Abstract
The RNA-dependent RNA polymerase (RdRP) of nonsegmented negative-sense RNA viruses consists of a large catalytic protein (L) and a phosphoprotein cofactor (P). During infection, the RdRP replicates and transcribes the viral genome, which resides inside an oligomer of nucleocapsid protein (N-RNA). The classical view of P as a cofactor for L assigns a primary role of P as a bridge mediating the access of L to the RNA template, whereby its N-terminal domain (PNTD) binds L and its C-terminal domain (PCTD) binds N-RNA. Recent biochemical and structural studies of a prototype nonsegmented negative-sense RNA virus, vesicular stomatitis virus, suggest a role for P beyond that of a mere physical link: P induces a structural rearrangement in L and stimulates polymerase processivity. In this study, we investigated the critical requirements within P mediating the functional interaction with L to form a fully functional RdRP. We analyzed the correlation between the impact of P on the conformation of L and its activity in RNA synthesis and the consequences of these events on RdRP function. We identified three separable elements of the PNTD that are required for inducing the conformational rearrangement of L, stimulating polymerase processivity, and mediating transcription of the N-RNA. The functional interplay between these elements provides insight into the role of P as a dynamic player in the RNA synthesis machine, influencing essential aspects of polymerase structure and function.
Keywords: large polymerase, replication and transcription, Mononegavirales, rhabdovirus
The functional unit necessary for transcription and replication of nonsegmented negative-sense (NNS) RNA viruses is a ribonucleoprotein (RNP) complex. The RNP complex comprises a genomic RNA encapsidated by a nucleocapsid protein oligomer (N-RNA), associated with the RNA-dependent RNA polymerase (RdRP) consisting of a complex of the large polymerase protein (L) and a phosphoprotein (P) (1–3). The template RNA is buried between the N- and C-terminal lobes of each N protomer; nevertheless, the overall integrity of the N-RNA structure is maintained during copying by the RdRP (4, 5). The L protein is the multifunctional catalytic core of the RNA synthesis machinery, harboring the RdRP as well as a capping enzyme and two methyltransferase activities that are required for mRNA synthesis (6–10). During RNA synthesis, L must gain access to the RNA, and its enzymatic activities must be regulated in accordance with a replicase or transcriptase mode of RNA synthesis. The access of L to the N-RNA is mediated by the noncatalytic cofactor P, which engages L and the N oligomer simultaneously (11, 12).
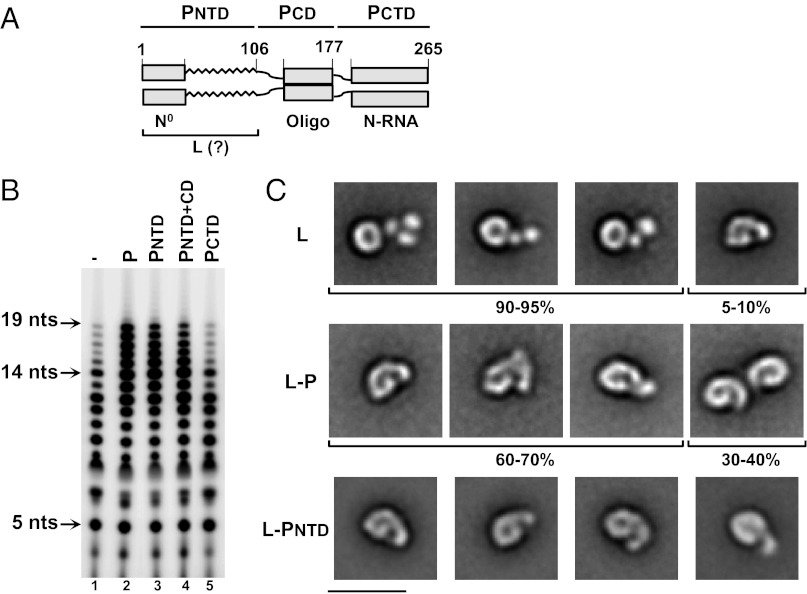
The functioning of an RNP as a highly regulated RNA synthesis machine requires an intricate, tight coordination of its individual components. The mechanisms that govern such functional coupling are largely unknown. Much of our understanding of the assembly, structure, and function of NNS RNA virus RNPs have come from studies of vesicular stomatitis virus (VSV). In part, this reflects the uniquely robust in vitro transcription that can be reconstituted from purified VSV N-RNA, P, and L (3, 13). The VSV P protein exhibits a modular organization (Fig. 1A), comprising an N-terminal domain (PNTD), a central domain (PCD), and a C-terminal domain (PCTD) (11, 14, 15). The roles of PNTD in binding L and of PCTD in binding N-RNA were first recognized by biochemical and genetic studies, establishing a primary role of P as a mediator of L’s access to the N-RNA template (11). A second essential role of P involves chaperoning the free N protein (N0) via the extreme N terminus of PNTD, maintaining N0 solubility and regulating its assembly on the nascent genomic RNA during replication (16, 17).
Fig. 1.
