Abstract
Early in the development of plant evolutionary biology, genetic drift, fluctuations in population size, and isolation were identified as critical processes that affect the course of evolution in plant species. Attempts to assess these processes in natural populations became possible only with the development of neutral genetic markers in the 1960s. More recently, the application of historically ordered neutral molecular variation (within the conceptual framework of coalescent theory) has allowed a reevaluation of these microevolutionary processes. Gene genealogies trace the evolutionary relationships among haplotypes (alleles) with populations. Processes such as selection, fluctuation in population size, and population substructuring affect the geographical and genealogical relationships among these alleles. Therefore, examination of these genealogical data can provide insights into the evolutionary history of a species. For example, studies of Arabidopsis thaliana have suggested that this species underwent rapid expansion, with populations showing little genetic differentiation. The new discipline of phylogeography examines the distribution of allele genealogies in an explicit geographical context. Phylogeographic studies of plants have documented the recolonization of European tree species from refugia subsequent to Pleistocene glaciation, and such studies have been instructive in understanding the origin and domestication of the crop cassava. Currently, several technical limitations hinder the widespread application of a genealogical approach to plant evolutionary studies. However, as these technical issues are solved, a genealogical approach holds great promise for understanding these previously elusive processes in plant evolution.
In the following succinct statements, G. L. Stebbins presents what would become the framework for the study of plant evolutionary mechanisms for the next 50 years (1).
Individual variation, in the form of mutation and gene recombination, exists in all populations; … the molding of this raw material … into variation on the level of populations by means of natural selection, fluctuation in population size, random fixation and isolation is sufficient to account for all of the differences, both adaptive and non-adaptive, which exist between related races and species … .
The problem of the evolutionist is … evaluating on the basis of all available evidence the role which each of these known forces has played in any particular evolutionary line … .
A central thesis of Stebbins' seminal book, Variation and Evolution in Plants (1), is the notion that to understand evolution we must examine its action at the level of populations within species. This reasoning may seem obvious to contemporary readers, but at the time of Stebbins' writing, the importance of population-level processes for evolution was far from apparent. Stebbins' elucidation of this connection is one of his most enduring contributions to plant evolutionary biology.
Fifty years ago, the study of plant evolution was necessarily concerned with the phenotype, much of which is subject to selection. Morphology, karyotypes, and fitness components are central traits for understanding evolution and adaptation, but they limit which evolutionary processes can be studied. In his book, Stebbins discusses such events as fluctuations in population size, random fixation (genetic drift), and isolation as all affecting the process of evolution (see passage quoted above; ref. 1). However, the study of these mechanisms requires markers that are not under selection.
In the years after the publication of Stebbins' book, among the first major technical advances in evolutionary biology were the development of protein electrophoresis and the identification of allozyme variation in natural populations. Many allozymes are selectively neutral, and thus, for the first time, evolutionary biologists could attempt to assess the amount of neutral genetic variation within species as well as its spatial distribution. Plant species were found to vary widely, both in levels of genetic variation and in the apportionment of this variation within and among populations. These observations spurred researchers to examine the mechanisms underlying the process of genetic differentiation in plants. One of the most common approaches for doing this analysis has been to look for correlations between the life history characteristics of a species (e.g., mechanisms of pollen and seed dispersal, system of mating, and generation length) and patterns of population genetic differentiation (2). Neutral allelic variation from allozymes also can be used to estimate levels of gene flow among populations. The population genetic theory developed by Wright, Fisher, and Malecot (among others) established that, for a group of populations at equilibrium, the level of genetic differentiation is roughly inversely proportional to the level of interpopulation gene flow per generation. This relationship is expressed by Wright's (3) familiar equation for estimating gene flow under an island model: FST ≈ 1/(4Nem + 1), where FST is the standardized variance in allele frequencies among populations, Ne is the effective population size, and m is the migration rate.
