency of ER-Src–transformed cells to form mammospheres (Fig. 1E). Similar results are observed in MDA-MB-436 and MDA-MB-231 breast cancer cell lines, for both miRNA inhibition and miRNA overexpression experiments (Fig. S1). In the cases tested, simultaneous inhibition of two miRNAs often results in higher levels of mammosphere formation than inhibition of either miRNA alone (Fig. 1E). Importantly, all miRNAs identified here that inhibit CSC and mammosphere growth are expressed at lower levels in CSCs than in nonstem transformed cells (12). Thus, the generation of CSCs from a transformed cell population involves the down-regulation of a set of miRNAs.
Differentially Regulated miRNAs Involved in CSC Function Regulate Common Target Genes.
The miR-200 family directly targets the Zeb1 and Zeb2 transcriptional repressors as well as the Suz12 and Bmi1 subunits of the PRC2 and PRC1 polycomb complexes. In addition, it has been suggested that miR-200 targets the stem cell transcription factor Klf4 (20). Interestingly, sequence complementarity suggests that these miR-200 target genes may be directly regulated by other miRNAs that are down-regulated in CSCs and important for CSC function (Fig. S2). Specifically, Suz12 is a putative target of the miR-103/7 and miR-15b/16 families, Zeb2 and Klf4 are putative targets of miR-145, and Bmi1 is a putative target of miR-128b.
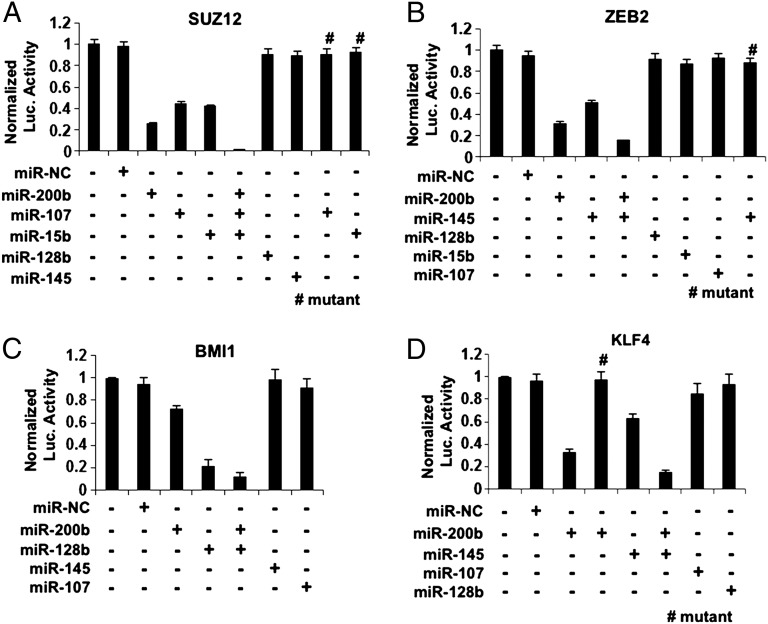
We validated these predictions by measuring the activities of luciferase reporter constructs containing the 3′ untranslated regions (3′ UTRs) of Suz12, Zeb2, Bmi1, and Klf4 in CSCs from ER-Src cells (Fig. 2). Overexpression of miR-107 or miR-15b reduces luciferase activity of the Suz12 3′UTR reporter construct, although to a slightly lesser extent than miR-200b (Fig. 2A). Inhibition by these miRNAs is direct because it is not observed upon mutation of the putative target sites in the Suz12 3′UTR. The combined overexpression of these three miRNAs nearly eliminates Suz12 luciferase activity. Overexpression of miR-145 inhibits the luciferase activity of both the Zeb2 (Fig. 2B) and Klf4 (Fig. 2D) reporter constructs, and this inhibition is not observed upon mutation of the putative target sites. For both Zeb2 and Klf4 reporters, inhibition by miR-145 is less pronounced than inhibition by miR-200b, and the combination of both miR-200b and miR-145 is more effective than either one alone. Finally, miR-128b inhibits the Bmi1 reporter more strongly than miR-200b, and again the combination is more effective than with either miRNA alone (Fig. 2C).
Fig. 2.