Effects of the N-terminal domain of P on the function and the structural organization of L. (A) Domain organization of VSV P. Schematic of a P homodimer showing the modular organization of P into three domains: N-terminal domain (PNTD, 1–106), central domain (PCD, 107–177), and C-terminal domain (PCTD, 178–265). Gray boxes depict regions of P with determined crystal structure, which were also shown to be involved in N0 binding, homo-oligomerization, and N-RNA binding. The zigzag line depicts the region of P of undetermined structure. PNTD has been shown to be involved in binding L; however, the precise binding site has not been determined. (B) PNTD is sufficient to stimulate the processivity of L on a nonencapsidated RNA template. RNA synthesis reactions were carried out with 0.2 μM Le19 template and 0.2 μM L, with 0.2 μM of either full-length P or fragments comprising the PNTD, PNTD+CD, or PCTD added when indicated. Reaction products were analyzed on a 20% polyacrylamide/7 M urea gel. (C) PNTD recapitulates the structural rearrangement in L induced by full-length P. EM characterization of L, L–P, and L–PNTD complexes showing five representative class averages of each, illustrating the structural similarity of the L–PNTD complex to the monomer population of the L–P complex. Shown is the percentage of the population of particles exhibiting similar conformations as the representative class averages for L and L–P. (Scale bar: 20 nm.)
An expanding structural map of P is beginning to provide additional functional insight into its role in RNA synthesis. Analysis of the crystal structure of the VSV PCTD has shown that PCTD makes direct contact with the C-terminal lobes of two adjacent N protomers (18). Structural and biophysical analyses have demonstrated that PCD is a homodimerization domain (14, 19). The role of P dimerization remains uncertain; however, it has been proposed to play a role in mediating the progression of L along the N-RNA template (20). The structure of N0 lacking an N-terminal arm, in complex with a peptide corresponding to the first 60 residues of P, supports a model in which residues 6–35 of P block the polymerization of adjacent N molecules as well as access of RNA to the RNA-binding groove (21). Biophysical and bioinformatic analyses have indicated the presence of multiple intrinsically disordered regions (IDRs) separating the different structured domains of P (15). These disordered regions have been suggested to enhance the dynamic properties of P function. Furthermore, a role for phosphorylation of P by cellular kinases has been proposed to play a role in regulating the function of P in viral transcription and replication (22, 23).
Until recently, functional interactions between L and P were largely uncharacterized. Recent biochemical and structural studies point to additional roles of P beyond serving as a mere physical bridge between L and the N-RNA template. EM characterization of L revealed its organization into a ring domain harboring the RNA polymerase, linked to a flexible appendage of three globular domains containing the cap-forming activities (24). After complex formation with P, the appendage rearranges into a more compact structure. Furthermore, reconstitution of RNA synthesis using purified L and a short synthetic RNA template demonstrated that P stimulates the initiation and processivity of L, indicating a role of P in stimulating L function independent of facilitation of access of L to the encapsidated RNA template (25).
In this study, we investigated the critical events resulting from the impact of P on the structure and function of L, and examined the contribution of these events to achieving a fully functional RdRP complex. We identified the elements of P necessary for inducing a conformational rearrangement in L and stimulating the processivity of L, and characterized the influence of these elements on RdRP function in copying the N-RNA template. Here we propose a model for an emerging role of P as a dynamic player in the RNA synthesis machine, influencing key aspects of polymerase structure and function.
Results
PNTD Is Sufficient to Induce Conformational Change in L and Stimulate L Processivity.
To probe the relationship between the conformational change in L on binding of P and processivity of the RdRP, we first defined the elements of P that mediate each of these functions. For the analysis of RdRP processivity, we used an RNA synthesis assay comprising a synthetic naked 19-nt template corresponding to the 3′ leader sequence of the VSV genome (Le19) (25). This assay specifically allowed us to study the function of P as a processivity factor independent of its role in facilitating L access to an encapsidated template. The L polymerase uses Le19 as a template to synthesize RNA products of 3–19 nt, and the addition of P resulted in a three- to fourfold increase in RNA synthesis and enhancement of the 13- to 19-nt products (Fig. 1B, lanes 1 and 2). To define the P domains that stimulate L activity in RNA synthesis, individual or combinations of the P domains were expressed in Escherichia coli and purified (Fig. S1A). We previously demonstrated that the phosphorylation of P did not influence polymerase activity on the Le19 template (25). PNTD (1–106) and PNTD+CD (1–177) stimulated L processivity to a similar extent as full-length P (1–265), whereas PCTD (178–265) had no effect on RNA synthesis (Fig. 1B, lanes 3–5). These data demonstrate that the PNTD is sufficient for stimulation of L processivity independent of the oligomeric state of P.
EM analysis revealed the organization of L into a ring-like domain containing the RNA polymerase activity and a flexible appendage consisting of three globular domains containing the cap-forming activities (24). The flexibility of the appendage makes only two globular domains visible in some classes (24) (Fig. 1C, Top). In ∼5–10% of particles, the appendage assumes a curved compact conformation (Fig. 1C, Top, rightmost). In the L–P complex, the flexible appendage is rearranged into a more compact conformation similar to that seen in the minor population of L alone (24) (Fig. 1C, Middle). Furthermore, a dimeric population constitutes 30–40% of the particles (Fig. 1C, Middle, rightmost). To examine the effect of PNTD on the molecular architecture of L, an L–PNTD complex was isolated using size-exclusion chromatography (Fig. S1B). EM of negatively stained L–PNTD revealed monodispersed particles (Fig. S1C); 5,842 particles were selected and classified into 20 classes (Fig. S1D). Four representative class averages (Fig. 1C, Bottom) illustrate that PNTD recapitulates the conformational rearrangement in L induced by P. The more compact structure resulting from the rearrangement of the flexible appendages of L is also reflected in the size-exclusion chromatography profile, in which L–PNTD elutes over a notably narrower range than L alone (Fig S1B). However, in contrast to the L–P complex, the L–PNTD complex consists solely of monomers, providing evidence that the dimers observed with full-length P are mediated by its oligomerization domain. Therefore, PNTD is sufficient to alter the conformation of L.