The use of allozymes has led to more than 30 years of insight into how plant populations evolve. However, inferring population structure solely from allele frequencies has its limitations. Allozyme alleles (or their DNA analogs, restriction fragment length polymorphisms, amplified fragment length polymorphisms, and microsatellites) are unordered, meaning that the genealogical pattern of relationships among alleles cannot be easily inferred. As a result, these data cannot be used in directly assessing genetic change over time but rather require indirect approaches based on models that often assume equilibrium conditions. For example, Wright's equation (above) for quantifying gene flow under the island model assumes that populations have reached an equilibrium between gene flow and random genetic drift. This equilibrium perspective can be biologically misleading, particularly for species in which recent history is a major determinant of population structure. We speculate that very few plant species have reached, or will ever reach, a gene flow-drift equilibrium. Many plants, both temperate and tropical, have altered their range subsequent to glaciations in the last 20,000 years, a recent event on an evolutionary time scale. Likewise, many plant species have a metapopulation structure with subpopulations continually being colonized, dispersing migrants, and going extinct. Such metapopulations may reach a system-wide equilibrium in which probabilities of extinction and recolonization are constant given enough time, but such a situation is unlikely considering the relatively short time frame of global climatic changes in the past. Equilibrium in plants is for the most part a theoretical construct with little relation to reality.
We would argue that the second major development since the publication of Stebbins' work has been the application of ordered, genealogical data to the study of population-level processes. This development, which has begun to reach its potential only in the last decade, was predicated on two major advances, one technical and the other conceptual. The technical advance has been the widespread availability of ordered genetic variation at the intraspecific level, typically in the form of DNA sequence variation or mapped restriction-site data. For such data, mutational differences among genetic variants indicate the patterns of relationship among variants. These data therefore provide the raw information needed to reconstruct genealogical relationships among alleles (i.e., gene trees).
The conceptual advance has been the application of coalescent theory to the study of microevolutionary processes (for a recent review, see ref. 4). For a population of constant size, new alleles are continually arising through mutation, and others are going extinct over successive generations (assuming neutrality). Therefore, the extant alleles of a gene in a population are all derived from (i.e., coalesce to) a single common ancestral allele that existed at some point in the past (Fig. 1). Coalescent theory provides a framework for studying the effects of population-level processes (e.g., population size fluctuations, selection, and gene flow) on the expected time to common ancestry of alleles within a gene tree. The application of gene genealogies to population genetics has allowed the study of population-level processes within a temporal, nonequilibrium framework. Thus, microevolution can be studied as a dynamic, historical process, changing over time within a species.
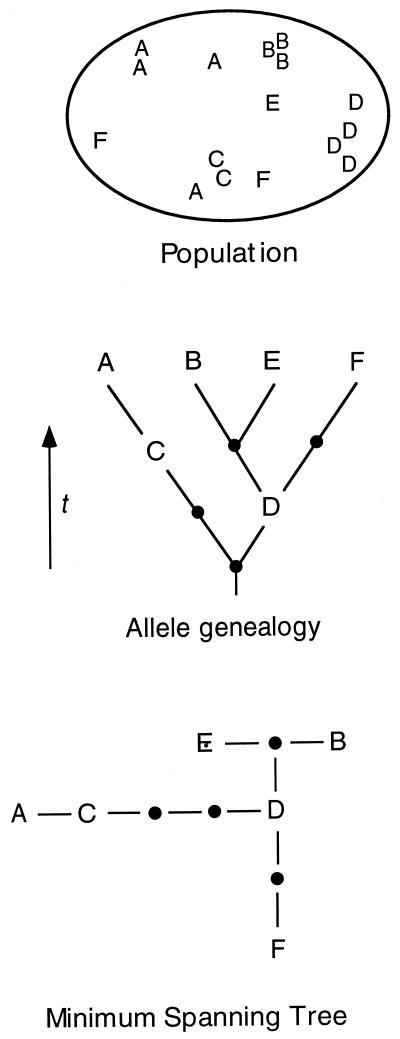
Figure 1.
Hypothetical population (Top) showing geographical distribution of alleles; allele genealogy (Middle) indicating true history of allelic divergence over time; unrooted minimum spanning tree (Bottom) showing inferred genealogical relationships among alleles.
At the foundation of all genealogical analyses is the gene tree, which represents the inferred genealogical relationships among alleles observed in a species. Most intraspecific gene trees are unrooted, because one often cannot determine the temporal polarity of mutations, even with an outgroup (5). A common means of representing the inferred genealogy is with a “minimum spanning tree” (e.g., ref. 6), for which the number of mutational changes among alleles is minimized (Fig. 1). If homoplasy (mutational convergence or reversal) is infrequent, then a single most parsimonious minimum spanning tree often can be inferred by using maximum parsimony search algorithms (7). For data showing high levels of homoplasy, more complicated tree estimation algorithms may be required (e.g., ref. 8). Extant alleles on a gene tree are often separated by more than one mutational step, and thus, the gene tree typically contains a number of inferred intermediate alleles; these unobserved alleles may be extinct, may have been missed during population sampling, or may never have existed at all (if mutations did not accumulate in single steps).