Target Identification of the CSC growth-inhibiting microRNAs. (A) Luciferase assays using a reporter construct containing the 3′UTR of Suz12 or a mutant carrying deletion of the miR-15b or miR-107 target sequence, 24 h after transfection with miR-NC, miR-200b, miR-107, miR-15b, miR-128b, and miR-145. (B) Luciferase assays using a reporter construct containing the 3′UTR of Zeb2 or a mutant carrying deletion of the miR-145 target sequence, 24 h after transfection with miR-NC, miR-200b, miR-145, miR-128b, miR-15b, and miR-107. (C) Luciferase assays using a reporter construct containing the 3′UTR of Bmi1, 24 h after transfection with miR-NC, miR-200b, miR-128b, miR-145, and miR-107. (D) Luciferase assays using a reporter construct containing the 3′UTR of Klf4 or a mutant carrying deletion of the miR-200b target sequence, 24 h after transfection with miR-NC, miR-200b, miR-145, miR-107, and miR-128b.
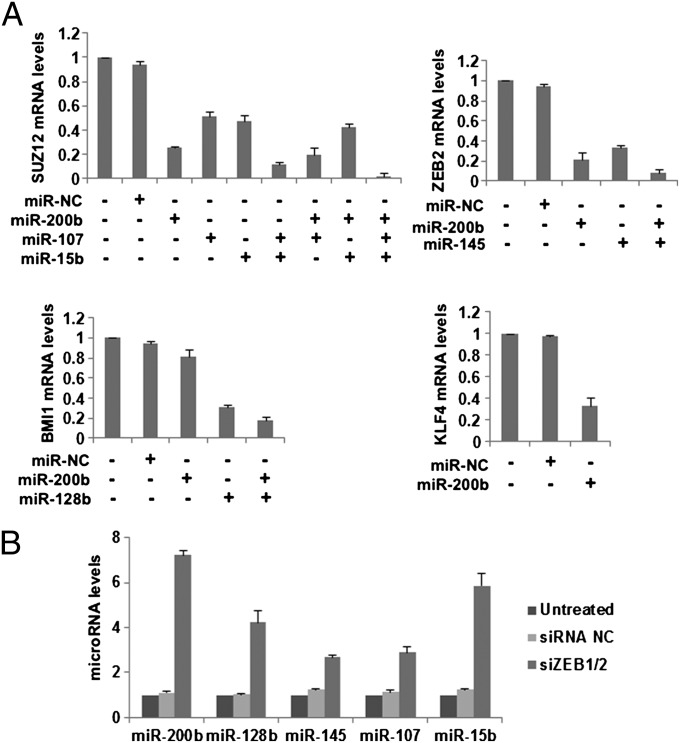
Further tests of the CSC growth-inhibiting miRNAs on the expression of the endogenous genes in CSCs from ER-Src cells verified our results using the luciferase reporters. Indeed, miR-200b down-regulated the mRNA levels of Suz12, Zeb2, and Klf4; miR-107 and miR-15b inhibited Suz12; miR-145 down-regulated Zeb2; and miR-128b blocked Bmi1 (Fig. 3A). In addition, combinational overexpression of the appropriate miRNAs targeting Suz12, Zeb2, Bmi1, or Klf4 resulted in more robust repression of the given targets than overexpression of individual miRNAs (Fig. S3). Thus, differentially regulated miRNAs that are important for CSC function have multiple targets in common.
Fig. 3.
Validation of the CSC growth-inhibiting microRNA targets. (A) Real-time PCR analysis for Suz12, Zeb2, Bmi1, and Klf4, 48 h post microRNA transfection in ER-Src cells. (B) MiR-200b, miR-128b, miR-145, miR-107, and miR-15b levels assessed by real-time PCR, 48 h post transfection with siRNAs against Zeb1 and Zeb2 in ER-Src cells.
Zeb1 and Zeb2 Inhibit Expression of Multiple miRNAs Involved in CSC Function.