PNTD Residues 81–106 Are Sufficient to Stimulate L Processivity on a Nonencapsidated RNA Template.
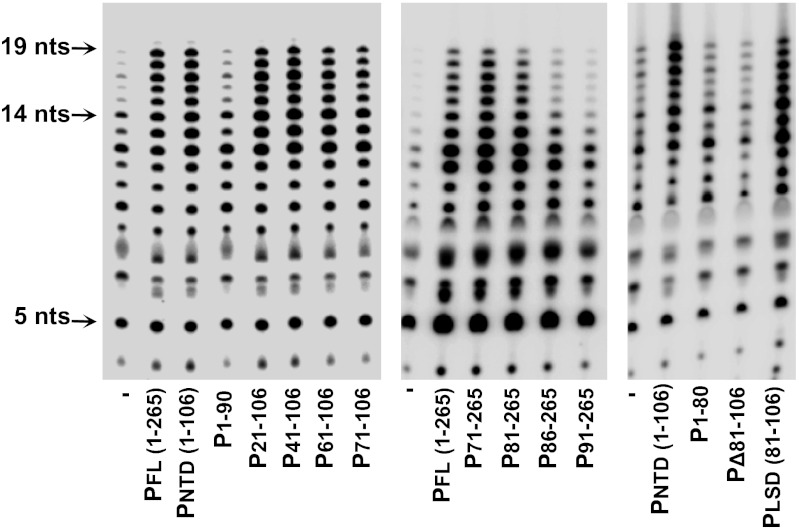
To further define the elements of PNTD that stimulate the processivity of L on the Le19 template, a series of sequential deletions of PNTD were expressed in E. coli and purified (Fig. S2A). Whereas P1–90 was inactive in stimulating L processivity, P21–106 and P41–106 retained full activity, and P61–106 and P71–106 exhibited a modest decrease in activity (Fig. 2, Left and Fig. S2B). To further refine the N-terminal boundary, a series of N-terminal truncations of P were expressed and purified (Fig. S2 A and B). Whereas P81–265 stimulated L processivity comparably to full-length P (1–265), P86–265 exhibited decreased L stimulation and P91–265 failed to stimulate L (Fig. 2, Middle). Collectively, these results indicate that P residues 81–106 constitute the element necessary for stimulation of L processivity. Consistent with this conclusion, P1-80 as well as an internal deletion in full-length P (PΔ81–106) lacked any stimulatory activity, and a synthetic peptide consisting of residues 81–106 retained a stimulatory activity comparable to that of full-length P (Fig. 2, Right and Fig. S2B). We termed the P81–106 peptide PLSD (for L stimulatory domain). Although PLSD (81–106) stimulated the activity of L similarly to full-length P, we noted the appearance of slightly shorter products (12–14 nt), which might reflect decreased binding efficiency of PLSD to L (Fig. 3).
Fig. 2.
Determining the region of PNTD required for stimulation of L processivity on the Le19 template. Here 0.2 μM P, PNTD, or each of the deletion mutants of the PNTD (Left) or N-terminal deletions of full-length P (Middle) were analyzed for stimulating L processivity as described in Fig 1B. (Right) An internal deletion of residues 81–106 in full-length P (PΔ81–106) and a chemically synthesized peptide comprising residues 81–106 of P (PLSD) (both 0.2 μM) were tested as well. Quantitative analysis of two independent experiments is presented in Fig S2B.
Fig. 3.
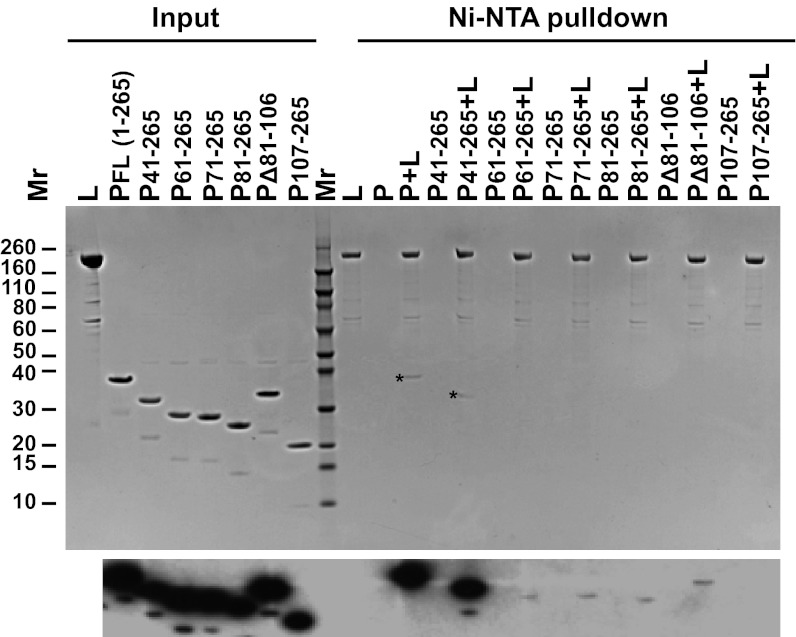
Delineating the region of P involved in L binding. 6xHis-tagged L was mixed with a fourfold molar excess of full-length P or with each of the N-terminal or internal deletions of P. The complexes were precipitated using Ni-NTA agarose beads, separated on 4–12% SDS/PAGE, and visualized by Coomassie blue staining (precipitated P proteins indicated by *) (Upper) or evaluated by Western blot analysis using a polyclonal antibody against P (Lower). For Western blot analysis, 20% of the input was loaded. FL, full-length. Quantitative analysis of this experiment is presented in Fig S3B.