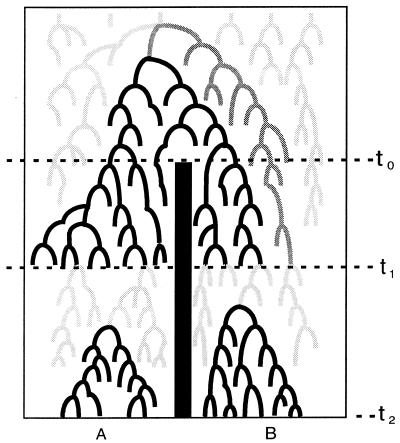
Allele genealogies can inform us about the effects of microevolutionary forces on organismal and population lineages. However, as has become well established in the last decade, a gene tree is far from equivalent to the population lineages through which it is transmitted (see review in ref. 9). Therefore, caution must be used in drawing inferences about population-level processes from genealogical data. The hypothetical allele genealogy in Fig. 2 illustrates the potential incongruity that can exist between a gene tree and the populations in which it exists. After two populations have become isolated from each other (and barring subsequent gene flow), the populations will diverge genetically until eventually all of the alleles within each population are more closely related to each other than to those from the other population (Fig. 2). At this point, the alleles show reciprocal monophyly with respect to the two populations and accurately reflect the history of population divergence. Before reaching reciprocal monophyly, however, alleles are expected to be polyphyletic, then paraphyletic, with respect to the populations (10). In these cases, genealogical relationships among alleles are not expected to correspond to population identity (Fig. 2). Thus, for recently diverged populations, inferences about the history of population divergence based on the gene tree may be misleading or erroneous. In some cases, ancestral allelic variation may actually persist in populations after population divergence. These shared ancestral polymorphisms can easily be misinterpreted as evidence of interpopulation gene flow.
Figure 2.
Hypothetical allele genealogy in populations A and B that became isolated from each other at time t0. At time t1, genealogical relationships show paraphyly with respect to the two populations. At time t2, alleles show reciprocal monophyly and are congruent with the history of population divergence.
Below, we present several studies that exemplify the usefulness of gene genealogies for studying population-level processes. We begin with several examples from Arabidopsis thaliana, including the homeotic loci APETALA3 and PISTILLATA and the disease-resistance locus RPS2, all of which are subject to selection. Then, we illustrate the utility of genealogies for tracing the postglacial range expansion in a variety of plant species. Finally, the usefulness of a genealogical approach for documenting crop origin is shown for cassava, a staple crop of the tropics.
Gene Genealogy: An Example from Arabidopsis.
The model plant, A. thaliana, is being used increasingly often for evolutionary studies. Arabidopsis offers many advantages as a study system, including its small size, simple genome, and rapid generation time. Molecular biologists have elucidated the function of many genes in Arabidopsis; mechanisms of development have been detailed, and the sequence of its genome is nearly complete. All of this work provides fertile ground for evolutionary biologists. There are an increasing number of excellent studies that use this information. For example, the role of homeotic genes in the development of floral structures has furthered understanding of tissue differentiation in plants; this work has provided the background for studies that investigate the evolutionary diversification of morphogenic pathways. Purugganan and Suddith (11) have examined the molecular evolution of the homeotic loci APETALA3 and PISTILLATA, which affect petal and stamen development in Arabidopsis flowers. They have compared the gene genealogies of these sequences with variation at five other nuclear loci of Arabidopsis. Based on an excess of low-frequency nucleotide polymorphisms and elevated within-species replacement polymorphisms, the authors conclude that A. thaliana has undergone rapid expansion in population numbers and size. Likewise, patterns of variation in restriction fragment length polymorphisms of several nuclear loci and the construction of a multilocus haplotype network have indicated that A. thaliana populations exhibit little to no geographical structuring (12), a conclusion that is consistent with rapid population expansion.
In the above examples from Arabidopsis, population history has strongly affected patterns of genealogy and molecular evolution. Other loci within the Arabidopsis genome may reflect different evolutionary processes, in particular selection. Arabidopsis has served as a model system for unraveling disease-resistance response, a trait presumed under strong selection. In Arabidopsis, the RPS2 gene is involved in the recognition of the plant pathogen Pseudomonas syringae pv. tomato. RPS2 interacts with an avirulence gene, avrRpt2, of the pathogen to initiate the cascade of events that led to disease resistance. Both the avirulence gene in the pathogen and the resistance gene in the host must be functional to elicit resistance. These genes interact in a specific “gene-for-gene” manner. The close relationship between avirulence genes and resistance genes as well as the obvious fitness consequences of resistance for a plant have led to speculation on the evolutionary dynamics of resistance genes.
