Next-generation sequencing (NGS) has revolutionized genome and transcriptome analyses in recent years. Now, a smart and simple modification termed duplex sequencing published in PNAS by Schmitt et al. (1) may pave the way to explore the full power of NGS in answering fundamental questions and particularly through its application to cancer research. This enthusiasm is warranted because duplex sequencing reportedly reduces the error rate of NGS up to 10 million-fold.
Duplex sequencing may therefore change the history of NGS, which began in 2005 when the first next-generation sequencer was launched (2). In the following years, the advent, development, and widespread deployment of NGS systems for many applications have fundamentally altered genome research and allowed investigators to address biological questions previously not conceivable or affordable (3). Further improvements in technology, reliability, and workflow standardization may enable translation into clinical diagnostics (4). Despite numerous success stories, critical readers could not escape feelings of discomfort raised by the inherently high sequencing error rate of 1%, which results in hundreds of millions of sequencing mistakes. The problem was recognized from the beginning, and several statistical approaches (5) were developed to discriminate between sequencing error-derived noise and real genetic variation. Although bioinformatic advances made limited improvements in the resolution of sequencing errors, they did not substitute for an experimental method correctly assigning the mutational landscape of individual tumors. Based on the data presented in the article by Schmitt et al. (1), the Gordian knot in sequencing accuracy has apparently been cut by the duplex sequencing approach.
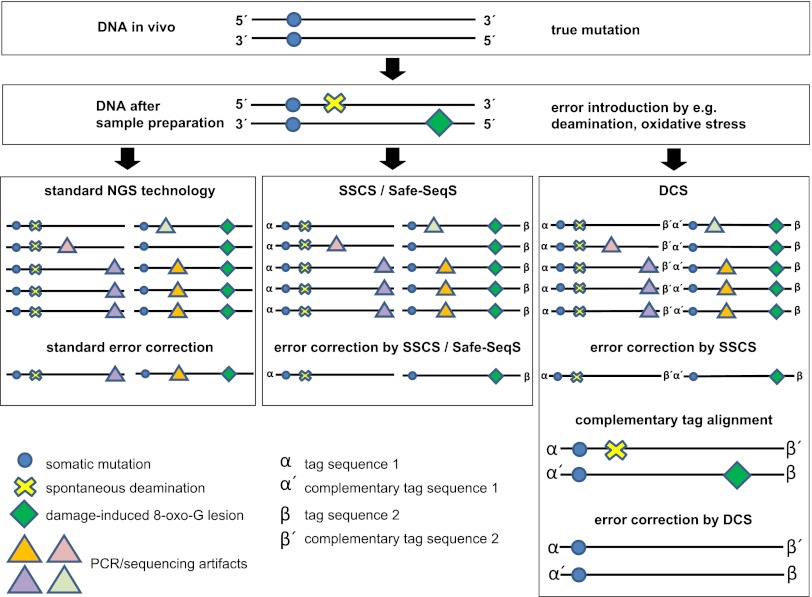
The authors do so by simply exploiting the redundant sequence information contained in the complementary DNA strand of double stranded genomic DNA molecule. They add a unique random yet complementary double-stranded nucleotide sequence to both strands of duplex DNA before amplification. This approach allows them to identify families of DNA molecules that share the same tag sequence on a single strand, called single-strand consensus sequence (SSCS), analogous to a recently published approach termed Safe-SeqS (6). However, it also identifies the complementary DNA strand by searching for the complementary tag sequences among the SSCS reads [duplex consensus sequences (DCS)]. A sequence base at a given position is then kept only if the read data from each of the two strands match perfectly (Fig. 1).
Fig. 1.
Sequence error correction by different methods. In the example given, the DNA has one somatic mutation (on both strands); both sample preparation and NGS add artifactual mutations as a consequence of deamination events, oxidative damage, and errors typical for the applied sequencing instrument. Standard methods for the correction of sequencing errors use the abundance of a sequence read to identify errors. SSCS and the analogous Safe-SeqS (6) use unique identifiers on single-strand molecules to group families of sequence reads and remove those that occur less frequently than in 95% of family reads. Duplex sequencing, in addition to assigning families of single-strand reads, exploits the information from the complementary strand. First, the complementary strand is identified via the complementary sequence tags on both strands, and only bases are kept that are present on both strands. Thereby, the correct sequence is identified.
Schmitt et al. (1) assess the power of their approach experimentally using phage M13mp2 DNA, which is a substrate that has been used extensively in sensitive genetic mutation assays and has a known mutation frequency. Standard sequencing methods predicted a 1,000-fold higher than expected mutation rate, which could be reduced 100-fold by SSCS, indicating that about 90% of mutations identified by SSCS are still artifacts. However, DCS resulted in a mutation frequency nearly identical to that of genetic methods, and the authors give good reasons as to why the DCS result may indeed be closer to the correct value than the genetic gold standard.