L Binding Encompasses the Region Between the N0-Binding Domain and the Oligomerization Domain of P.
Earlier work determined that PNTD binds L (11); however, the region within P that binds L was not mapped precisely. Given that single amino acid changes in P that disrupt L binding have not been identified, we elected to use deletion mutants in P to map the L-binding domain. To delineate the region within the PNTD that binds L, we used His-tagged L as bait to precipitate untagged deletion mutants of full-length P (Fig. 3 and Fig. S3). As expected, deletion of the entire PNTD (P107–265) completely abolished binding. Deletion of the N0-binding domain of P (P41–265) had no effect, whereas larger N-terminal deletions (P61–265, P71–265, and P81–265) significantly decreased P binding to L. Consistent with the critical nature of the PLSD in L stimulation, an internal deletion (PΔ81–106) also significantly decreased L binding. These results indicate that whereas PLSD is sufficient for stimulation of L processivity on Le19, the L-binding region on P spans a larger interval between the N0-binding domain and the oligomerization domain.
PNTD Region Required to Induce Conformational Rearrangement in L.
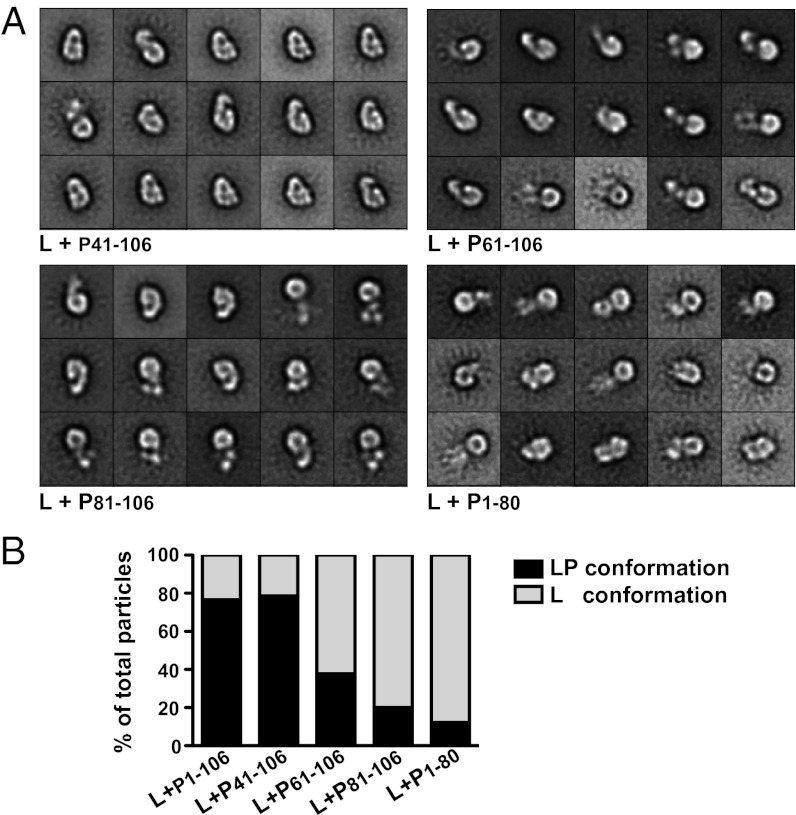
We next used EM to examine the contribution of elements of PNTD to the observed structural rearrangement in L. To do this, we mixed L with PNTD mutants harboring the indicated deletions and calculated 20 class averages for each sample (Fig. 4A and Fig. S4). In Fig. 4A, the 15 classes shown for each sample are arranged in order of decreasing abundance from left to right and top to bottom. Individual class averages from each sample were visually scored as L conformation or L–P conformation, and the percentage of each conformation relative to the total number of particles was calculated (Fig. 4B). The class averages illustrate that deletions up to the N0-binding domain of PNTD (P 41–106) retained the ability to induce the conformational rearrangement in L to a similar extent as PNTD. Further deletions in the PNTD (P61–106 and P81–106) decreased the population of L exhibiting a conformational rearrangement. Similarly, deletion of PLSD (P1–80) inhibited the conformational rearrangement of L, and this inhibition could not be rescued by supplementation of PLSD (P1–80 + P81–106) (Fig. S4). Collectively, these results indicate that the entire L-binding region of P (41–106) appears to be necessary to elicit a conformational rearrangement in L.
Fig. 4.
EM characterization of the effect of PNTD deletion mutants in inducing a conformational rearrangement in L. (A) L was mixed with each PNTD deletion mutant at a 1:10 molar ratio, and the resulting complexes were analyzed by EM. The averages of the 15 most populous classes obtained by classification of the particles into 20 classes are shown. The side length of the individual panels is 29 nm. (B) Individual class averages from two independent experiments (Fig. S3) were scored as L conformation or LP conformation. The average percentage of each conformation relative to the total number of particles in each sample is plotted.
Level of Stimulation of L Activity on N-RNA Correlates with Extent of Conformational Rearrangement in L Induced by Elements of PNTD.