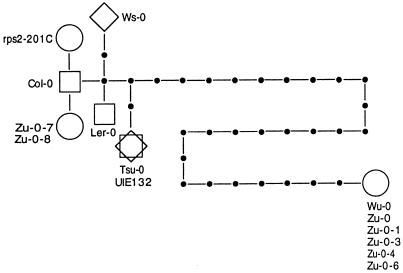
RPS2 encodes a 909-amino acid gene product. The gene contains several motifs that suggest it is part of a signaling pathway, including a leucine zipper, leucine-rich repeats, a hydrophobic region, and a nucleotide-binding site. A gene genealogy for the RPS2 locus has been constructed to investigate the molecular evolution of the gene (13); 17 accessions of A. thaliana, representing a diversity of ecotypes, were sequenced for RPS2, and their resistance to Pseudomonas was determined. The resulting genealogy reveals an intriguing pattern (Fig. 3). Disease-resistance haplotypes (alleles) are clustered on the gene tree, indicating that resistance haplotypes are closely related. In contrast, a susceptible ecotype is 23 mutational steps from this cluster (the other two susceptible ecotypes represent a mutation to a stop codon and a strain created by mutagenesis). Silent mutations are distributed more often on the long branch of the tree, whereas nonsilent mutations occur more frequently on the short branches of the genealogy. Such genealogies have potential for inferring gene function as well as unraveling the dynamics of molecular evolution.
Figure 3.
Gene tree for the RPS2 locus of A. thaliana. Open circles represent susceptible haplotypes; open squares are resistant haplotypes; and open diamonds are haplotypes intermediate in resistance. Closed circles are haplotypes not present in the sample but inferred from single-step mutations. The figure is modified from Caicedo et al. (13).
Phylogeography.
One of the most successful applications of genealogical methods in natural populations has been in field of phylogeography. The conceptual approach of phylogeography was pioneered by John Avise and colleagues (14). Avise (9) defines phylogeography as “a field of study concerned with the principles and processes governing the geographic distribution of genealogical lineages, especially those within and among closely related species.” Phylogeographic studies draw inferences about the history of population divergence based on associations between the geographical distribution of the alleles and their genealogical relationships. Because these studies are not based on equilibrium assumptions of gene flow and genetic drift, they have proved insightful in studying historical changes in patterns of gene flow, isolation, and secondary contact among divergent populations. The vast majority of phylogeography studies have focused on animal systems, and most of these have relied on the rapidly evolving regions of the mitochondrial genome as a source of genetic variation. Phylogeographic studies in plants have lagged behind those of animal studies, primarily because of difficulties in finding ordered, neutral intraspecific variation required for constructing gene trees (see Conclusions below).
Postglacial Migration.
Some of the most elegant studies of phylogeography in plants have examined the postglacial migration of species from Pleistocene refugia. A series of studies, using polymorphism in the chloroplast genome, on European trees, such as oaks (Quercus spp.), beech (Fagus sylvatica), and black alder (Alnus glutinosa), have shown similar patterns of variability; these species show a strong east–west cline in variation. Investigators interpret this cline as a result of postglacial migration from the same glacial refugia, leading to the concordance of variation patterns among species (15). Phylogeographic studies of eight oak species have demonstrated that recolonization of Europe subsequent to the last glaciation was from several refugia in the peninsulas of Iberia, Italy, and the Balkans (16). In this case, each refugium was represented by a distinct haplotype lineage. Fine-scale phylogeographic analysis further indicated that chloroplast DNA polymorphisms are shared between several oak species and, in this case, are attributed to hybridization and introgression subsequent to the recolonization of Europe. Similarly, chloroplast DNA analysis has shown concordance between beech (17) and black alder (18) phylogeography. Both species are believed to have colonized Europe after glaciation from a refugium in the Carpathian Mountains. Moreover, the data indicate that an additional refugium for these species in Italy did not contribute to the recolonization of Europe. Similar concordance in phylogeographic patterns associated with postglacial spread is observed between plant species in the Pacific Northwest of North America. Soltis et al. (19) have shown, via chloroplast DNA phylogenies, similar patterns in the structuring of variation among several different types of plants, including ferns, trees, and several members of the Saxifragaceae, suggesting that the present genetic structure of these species is strongly affected by their postglacial pattern of colonization.