Depletion of Zeb1 and Zeb2 decreases the proportion of CSCs in the population of transformed ER-Src cells (11), and Zeb1 and Zeb2 repress the expression of miR-200 family members (24). As the miRNAs involved in CSC function are coordinately down-regulated in CSC cells, we examined whether Zeb1 and Zeb2 affect the expression of the CSC-regulating miRNAs (Fig. 3B). Simultaneous siRNA-mediated depletion of Zeb1 and Zeb2 results not only in the expected up-regulation of miR-200b (sevenfold), but also in the up-regulation of miR-15b (sixfold), miR-128b (fourfold), and to a lesser extent, miR-107 and miR-145 (two- to threefold). Thus, in CSCs, Zeb1 and Zeb2 are important for repressing the miRNAs involved in inhibiting CSC function. Conversely, the reduction of Zeb1 and Zeb2 that occurs in nonstem cancer cells is linked to increased expression of miRNAs that inhibit CSC function.
miRNA-Target Interactions Important for CSC Function Are Relevant to Triple-Negative Breast Cancer.
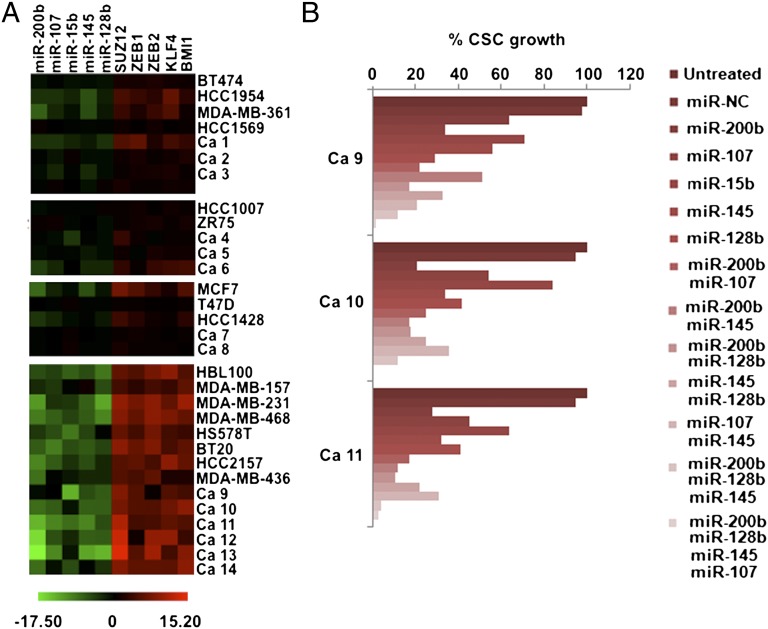
To address the relevance of the CSC-regulating miRNAs to human cancer, we measured the expression levels of miR-200b, miR-107, miR-15b, miR-145 and miR-128b in CSCs and NSCCs derived from 18 breast cancer cell lines and in tumor tissues from 14 patients with breast cancer. In parallel, we measured the levels of Suz12, Zeb2, Bmi1, and Klf4 in the same subpopulations. Data were clustered in the four molecular subtypes of breast cancer based on the molecular and genetic characteristics of each cell line and tumor tissue (Fig. S4).
The results are presented as the ratio of RNA levels in CSCs vs. the corresponding NSCCs (Fig. 4A). Expression levels of CSC-regulating miRNAs are significantly reduced in the CSCs derived from the triple-negative/basal-like cell lines and tumor tissues. In the same samples, there is a striking inverse relationship between CSC-regulating miRNAs and the levels of their respective targets (Fig. 4A). The same trend was observed in the three other types of breast cancer (ER-positive, HER2-positive, ER- and PR-positive), but the effects are much less pronounced both quantitatively and qualitatively. To test the role of these miRNAs in the context of human cancer, we overexpressed them either alone or in combination in CSCs derived from three triple-negative breast tumors (ca9-11). All of the tested miRNAs partially inhibited CSCs growth whereas their combinations had more robust inhibitory effects (Fig. 4B). These observations provide an explanation for the coordinate down-regulation of specific microRNAs in highly aggressive breast tumors and their mechanistic relationship to the up-regulation of their targets that encode transcription factors and polycomb complex components.
Fig. 4.
CSC-specific microRNA-chromatin signature is active in basal-like breast cancer cell lines and tissues. (A) miR-200b, miR-107, miR-15b, miR-145, miR-128b, Suz12, Zeb1, Zeb2, Klf4, and Bmi1 expression levels assessed by real-time PCR in CSCs and NSCCs derived from breast human tumors and cell lines. (B) Growth of CSCs derived from three triple-negative/basal-like breast tumors 48 h after transfection with the respective microRNAs.