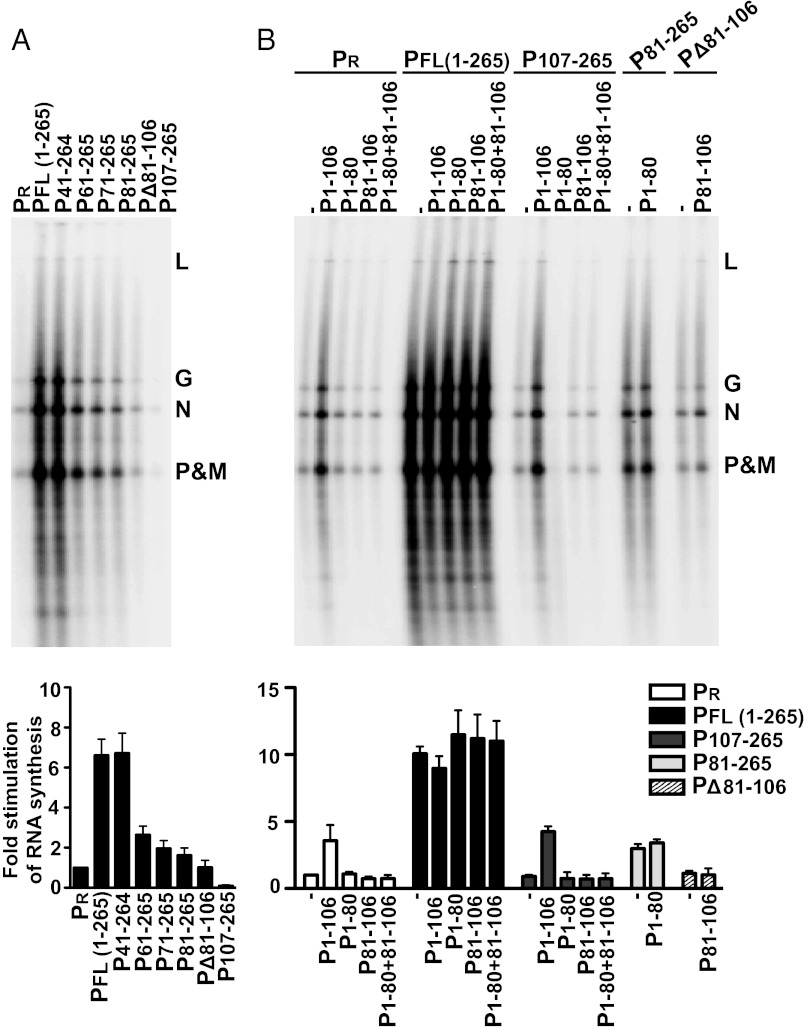
To determine the impact of altering the conformational state of L on its RdRP function during RNA synthesis, we used an in vitro transcription assay reconstituted with individually purified N-RNA, L, and P or sequential N-terminal deletions of full-length P. We analyzed deletions of the PNTD in the context of full-length P, because PCTD is required to bind the template-associated N. We isolated N-RNA from detergent-disrupted purified virus using a standard protocol of high-salt treatment followed by double isolation on a CsCl gradient (26). This purification removes virtually all of the endogenous P protein; nevertheless, trace amounts of P (PR) remained bound, which we estimated as ∼16.67 nM (Fig. S5A). PR resulted in a basal level of transcription that was stimulated up to 8- to 10-fold by 333.4 nM exogenous purified P (Fig. S5B). We tested each of the P deletion mutants at a saturating concentration of 1,667 nM (Fig. 5A). Deletion of the N0-binding domain (P41–265) resulted in transcription levels indistinguishable from those achieved by full-length P, indicating that the role of this domain is restricted to genome replication. Larger N-terminal deletions (P61–265, P71–265, and P81–265) resulted in a gradual decrease in stimulation of transcription relative to full-length P. The N-terminal deletions in P resulted in a more pronounced reduction of RNA synthesis on N-RNA than on Le19. This indicates that the conformational change in L mediated by P41–106 is critical for efficient use of the N-RNA template. Internal deletion of the PLSD (PΔ81–106) resulted in further decrease in stimulation, whereas deletion of the entire PNTD (P107–265) served as a dominant negative, inhibiting the basal levels of transcription resulting from PR. This likely reflects the ability of P107–265 to oligomerize with PR or to compete it off the template. The consequences of the deletion of elements of the PNTD on stimulation of RNA synthesis are correlated with the effect of these deletions on L binding and their ability to induce a conformational rearrangement in L.
Fig. 5.
Functional analysis of PNTD in transcription of an encapsidated N-RNA template. (A) 50 μL in vitro transcription reactions were reconstituted with 5 μg of N-RNA, 1 μg (4.16 pmol) of L, and 83.2 pmol P or each of the N-terminal or internal deletions of P. The N-RNA template was isolated from purified virus and retained 0.22 pmol of residual P (PR)/μg of purified N-RNA, resulting in a final concentration of 16.67 nM per reaction (Fig S5A). Transcription reactions were performed in the presence of [α-32P] GTP. The products were separated by electrophoresis on acid-agarose gels and analyzed with a PhosphorImager. The five VSV mRNAs P, M (matrix), N, G (glycoprotein), and L are indicated to the right. The total amount of synthesis was quantified by summing the band intensities, normalized to levels of RNA synthesis produced by PR, and graphed. Error bars represent the SD from the mean of three independent experiments. (B) In vitro transcription reactions were reconstituted with 5 μg of N-RNA, 1 μg (4.16 pmol) of L, and 16.64 pmol of P, P107–265, P81–265, or PΔ81–106, with 83.2 pmol of P1–106, P1–80, or P81–106 added to reactions when indicated. RNA synthesis was analyzed as in A.
P-Induced Conformational Change in L Stimulates Transcription Independent of P Oligomerization.