Phylogeography and Plant Domestication: Manihot esculenta.
We have used a genealogical approach in examining two questions involving the species M. esculenta (Euphorbiaceae): the origin of the staple root crop cassava (M. esculenta subsp. esculenta) and the phylogeography of cassava's closest wild relative (M. esculenta subsp. flabellifolia). Cassava (manioc) is the sixth most important crop in the world (20). It is the primary source of calories in sub-Saharan Africa and serves as the main carbohydrate source for over 500 million people in the tropics worldwide (21, 22). Cassava is mostly grown by subsistence farmers, and despite its global importance as a food crop, it has traditionally received less attention by researchers than have temperate cereal crops. One fundamental question that has remained unresolved concerns the crop's geographical and evolutionary origins. Cassava was traditionally proposed to be a “compilospecies” derived from multiple hybridizing progenitor species in the genus Manihot (23, 24). Manihot includes ≈98 species occurring in both northern South America (≈80 spp.) and in Mexico/Central America (≈17 spp.); sites of domestication were proposed from much of this vast geographical area.
Traditional phylogenetic approaches were only partially successful in determining cassava's origin. Species of Manihot show low levels of divergence in both morphological and molecular characters, probably reflecting a recent diversification of the genus (25, 26). A phylogeny of the genus based on DNA sequences in the nuclear ribosomal ITS region (B.A.S., unpublished data) is not highly resolved but does place cassava in a clade of South American species. This finding was consistent with the proposition, based on morphological characters (27), that cassava is derived from a single wild South American progenitor (referred to as M. esculenta subsp. flabellifolia under present taxonomy).
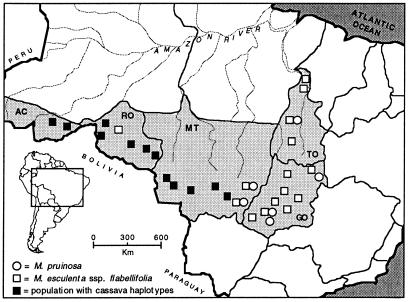
To test the hypothesis that cassava is derived from flabellifolia, we examined DNA sequence variation in 20 crop accessions, 27 populations of flabellifolia, and 6 populations of a closely related species, Manihot pruinosa, which has been proposed to hybridize with flabellifolia (28, 29). Populations of flabellifolia occur in mesic transitional forest patches in the ecotone between the lowland rainforest of the Amazon basin and the seasonally dry cerrado (savanna–scrub) found to the south and east on the Brazilian Shield plateau (Fig. 4). Populations were sampled in two transects, one along the southern border of the Amazon and the other along the eastern border. M. pruinosa is a cerrado species that occurs within the eastern range of flabellifolia. We included this species to test whether it has contributed to the genetic diversity of cassava through hybridization with flabellifolia.
Figure 4.
Locations of populations of M. esculenta subsp. flabellifolia (squares) and M. pruinosa (circles) sampled for the G3pdh phylogeography study. Shaded squares indicate populations containing one or more haplotypes found in domesticated cassava accessions. The figure is modified from Olsen and Schaal (26).
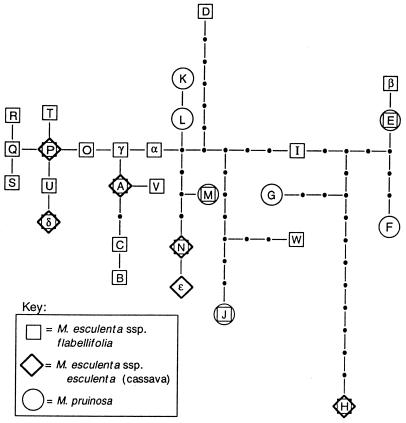
The study was based on sequence variation within a portion of the low-copy nuclear gene G3pdh, which encodes glyceraldehyde 3-phosphate dehydrogenase (26). Using primers designed by Strand et al. (30), we PCR amplified and sequenced a 962-bp region that spanned three exons, four introns, and parts of two flanking exons. From the 424 alleles (212 individuals) that were sequenced, we observed a total of 63 nucleotide polymorphisms, which characterized 28 different haplotypes. Maximum parsimony analysis yielded two negligibly different gene tree topologies, one of which is shown in Fig. 5.
Figure 5.