Discussion
CSCs and NSCCs can be interconverted in cell lines and tumors, and they are characterized by distinct transcriptional profiles. The miR-200 family of miRNAs is strongly down-regulated in CSCs, and the near absence of these miRNAs is critical for CSC growth and function. The miR-200 miRNAs inhibit CSC function by directly inhibiting the expression of genes encoding the Bmi1 and Suz12 components of the PRC1 and PRC2 polycomb complexes, the Klf4 stem-cell transcription factor, and the Zeb1 and Zeb2 transcriptional repressors. Conversely, the Zeb1 and Zeb2 repressors directly inhibit expression of the miR-200 family via binding to target sites in the promoter (16, 18), and hence these repressors are important for CSC growth and function. As such, the Zeb repressors and miR-200 miRNAs form a negative feedback loop that reinforces the CSC phenotypes.
Here, we use a genetic screen to identify miRNAs that inhibit CSC function, and we show how these correspond to miRNAs identified previously as having lower levels in CSCs. Aside from the miR-200 and Let-7 families, which were previously known in this regard, these include the miR-15/16 and miR-103/107 families as well as miR-145, miR-335, and miR-128b. In accord with the defining high tumor formation properties of CSCs, all of these miRNAs have been have generally been described as tumor suppressors (e.g., see refs. 26–31).
Strikingly, the miRNAs that are down-regulated in CSCs share many common targets. Each of the well-described miR-200 targets (Suz12, Bmi1, Zeb1, Zeb2, and Klf4) is also targeted by at least one other miRNA that is coordinately down-regulated in CSCs. Conversely, the Zeb1 and Zeb2 repressors, which directly inhibit expression of the miR-200 family, also inhibit expression of the other miRNAs that are down-regulated in CSCs. Direct binding of the Zeb repressors to these other miRNA promoters has yet to be demonstrated. Thus, the miRNAs and targets described here represent an integrated transcriptional regulatory circuit (Fig. 5), not a simple linear pathway.
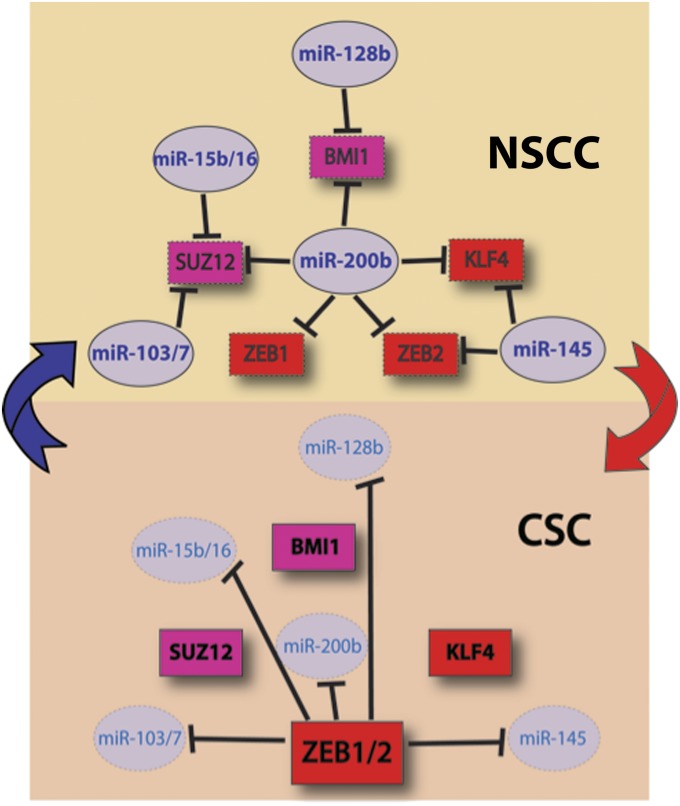
Fig. 5.
Schematic representation of the bistable switch that drives cells into either NSCCs and CSCs. In NSCCs, microRNAs suppress the expression of Suz12, Zeb1, Zeb2, Klf4, and Bmi1. Furthermore, the Zeb repressors are less active, thereby leading to higher levels of the miRNAs, and hence to further reduced levels of the Zeb repressors, Klf4, and polycomb complex components (Upper). In the CSC state, the expression of the Zeb repressors inhibits expression of the key miRNAs, thereby leading to up-regulation of the target genes. Repression is enhanced by the increased expression of both Zeb repressors as well as Klf4 and the polycomb complex components (Lower).