To further separate the impacts of conformational change and oligomerization status of P on RdRP function, we tested the effect of the isolated PNTD (P1–106) on transcription levels (Fig. 5B and Fig. S5B). P1–106 stimulated the basal level of transcription resulting from PR by fourfold to fivefold at concentrations ranging of 333.4–1,667 nM (Fig S5B). This stimulatory effect is likely reflective of a dynamic exchange of L between PNTD and full-length P, where high concentrations of PNTD stimulate the RdRP that is already engaged with the N-RNA via full-length P. Consistent with this idea, no further stimulation by PNTD was observed in the presence of optimal amounts of exogenous P (Fig. 5B). Furthermore, PNTD stimulated transcription between fourfold and fivefold in the presence of 333.4 nM of P107–265. Because P107–265 contains the oligomerization domain but lacks the L-binding domain, the conformational change induced in L by the isolated PNTD is sufficient to produce the enhanced activity. Fragments of P that were insufficient to induce the conformational change of L (P1–80, PLSD, or a combination of the two) had no effect on transcription in the presence of PR, exogenous P, or P107–265 (Fig. 5B). Furthermore, P1–80 and PLSD failed to complement the corresponding deletion mutants of full-length P (P81–265 and PΔ81–106, respectively). These data indicate that the stimulatory effect of P on L function in transcription is independent of the oligomeric status of P. Consistent with this interpretation, PNTD+CD stimulated basal transcription to a similar degree as PNTD, and this stimulation was not enhanced by the addition of PCTD or PCD+CTD (Fig. S6). Collectively, these results demonstrate that PNTD, a domain that is sufficient for inducing a conformational rearrangement in L, can stimulate L-mediated transcription of an encapsidated RNA template.
Discussion
This study provides a detailed analysis of the elements of P that are involved in the functional interaction with L and their effect on the architectural organization and RNA synthesis activity of the RdRP. The region of P that mediates the functional interaction with L spans residues 41–106 of the PNTD. Whereas this entire region appears to be necessary for inducing a conformational rearrangement in L and stimulating RNA synthesis on the viral encapsidated N-RNA template, a subdomain of this region (PLSD, residues 81–106) is sufficient for stimulating L processivity on a short naked RNA template. We posit a model in which the effects of P on RdRP are separated into (i) stimulation of L processivity in directly copying an RNA substrate that is likely mediated through a local effect on the polymerase active site and (ii) stimulation of L activity on the encapsidated template, which requires a global rearrangement in the architecture of L that may be necessary for correct positioning of L in coordination with the N molecules in its vicinity and/or for coordination of the various enzymatic activities of L during viral RNA synthesis.
L-Binding Region of P.
The results of this study demonstrate that the L-binding region of P resides within P residues 41–106. There are no available structural data for this region. Bioinformatic analysis predicts that residues 40–90 constitute an IDR, and that residues 91–106 constitute a structured domain speculated to be the binding site for L (15). Further analysis using the PHYRE protein homology/analogy recognition engine (http://www.sbg.bio.ic.ac.uk/∼phyre2) predicted an α-helical region at residues 79–100. These analyses suggest that PLSD (P81–106) is likely a structured domain of PNTD.
Recent reports have described a disorder-to-order transition governing the binding of N and P in paramyxoviruses, suggesting that IDRs play a role in establishing multiple molecular partnerships (27). These mechanisms are proposed to be advantageous for achieving pleiotropy and genetic compaction for the virus. Such molecular flexibility also might be required to support the roles of PNTD as a chaperone for N0 and as a binding site for L. The atomic structure of the N0–P1–60 complex reveals that P residues 6–40 form a molecular recognition element that engages N0, with amino acids 1–5 and 41–60 remaining flexible. Those flexible regions of P were suggested to act as “entropic bristles” that repel incoming RNA or N molecules or mask their binding interfaces (21). In the present study, the dispensability of the N0-binding molecular recognition element during transcription (P41–265) demonstrates the lack of effect of this region on polymerase function and is consistent with a primary role in the chaperoning N0 during replication. Whether a single PNTD can simultaneously bind N0 and L remains unknown; however, the close proximity of the two binding regions makes such a scenario unlikely. Perhaps this need for PNTD to interact with two molecular partners explains one of the requirements for oligomerization of P, with one PNTD binding L and a second PNTD binding the N0 necessary for encapsidation of the nascent RNA during replication.
The prediction that P81–106 is a structured domain is supported by the demonstration that PLSD (P81–106) as a discrete domain stimulates processivity of L on the naked RNA template. Independent folding of PLSD would be consistent with the retention of its functionality when separated from the rest of the L-binding region. The significant decrease in L binding resulting from N-terminal deletions downstream of amino acid 41, as well as from an internal deletion of PLSD, suggests that the binding of P41–106 to L is likely a cooperative mechanism involving the IDR (P41–80) and PLSD. Such a mechanism is also consistent with the failure to restore the conformational change and the stimulation of RNA synthesis on N-RNA induced by P1–106 by complementation of P1–80 and PLSD.
Stimulation of L Processivity by PLSD.
PLSD is sufficient for stimulating L processivity on a short naked RNA template, independent of the rest of the L-binding region. The mechanism by which PLSD stimulates L processivity is not clear. EM analysis of L in the presence of PLSD showed no significant change in the conformation of L, suggesting a lack of correlation between stimulation of L processivity by PLSD and the global rearrangement in L conformation. However, the possibility of a more subtle or local alteration of L conformation that is not discernible by EM at the current resolution cannot be ruled out. For a mechanistic understanding of the effect of PLSD on polymerase processivity, kinetic analysis of the effects of P and PLSD on the affinity of the polymerase for the template and on the rate of polymerase elongation is needed. However, the retained ability of PLSD to stimulate L processivity on Le19 indicates that such a mechanism does not require a tight association between L and P. In contrast, the entire L-binding region is required for stimulation of polymerase activity on N-RNA, indicating a requirement for a tighter binding in this latter case.