G3pdh gene tree for M. esculenta and M. pruinosa. Letters correspond to haplotype designations in GenBank accession numbers (AF136119–AF136149). Shapes around letters indicate the taxon or taxa in which a haplotype was found, as indicated in the key. The figure is modified from Olsen and Schaal (26).
Because the domestication of cassava is an extremely recent event evolutionarily speaking, one would not expect the divergence between cassava and flabellifolia to be reflected in the G3pdh gene tree. However, by looking at the haplotypes that are shared between cassava and the wild taxa and by examining the geographical locations of these haplotypes in the wild populations, we were able to draw several insights into the origin of the crop. First, we found that genetic variation in the crop is almost entirely a subset of that found in flabellifolia. Flabellifolia contains 24 haplotypes, of which 6 are found in cassava; cassava's haplotype diversity, therefore, represents 25% of that found in M. esculenta overall. Thus, the crop is most likely derived directly from flabellifolia, rather than from several hybridizing progenitor species as traditionally thought. In addition, we found that the cassava haplotypes occur in flabellifolia populations along the southern border of the Amazon basin and not along the eastern border. This finding points to the southern Amazonian region as the likely site of domestication of cassava. Interestingly, paleobotanical and other anthropological data indicate this region as a probable zone of domestication shared with peanut, two species of chili pepper, and jack bean (31). Finally, we found that none of the cassava haplotypes occur in M. pruinosa, suggesting that this species is not a progenitor of the crop. All of these conclusions are corroborated by an analysis of this same study system with microsatellite markers (K.M.O. and B.A.S., unpublished work).
The phylogeographic aspect of the study has focused on historical patterns of population divergence in flabellifolia and between flabellifolia and pruinosa. The distribution of the rainforest–cerrado ecotone where these species occur is likely to have shifted during the climatic changes of the Pleistocene (32, 33). Although there is not yet a consensus on the pattern or extent of habitat shifts, cooler/drier periods (associated with glaciations in temperate latitudes) are expected to have favored the expansion of cerrado and transitional forest; during warmer, humid periods (including the present), these habitats would be expected to be more restricted and fragmented as rainforest expanded. The repeated climate fluctuations of the Pleistocene are therefore predicted to have led to cycles of population fragmentation followed by range expansions and secondary contact in populations of flabellifolia and pruinosa. If these events have occurred, they should be reflected in the present phylogeographic structure of these taxa.
These hypotheses are being tested currently through a nested cladistic analysis (34) of the G3pdh data set (K.M.O., unpublished data). Although the statistical analyses are not complete, some preliminary insights are possible by visual inspection of the G3pdh gene tree. One interesting finding is that three haplotypes are shared between flabellifolia and pruinosa, suggesting interspecific introgression and/or shared ancestral polymorphisms that predate the divergence of these species. Two of the shared haplotypes (E and J) are common in eastern flabellifolia populations, and each is found in a single pruinosa individual from a population in close proximity to flabellifolia populations. This pattern suggests introgression from flabellifolia into M. pruinosa. The position of these haplotypes near the tips of the gene tree also favors this explanation over shared ancestral polymorphisms; tip haplotypes are likely to be younger than interior haplotypes (5) and therefore would be less likely to represent ancestral variation. The third shared haplotype (M) is also a tip haplotype. However, this haplotype is common in M. pruinosa and is found in a single flabellifolia population approximately 1,000 km west of the current range of M. pruinosa. Although clearly not the result of contemporary gene flow, this pattern could possibly have arisen through hybridization in the recent past. Palynological data indicate that during the last glacial maximum (<18,000 years B.P.), cerrado vegetation expanded into areas along the southern border of the Amazon basin that are presently rainforest (reviewed in ref. 33). Thus, hybridizing pruinosa populations could have existed in this region as recently as 11,000 years B.P.
Haplotypes on the G3pdh tree are not clustered by species (Fig. 4). Because flabellifolia and pruinosa are closely related taxa within a recently radiated genus, they would not necessarily be expected to have reached a pattern of reciprocal monophyly with respect to G3pdh haplotypes (Fig. 2). The phylogeographic structure within each species is also complex. However, although there is no simple concordance between the geographical distributions of haplotypes and their genealogical relationships, contingency analyses (35) reveal that nested clades within the gene tree are geographically structured. Thus, the phylogeographic structure reflects more than just the random sorting of ancestral polymorphisms among populations. Detailed phylogeographic analysis (34) and the analysis of DNA sequence data from two additional nuclear genes (K.M.O., unpublished data) will be useful in elucidating the historical processes that have led to the current phylogeographic structure in this study system.