The integrated regulatory circuit can act as a bistable switch that drives cells into either NSCCs and CSCs, with intermediate states being minimized. In the CSC state, the Zeb repressors inhibit expression of the key miRNAs, thereby leading to up-regulation of the target genes. Repression is enhanced by the increased expression of both Zeb repressors as well as Klf4 and the polycomb complex components. In the NSCC state, the Zeb repressors are less active, thereby leading to higher levels of the miRNAs, and hence to further reduced levels of the Zeb repressors, Klf4, and polycomb complex components. Genetic experiments described here and elsewhere indicate that all of these components are relevant for the discrimination between NSCCs and CSCs. Importantly, the transcription factors, miRNAs, and chromatin-modifying activities that constitute the integrated regulatory circuit also have many other targets that undoubtedly contribute to the NSCC and CSC states.
In standard cell culture medium, CSCs and NSCCs are not epigenetically stable, but rather exist in a dynamic equilibrium mediated by IL6 secretion into the medium (12). Under these conditions, CSCs differentiate back into NSCCs, but this is balanced out by conversion of NSCCs to CSCs due to the high secretion of IL6 into the medium by CSCs. As such, there must be an IL6-mediated step that is critical for bistable switch. In principle, IL6 could affect one or more steps in the circuit, either by altering gene expression via transcriptional regulatory proteins or by altering the activity of the Zeb repressors and/or Klf4. Given the central role of the Zeb repressors in controlling miRNA expression, it is tempting to speculate that they are major targets of IL6 action. In this regard, after this work was completed, it has been reported that IL6 suppresses miR-200c expression to drive activation of inflammatory signaling and tumorigenesis (32).
The concept and terminology of cancer stem cells has been controversial. The dynamic equilibrium term between breast CSCs and NSCCs strongly argues that breast CSCs behave very differently than classic stem cells (12). On the other hand, the transcriptional profile of CSCs resembles that of embryonic stem cells, suggesting that CSCs represent a more dedifferentiated form of cancer cells (7–9). In particular, polycomb complexes directly regulate key developmental factors that maintain ESC self-renewal and pluripotency (33, 34); are commonly up-regulated in cancer types in a manner associated with the aggressiveness of the tumor (35–38); and, via the Bmi1 and Suz12 subunits (9, 11, 20), are targets of the miR-200 family. Our observations that Bmi1 and Suz12 are targets of additional miRNAs that are down-regulated in CSCs in a Zeb1/Zeb2-dependent manner provide further support for the link of polycomb repression to CSC function. Furthermore, the Klf4 stem cell transcription factor is inhibited by miR-145, whose expression is inhibited by the Zeb repressors in CSCs. These observations prompt the speculations that polycomb-mediated repression of genes involved in differentiation is a common feature in the biology of cancer and embryonic stem cells.
As active players in important human oncogenic signaling pathways, miRNAs may affect the development of strategies for cancer therapy. Given the importance of CSCs in tumor formation, miRNAs that inhibit CSC formation are of potential interest for cancer therapy. For example, we previously showed that, in mouse xenografts, the combination of miR-200 and doxorubicin stimulated tumor regression and prolonged remission beyond that obtained with either agent alone (11). Thus, the identification of a miRNA network/transcription factor network that governs breast CSC growth and whose signature appears frequently in triple-negative/basal-like breast cancer may have therapeutic implications. Reintroduction of miR-200 and other miRNAs that target CSCs in combination with conventional chemotherapy, which targets the non-CSCs, may serve as an effective strategy to treat aggressive breast cancer.
Materials and Methods
Cell Culture.
The nontransformed breast cell line MCF-10A (39) contains an integrated fusion of the v-Src oncoprotein with ER-Src (40). These cells were grown in DMEM/F12 medium supplemented with 5% (vol/vol) donor horse serum, 20 ng/mL epidermal growth factor (EGF), 10 μg/mL insulin, 100 μg/mL hydrocortisone, 1 ng/mL cholera toxin, 50 units/mL penicillin/streptomycin, with the addition of puromycin. Src induction and cellular transformation was achieved by treatment with 1 μM 4-OH tamoxifen, typically for 36 h as described (10, 25, 41). Breast cancer cell lines were grown in DMEM, 10% (vol/vol) FBS, and penicillin/streptomycin.