Role of PNTD in Stimulating L Activity on the Encapsidated N-RNA Template.
Our results point to a correlation between the conformational rearrangement in L and stimulation of its activity on N-RNA. Specifically we found that (i) N-terminal deletions in full-length P downstream of the N0-binding site resulted in a gradual decrease in stimulation of the RNA synthesis activity on the N-RNA template; (ii) an internal deletion of PLSD also significantly reduced synthesis; and (iii) similar deletions in PNTD resulted in a gradual decrease in the population of L exhibiting a conformational rearrangement similar to that induced by PNTD. However, a conformational rearrangement in L per se is not necessary for stimulation of processivity, given that L processivity on an nonencapsidated 19-nt RNA was dependent only on the PLSD, which did not induce a significant alteration in the molecular architecture of L.
Collectively, our data support a model in which the conformational rearrangement in L is necessary for stimulation of polymerase activity on the encapsidated RNA. A notable feature of NNS virus genomic RNA is its resistance to digestion by nucleases even during the process of RNA synthesis (28), indicating a tight spatial regulation coordinating the release of a stretch of RNA from N to be copied by L. Thus, a global change in the conformation of L controlling the flexibility of the appendage of L might be required for precise positioning of L on the template in coordination with the nucleocapsid. The conformational rearrangement also could be necessary for spatial and temporal control of the cap-modifying activities during replication and transcription. On a short naked RNA such as Le19, the conformational rearrangement has no effect on RNA synthesis, because these levels of control are irrelevant. Therefore, this independent role of PLSD in stimulating RNA synthesis on Le19 might reflect an evolutionary pathway in which a processivity factor sufficient to stimulate polymerase activity on naked RNA is concatenated to additional regions that mediate RdRP activity on an encapsidated template. Finally, although the phosphorylation status of P does not impact polymerase activity in the in vitro assays used in this study, phosphorylation of P is essential for virus recovery from infected cells (29), indicating a key role for phosphorylation in regulating P function.
In summary, our work provides insight into the mechanisms by which the VSV P protein facilitates RdRP function of the L protein. A full understanding of the dynamic role of P in RNA synthesis will require knowledge not only of the atomic-level structure of L, but also of how the L–P complex accesses the N-RNA. This work provides important tools for examining these issues by providing a better molecular understanding of the function of specific regions of P and how they influence the architecture of L. Because the process of RNA synthesis involves dynamic interactions between multiple molecular partners, further studies of the spatial and temporal regulation of this complex RNA synthesis machinery also will likely benefit from the application of single-molecule enzymology approaches.
Materials and Methods
Protein Expression and Purification.
Proteins were expressed and purified as detailed in SI Materials and Methods.
Peptide Synthesis.
PLSD (AEQVEGFIQGPLDDYADEDVDVVFTS) was synthesized with standard Fmoc chemistry on ABI 431 Peptide Synthesizers at the Tufts University Core Facility, purified using reverse-phase HPLC, and analyzed by MS.
RNA Synthesis Assays.
RNA synthesis on Le19.
Le19 was chemically synthesized and PAGE-purified (Integrated DNA Technologies). Polymerase reactions were performed as described previously (25) using 0.2 μM Le19 and 0.2 μM L, with 0.2 μM P or P deletions added when indicated. Reactions were incubated for 3 h, and products were resolved by 20% (wt/vol) polyacrylamide/7 M urea gel electrophoresis and analyzed with a PhosphorImager (GE Healthcare).
In vitro transcription of N-RNA.
Standard in vitro transcription reactions were carried out as described previously (24) using 5 μg of N-RNA, 1 μg (4.16 pmol) of L, and the indicated concentrations of P or P deletions. The products were purified by RNeasy (Qiagen), separated by acid-agarose urea gel electrophoresis, and analyzed with a PhosphorImager.
EM and Image Processing.
L (0.14 μM) was mixed with PNTD deletions (1.4 μM) in buffer containing 50 mM Tris-HCl (pH 7.4), 280 mM NaCl, 3% (wt/vol) glycerol, and 1 mM DTT and incubated on ice for 1 h. Samples were diluted 1:30–1:50 in binding buffer without glycerol and adsorbed to glow-discharged, carbon-coated EM grids, then stained with 0.75% (wt/vol) uranyl formate as described previously (30). Images were collected and processed as described in detail in SI Materials and Methods.
Ni-Nitrilotriacetic Acid Pulldown Assay.
For this assay, 3 μg (12.5 pmol) of His-tagged L was incubated with 50 pmol of untagged P or P deletions, and the formed complex was pulled down by Ni-nitrilotriacetic acid (NTA) agarose, as described in SI Materials and Methods. The precipitated proteins were separated on 4–12% (wt/vol) SDS/PAGE gels and visualized by Coomassie blue staining or transferred to a nitrocellulose membrane for Western blot analysis with an anti-P antibody, as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Robin Ross and Benjamin Seiler for their exceptional support in protein production as part of Core D of the New England Regional Center of Excellence–Biodefense and Emerging Infectious Diseases. We thank Amy Lee, Silvia Piccinotti, Matthijs Raaben, and Philip Kranzusch for their critical review of the manuscript. This study was supported by National Institutes of Health Grants AI059371 and AI057159. S.P.J.W. is the recipient of a Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award. A.D.S. is supported by a Swiss National Science Foundation fellowship. T.W. is a Howard Hughes Medical Institute investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209147109/-/DCSupplemental.