Conclusions.
Gene genealogies have lead to several important insights into plant evolution and have the potential for far greater contributions. Many of the processes that affect the evolution of plant populations, such as selection, isolation, size fluctuations, and gene flow, are amenable to genealogical analysis. In particular, the use of genealogies within the framework of coalescence theory will allow us to understand in greater detail the role of historical fluctuations in population size, colonization, and range expansion. Although the large-scale metapopulation structure of many plants is clearly documented, there are relatively few studies of the genetic dynamics of this structure: colonization and establishment of subpopulations, gene migration, and extinction. The genetic aspects of such processes largely remain to be explored, and a historical genealogical approach will be particularly instructive.
The major impediment to wide application of gene genealogies for phylogeographic studies in plants is identifying DNA sequences with appropriate levels of ordered variation within chloroplast, mitochondrial, or nuclear genomes (36). In many cases, the chloroplast spacer regions that have been informative for some species (see above) show little to no intraspecific variation in other plant species. Moreover, chloroplast restriction fragment length polymorphism genealogies based on length variation alone can be confounded by homoplasy. The nuclear genome remains problematic because of the difficulty in finding regions that have sufficient levels of neutral variation and that are not involved in intragenic recombination. Moreover, the effective population size of a nuclear gene is four times that of an organelle gene, because it is diploid and biparentally inherited. The larger effective population size results in increased coalescent times, which in turn, increases the likelihood of encountering ancestral polymorphisms. High-resolution nuclear markers such as random amplified polymorphic DNAs and amplified fragment length polymorphisms are historically unordered, and variants cannot be related easily in a genealogical manner. Because of the difficulty in finding genealogically informative markers, many plant studies have been phylogeographic only in the broad sense, meaning that they detect an association between patterns of genetic variation and geography. Such studies do not incorporate a genealogical perspective.
The search for appropriate markers has turned to nuclear genes that are increasingly the focus for genealogical studies. Nuclear genes often contain multiple introns, and many of the introns contain high levels of neutral variation. This approach has been applied successfully in several animal species: e.g., oysters (37), fish (38), and birds (39). Nuclear sequences of plants have been used to understand the genetic relationships of wild populations of A. thaliana (12), selection, and evolution of homeotic genes (11), as well as in the example from cassava above. Numerous studies of nuclear gene genealogies are currently underway and promise to provide new insights into the processes identified by Stebbins a half century ago as central for the evolution of plants.
Acknowledgments
This work was supported in part by a grant from the Explorer's Club, by National Science Foundation Doctoral Dissertation Improvement Grant DEB 9801213 to K.M.O., and by grants from the Rockefeller and Guggenheim Foundations to B.A.S.
Footnotes
This paper was presented at the National Academy of Sciences colloquium “Variation and Evolution in Plants and Microorganisms: Toward a New Synthesis 50 Years After Stebbins,” held January 27–29, 2000, at the Arnold and Mabel Beckman Center in Irvine, CA.
References
- 1.Stebbins G L. Variation and Evolution in Plants. New York: Columbia Univ. Press; 1950. [Google Scholar]
- 2.Hamrick J, Godt M. In: Plant Population Genetics, Breeding, and Genetic Resources. Brown A, Clegg M, Kahler A, Weir B, editors. Sunderland, MA: Sinauer; 1989. [Google Scholar]
- 3.Wright S. Ann Eugen. 1951;15:322–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 4.Fu Y, Li W-H. Theor Popul Biol. 1999;56:1–10. doi: 10.1006/tpbi.1999.1421. [DOI] [PubMed] [Google Scholar]