CSC Purification.
Purification of CD44high/CD24low cells derived from breast cancer cell lines has been performed as described previously (11). Briefly, flow cytometric cell sorting was performed on single-cell suspensions that were stained with CD44 antibody (FITC-conjugated) (555478; BD Biosciences) and with CD24 antibody (phycoerythrin-conjugated) (555428; BD Biosciences) for 30 min (10, 13, 41). As used throughout this work, CSCs are defined by the CD44high/CD24low population.
Purification of CSCs from Human Breast Tissues.
Purification of CD44high/CD24low cells derived from human breast tumors was performed as described previously (12, 20). Briefly, the breast tissues were minced into small pieces (1 mm) by using a sterile razor blade. The tissues were digested with 2 mg/mL collagenase I (C0130; Sigma) and 2 mg/mL hyalurinidase (H3506; Sigma) in 37 °C for 3 h. Cells were filtered, washed with PBS, and followed by Percoll gradient centrifugation. The first purification step was to remove the immune cells by immunomagnetic purification by using an equal mix of CD45 (leukocytes), CD15 (granulocytes), CD14 (monocytes), and CD19 (B cells) Dynabeads (Invitrogen). The second purification step was to isolate fibroblasts from the cell population by using CD10 beads for magnetic purification. The third step was to isolate the endothelial cells by using “endothelial cocktail” beads (CD31: BD Pharmingen, catalog no. 555444; CD146 P1H12 MCAM: BD Pharmingen, catalog no. 550314; CD105: Abcam, catalog no. Ab2529; Cadherin 5: Immunotech, catalog no. 1597; and CD34: BD Pharmingen, catalog no. 555820). In the final step, from the remaining cell population, only the CD44high cells were purified by using CD44 beads. These cells were sorted for CD44high/CD24low (CSC) cells (purity >99.2%).
Mammosphere Formation Assay.
Mammospheres were generated by placing transformed cell lines in suspension (1,000 cells/mL) in serum-free DMEM/F12 media, supplemented with B27 (1:50; Invitrogen), 0.4% BSA, 20 ng/mL EGF, and 4 μg/mL insulin (12). After 6 d of incubation, mammospheres were typically >75 μm in size with ∼97% being CD44high/CD24low.
miRNA and siRNA Transfection Experiments.
All miRNAs and siRNAs were obtained from Ambion, Inc., and are listed by their catalog numbers in the following sentence. Cells were transfected with 100 nM miRNA negative control (miR NC; AM17110), miR-200b (002251), miR-107 (000443), miR-15b (000390), miR-128b (002216), and miR-145 (002278) or with 20 nM of siRNA for Zeb1 (s229972) and Zeb2 (s19033) or the siRNA negative control (siRNA NC; AM4611) by using siPORT NeoFX transfection agent.
miRNA Library Screen.
A miRNA library, consisting of 355 miRNA mimics and 2 miRNA negative controls (100 nM) (Dharmacon) was transfected in CD44low/CD24high ER-Src cells plated in 96-well plates. Forty-eight hours post transfection, the number of cells was assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). miRNAs that inhibited the luminescence by more than 50% were considered as positive hits. The positive hits identified in the primary screen were validated in a secondary screen. Specifically, 100 nM of these miRNA mimics were transfected in CD44low/CD24high ER-Src cells plated in six-well plates, and 48 h post transfection, cells were directly counted. The transfection dose used for the miRNA mimics was assessed through control experiments performed to identify the maximum dose without any cytotoxic effects.
miRNA Target Prediction and Validation and RNA Analysis.