References
- 1.Emerson SU, Wagner RR. Dissociation and reconstitution of the transcriptase and template activities of vesicular stomatitis B and T virions. J Virol. 1972;10:297–309. doi: 10.1128/jvi.10.2.297-309.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee AK, Rhodes DP. In vitro synthesis of RNA that contains polyadenylate by virion-associated RNA polymerase of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1973;70:3566–3570. doi: 10.1073/pnas.70.12.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emerson SU, Wagner RR. L protein requirement for in vitro RNA synthesis by vesicular stomatitis virus. J Virol. 1973;12:1325–1335. doi: 10.1128/jvi.12.6.1325-1335.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green TJ, Zhang X, Wertz GW, Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- 5.Albertini AA, et al. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313:360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 6.Sleat DE, Banerjee AK. Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J Virol. 1993;67:1334–1339. doi: 10.1128/jvi.67.3.1334-1339.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogino T, Banerjee AK. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol Cell. 2007;25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Fontaine-Rodriguez EC, Whelan SP. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J Virol. 2005;79:13373–13384. doi: 10.1128/JVI.79.21.13373-13384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barr JN, Whelan SP, Wertz GW. Transcriptional control of the RNA-dependent RNA polymerase of vesicular stomatitis virus. Biochim Biophys Acta. 2002;1577:337–353. doi: 10.1016/s0167-4781(02)00462-1. [DOI] [PubMed] [Google Scholar]
- 10.Rahmeh AA, Li J, Kranzusch PJ, Whelan SP. Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J Virol. 2009;83:11043–11050. doi: 10.1128/JVI.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerson SU, Schubert M. Location of the binding domains for the RNA polymerase L and the ribonucleocapsid template within different halves of the NS phosphoprotein of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1987;84:5655–5659. doi: 10.1073/pnas.84.16.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellon MG, Emerson SU. Rebinding of transcriptase components (L and NS proteins) to the nucleocapsid template of vesicular stomatitis virus. J Virol. 1978;27:560–567. doi: 10.1128/jvi.27.3.560-567.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Rahmeh A, Morelli M, Whelan SP. A conserved motif in region v of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J Virol. 2008;82:775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding H, Green TJ, Lu S, Luo M. Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J Virol. 2006;80:2808–2814. doi: 10.1128/JVI.80.6.2808-2814.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leyrat C, et al. Structural disorder in proteins of the rhabdoviridae replication complex. Protein Pept Lett. 2010;17:979–987. doi: 10.2174/092986610791498939. [DOI] [PubMed] [Google Scholar]
- 16.Howard M, Wertz G. Vesicular stomatitis virus RNA replication: A role for the NS protein. J Gen Virol. 1989;70:2683–2694. doi: 10.1099/0022-1317-70-10-2683. [DOI] [PubMed] [Google Scholar]
- 17.Chen M, Ogino T, Banerjee AK. Interaction of vesicular stomatitis virus P and N proteins: Identification of two overlapping domains at the N terminus of P that are involved in N0-P complex formation and encapsidation of viral genome RNA. J Virol. 2007;81:13478–13485. doi: 10.1128/JVI.01244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green TJ, Luo M. Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. Proc Natl Acad Sci USA. 2009;106:11713–11718. doi: 10.1073/pnas.0903228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerard FC, et al. Unphosphorylated rhabdoviridae phosphoproteins form elongated dimers in solution. Biochemistry. 2007;46:10328–10338. doi: 10.1021/bi7007799. [DOI] [PubMed] [Google Scholar]
- 20.Kolakofsky D, Le Mercier P, Iseni F, Garcin D. Viral DNA polymerase scanning and the gymnastics of Sendai virus RNA synthesis. Virology. 2004;318:463–473. doi: 10.1016/j.virol.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 21.Leyrat C, et al. Structure of the vesicular stomatitis virus N0-P complex. PLoS Pathog. 2011;7:e1002248. doi: 10.1371/journal.ppat.1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spadafora D, Canter DM, Jackson RL, Perrault J. Constitutive phosphorylation of the vesicular stomatitis virus P protein modulates polymerase complex formation but is not essential for transcription or replication. J Virol. 1996;70:4538–4548. doi: 10.1128/jvi.70.7.4538-4548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chattopadhyay D, Banerjee AK. Phosphorylation within a specific domain of the phosphoprotein of vesicular stomatitis virus regulates transcription in vitro. Cell. 1987;49:407–414. doi: 10.1016/0092-8674(87)90293-5. [DOI] [PubMed] [Google Scholar]
- 24.Rahmeh AA, et al. Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc Natl Acad Sci USA. 2010;107:20075–20080. doi: 10.1073/pnas.1013559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morin B, Rahmeh AA, Whelan SP. Mechanism of RNA synthesis initiation by the vesicular stomatitis virus polymerase. EMBO J. 2012;31:1320–1329. doi: 10.1038/emboj.2011.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emerson SU, Yu Y. Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J Virol. 1975;15:1348–1356. doi: 10.1128/jvi.15.6.1348-1356.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Habchi J, Longhi S. Structural disorder within paramyxovirus nucleoproteins and phosphoproteins. Mol Biosyst. 2012;8:69–81. doi: 10.1039/c1mb05204g. [DOI] [PubMed] [Google Scholar]
- 28.Green TJ, et al. Access to RNA encapsidated in the nucleocapsid of vesicular stomatitis virus. J Virol. 2011;85:2714–2722. doi: 10.1128/JVI.01927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das SC, Pattnaik AK. Phosphorylation of vesicular stomatitis virus phosphoprotein P is indispensable for virus growth. J Virol. 2004;78:6420–6430. doi: 10.1128/JVI.78.12.6420-6430.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification: Powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.