- 5.Castelloe J, Templeton A R. Mol Phylogenet Evol. 1994;3:102–113. doi: 10.1006/mpev.1994.1013. [DOI] [PubMed] [Google Scholar]
- 6.Smouse P. Mol Ecol. 1998;7:399–412. [Google Scholar]
- 7.Swofford D. paup, Phylogenetic Inference Using Parsimony. Champaign, IL: Illinois Nat. Hist. Surv.; 1993. , Version 3.1. [Google Scholar]
- 8.Templeton A, Crandall K, Sing C. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avise J. Phylogeography: The History and Formation of Species. Cambridge, MA: Harvard Univ. Press; 2000. [Google Scholar]
- 10.Neigel J, Avise J. In: Evolutionary Processes and Theory. Karlin S, Nevo E, editors. New York: Academic; 1986. [Google Scholar]
- 11.Purugganan M, Suddith J. Genetics. 1999;151:839–848. doi: 10.1093/genetics/151.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergelson J, Stahl E, Dudek S, Kreitman M. Genetics. 1998;148:1289–1323. doi: 10.1093/genetics/148.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caicedo A, Schaal B, Kunkel B. Proc Natl Acad Sci USA. 1999;96:302–306. doi: 10.1073/pnas.96.1.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avise J, Arnold J, Ball R, Bermingham E, Lamb T, Neigel J E, Reeb C A, Saunders N C. Annu Rev Ecol Syst. 1987;18:489–522. [Google Scholar]
- 15.Newton A, Allnutt T, Gilles A, Lowe A, Ennos R. Trends Ecol Evol. 1999;14:140–145. doi: 10.1016/s0169-5347(98)01555-9. [DOI] [PubMed] [Google Scholar]
- 16.Dumolin-Lapegue S, Demesure B, Fineschi S, Le Corre V, Petit R. Genetics. 1997;146:1475–1487. doi: 10.1093/genetics/146.4.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demesure B, Comps B, Petit R. Evolution. 1996;50:2515–2520. doi: 10.1111/j.1558-5646.1996.tb03638.x. [DOI] [PubMed] [Google Scholar]
- 18.King R, Ferris C. Mol Ecol. 1998;7:1151–1163. [Google Scholar]
- 19.Soltis D, Gitzendanner M, Strenge D, Soltis P. Plant Syst Evol. 1997;206:353–373. [Google Scholar]
- 20.Mann C. Science. 1997;277:1038–1043. [Google Scholar]
- 21.Cock J. Cassava: New Potential for a Neglected Crop. London: Westfield; 1985. [Google Scholar]
- 22.Best R, Henry G. In: Report of the First Meeting of the International Network for Cassava Genetic Resources. Roca W M, Thro A M, editors. Cali, Colombia: Cent. Int. Agric. Trop.; 1992. [Google Scholar]
- 23.Sauer J. Historical Geography of Crop Plants. Boca Raton, FL: CRC; 1993. [Google Scholar]
- 24.Jennings D. In: Evolution of Crop Plants. Smartt J, Simmonds N, editors. New York: Wiley; 1995. [Google Scholar]
- 25.Rogers D, Appan S. Manihot and Manihotoides (Euphorbiaceae): A Computer Assisted Study. New York: Hafner; 1973. [Google Scholar]
- 26.Olsen K M, Schaal B A. Proc Natl Acad Sci USA. 1999;96:5586–5591. doi: 10.1073/pnas.96.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allem A. Genet Res Crop Evol. 1994;41:133–150. [Google Scholar]
- 28.Allem A. In: Report of the First Meeting of the International Network for Cassava Genetic Resources. Roca W M, Thro A M, editors. Cali, Colombia: Cent. Int. Agric. Trop.; 1992. [Google Scholar]
- 29.Allem A. Euphytica. 1999;107:123–133. [Google Scholar]
- 30.Strand A, Leebens-Mack J, Milligan B. Mol Ecol. 1997;6:113–118. doi: 10.1046/j.1365-294x.1997.00153.x. [DOI] [PubMed] [Google Scholar]
- 31.Piperno D, Pearsall D. The Origins of Agriculture in the Lowland Neotropics. New York: Academic; 1998. [Google Scholar]
- 32.Behling H. Rev Palaeobot Palynol. 1998;99:143–156. [Google Scholar]
- 33.Burnham R, Graham A. Ann Mo Bot Gard. 1999;86:546–589. [Google Scholar]
- 34.Templeton A, Routman E, Phillips C. Genetics. 1995;140:767–782. doi: 10.1093/genetics/140.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Posada D, Crandall K, Templeton A. geodis. Provo, UT: Brigham Young Univ.; 1999. , Version 2.0. [Google Scholar]
- 36.Schaal B A, Hayworth D A, Olsen K M, Rauscher J T, Smith W A. Mol Ecol. 1998;7:465–474. [Google Scholar]
- 37.Hare M, Avise J. Mol Biol Evol. 1998;15:119–128. doi: 10.1093/oxfordjournals.molbev.a025908. [DOI] [PubMed] [Google Scholar]
- 38.Bagley J, Gall G. Mol Ecol. 1998;7:945–961. [PubMed] [Google Scholar]
- 39.Degnan S. Mol Ecol. 1993;2:219–225. [Google Scholar]