These numbers provide a frame for the assessment of recently published NGS analyses of the mutational load of human breast cancers (7–9), which, in turn, were used to infer patterns of DNA damage and repair processes (8) as well as tumor evolutionary histories (9). In one example of a patient with a rapidly fatal disease course, the primary tumor, a xenograft from the primary tumor, and a brain metastasis were sequenced and harbored 27,173, 109,078, and 51,710 “novel somatic single-nucleotide variants” (7), respectively, of which most were C > T mutations. Fifty somatic point mutations and small insertions and deletions in coding sequences were validated. These mutations were largely shared by all three samples (7). Another case of breast cancer was sequenced with 188-fold depth: 70,690 mutations were identified. Here, it was calculated that 100% of cancer cells shared 27,000 mutations and that from the most recent common ancestor, several subclones diverge that share other sets of thousands of mutations (9). The assumption that these were all true somatic mutations was taken from the dominance of C > A, C > G, and C > T mutations (8, 9).
Although various types of filters to correct for sequencing and mapping errors were used, neither study utilized an SSCS-like approach to reduce single-strand errors. From the studies of Schmitt et al. (1) and Kinde et al. (6), one may conclude that the true mutational load is overestimated at least 10- to 20-fold, assuming that the applied algorithms reach the level of SSCS/Safe-SeqS; if not, up to 99.9% would be erroneous. The suspicion that mutation numbers were overestimated is further supported by an interesting observation of Schmitt et al. (1) when they analyzed the spectrum of mutations. Even the single-strand correction method of SSCS identified a large excess of C > T and G > T mutations. DCS corrected for these errors, suggesting that they are derived from first-round de novo PCR errors generated during the preparation of the library for sequencing. In addition, C > T transitions are often caused by spontaneous deamination of cytosine to uracil and an adenosine is inserted into the nascent DNA strand across from the damaged base. This results in a C > T transition. It is also of concern that the type of error apparently depends on the source of DNA. In an experiment on human mtDNA, which is extensively exposed to free radicals during metabolism, Schmitt et al. (1) find a 130-fold excess of G > T relative to C > A mutations, consistent with oxidative damage of the DNA. Such uncorrected errors in DNA replication would result in first-round PCR errors. Although the importance of damage bypass-related mutations is currently not
The Gordian knot in sequencing accuracy has apparently been cut by the duplex sequencing approach.
well studied, one may wonder whether the heterogeneous vascularization and oxidation in cancer tissues before surgery and the often uncontrolled ischemic periods during and after surgery or other factors of tissue processing may generate different patterns of mutational spectra, albeit unrelated to cancer mutational history and selection. Such errors would then reflect artifacts of sample preparation. The probability that duplex sequencing would pick up alterations that are solely related to sequencing errors or to the preparatory damage of the sample is calculated to be lower than 3.8 × 10−10, which is the theoretical chance that they occur in both complementary bases of a duplex DNA molecule. In summary, the differences between standard methods for the correction of sequencing errors, SSCS-like methods, and DCS place caveats on the deduction of mutational landscapes (9) and processes (8) from data that did not apply DCS.
What happens with rare true mutations if they are buried among thousands of sequencing errors? Schmitt et al. (1) also address this question experimentally. They constructed a series of M13mp2 variant genomes containing specific nucleotide substitutions, mixed the variants at known ratios, and performed duplex sequencing. With standard methods, variants present at less than 1% were not detected because artifactual mutations manifest at the same level obscured them. In contrast, duplex sequencing with very high sequencing depth allowed accurate recovery of mutant sequences down to the lowest tested level: 1 mutant molecule per 10,000 WT molecules.
The apparent power of duplex sequencing and caveats regarding NGS methods raise questions about whether and how previously generated sequencing information on human cancers should be used. Ideally, selected samples would be processed in parallel with and without DCS or, if material is still available, published samples resequenced with duplex sequencing. For a rapid retrospective confirmation of candidate mutations, Schmitt et al. (1) suggest using the randomly sheared ends as unique identifiers. Then, elegant methods to deduce the life history of human cancers (9) could be reapplied and the results compared. Otherwise, it may remain unclear to what extent non-DCS approaches reflect the mutational history of cancer rather than NGS artifacts. This concern does not include reports on cancer mutations that have been validated and confirmed on many samples by independent methods. On the other hand, duplex sequencing now has to be applied to “real” heterogeneous and highly complex samples other than M13mp2. If its power is confirmed, duplex sequencing will likely improve our understanding of the clonal substructure of human cancers, modify the catalog of rare mutations, help to pinpoint mechanisms of mutation generation, and potentially identify mutator phenotypes (10). Eventually, it may open doors to clinical applications in which diagnostic accuracy is the sine qua non for ethical treatment decisions.
Footnotes
The authors declare no conflict of interest.
See companion article on page 14508.
References
- 1.Schmitt MW, et al. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci USA. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hawkins RD, Hon GC, Ren B. Next-generation genomics: An integrative approach. Nat Rev Genet. 2010;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desai AN, Jere A. Next-generation sequencing: Ready for the clinics? Clin Genet. 2012;81:503–510. doi: 10.1111/j.1399-0004.2012.01865.x. [DOI] [PubMed] [Google Scholar]
- 5.Yang X, Chockalingam SP, Aluru S. A survey of error-correction methods for next-generation sequencing. Brief Bioinform. 2012 doi: 10.1093/bib/bbs015. 10.1093/bib/bbs015. [DOI] [PubMed] [Google Scholar]
- 6.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nik-Zainal S, et al. Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nik-Zainal S, et al. Breast Cancer Working Group of the International Cancer Genome Consortium The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]