The miRNA database TargetScan version 5.1 (http://www.targetscan.org/index.html) was used to identify potential miRNA targets and to compare the seed sequences with the 3′UTRs of Suz12, Zeb2, Bmi1, or Klf4. Untreated miR-NC or miRNA (100 nM) treated cells were transfected with luciferase reporter gene constructs containing the 3′UTR of Suz12, Zeb2, Bmi1, or Klf4 (Switchgear Genomics Inc.). Cell extracts were prepared 24 h after transfection of the luciferase vector, and the luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega). RNA purified from cancer cell lines, tissues, and miR-NC or miRNA (100 nM) treated cells was reverse-transcribed to form cDNA, which was subjected to quantitative PCR in real-time using β-actin levels as a normalization control. The primers used to test Klf4 mRNA levels were ACCAGGCACTACCGTAAACACA and GGTCCGACCTGGAAAATGCT. The primers for Suz12, Zeb2, and Bmi1 have been described previously (10). miRNA expression levels were tested using the mirVana qRT-PCR miRNA Detection Kit and qRT-PCR Primer Sets according to the manufacturer’s instructions (Ambion Inc.). RNU48 expression was used as an internal control. The experiments have been performed in triplicate, and data are presented as mean ± SE.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant CA 107486 (to K.S.) and a Sidney Kimmel Foundation grant (to D.I.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212811109/-/DCSupplemental.
References
- 1.Ailles LE, Weissman IL. Cancer stem cells in solid tumors. Curr Opin Biotechnol. 2007;18:460–466. doi: 10.1016/j.copbio.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 3.Grimshaw MJ, et al. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008;10(3):R52. doi: 10.1186/bcr2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marotta LL, Polyak K. Cancer stem cells: A model in the making. Curr Opin Genet Dev. 2009;19(1):44–50. doi: 10.1016/j.gde.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: Cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 7.Gotoh N. Control of stemness by fibroblast growth factor signaling in stem cells and cancer stem cells. Curr Stem Cell Res Ther. 2009;4(1):9–15. doi: 10.2174/157488809787169048. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimono Y, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iliopoulos D, et al. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39:761–772. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci USA. 2011;108:1397–1402. doi: 10.1073/pnas.1018898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res. 2009;69:7507–7511. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iliopoulos D, Hirsch HA, Struhl K. Metformin decreases the dose of chemotherapy for prolonging tumor remission in mouse xenografts involving multiple cancer cell types. Cancer Res. 2011;71:3196–3201. doi: 10.1158/0008-5472.CAN-10-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bracken CP, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 17.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 18.Burk U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wellner U, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 21.Korpal M, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howe EN, Cochrane DR, Richer JK. Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res. 2011;13(2):R45. doi: 10.1186/bcr2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eades G, et al. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J Biol Chem. 2011;286:25992–26002. doi: 10.1074/jbc.M111.229401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brabletz S, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011;30:770–782. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirsch HA, et al. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell. 2010;17:348–361. doi: 10.1016/j.ccr.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bandi N, et al. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553–5559. doi: 10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- 27.Datta J, et al. 2011. microRNA-107 functions as a candidate tumor-suppressor gene in head and neck squamous cell carcinoma by downregulation of protein kinase CE. Oncogene, 10.1038/onc.2011.565.
- 28.Sachdeva M, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Png KJ, et al. MicroRNA-335 inhibits tumor reinitiation and is silenced through genetic and epigenetic mechanisms in human breast cancer. Genes Dev. 2011;25:226–231. doi: 10.1101/gad.1974211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katada T, et al. microRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34:537–542. [PubMed] [Google Scholar]
- 32.Rokavec M, Wu W, Luo J-L. IL6-mediated suppression of miR-200c directs constitutive activation of inflammatory signaling circuit driving transformation and tumorigenesis. Mol Cell. 2012;45(1):1–13. doi: 10.1016/j.molcel.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 35.Squazzo SL, et al. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong X, et al. Identification of tumorigenic retinal stem-like cells in human solid retinoblastomas. Int J Cancer. 2007;121:2125–2131. doi: 10.1002/ijc.22880. [DOI] [PubMed] [Google Scholar]
- 37.Hoenerhoff MJ, et al. BMI1 cooperates with H-RAS to induce an aggressive breast cancer phenotype with brain metastases. Oncogene. 2009;28:3022–3032. doi: 10.1038/onc.2009.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bracken AP, Helin K. Polycomb group proteins: Navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 39.Soule HD, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 40.Aziz N, Cherwinski H, McMahon M. Complementation of defective colony-stimulating factor 1 receptor signaling and mitogenesis by Raf and v-Src. Mol Cell Biol. 1999;19:1101–1115. doi: 10.1128/mcb.19.2.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.