Abstract
p73, a p53 family protein, shares significant sequence homolog and functional similarity with p53. However, unlike p53, p73 has at least seven alternatively spliced isoforms with different carboxyl termini (p73α-η). Moreover, the p73 gene can be transcribed from a cryptic promoter located in intron 3, producing seven more proteins (ΔNp73α-η). ΔNp73, which does not contain the N-terminal activation domain in p73, has been thought to be transcriptionally inactive and dominant negative over p53 or p73. To systemically analyze the activity of the ΔN variant, we generated stable cell lines, which inducibly express ΔNp73α, ΔNp73β, and various ΔNp73β mutants by using the tetracycline-inducible expression system. Surprisingly, we found that ΔNp73β is indeed active in inducing cell cycle arrest and apoptosis. Importantly, we found that, when ΔNp73β is expressed at a physiologically relevant level, it is capable of suppressing cell growth. We then demonstrated that these ΔNp73β activities are not cell type specific. We showed that the 13 unique residues at the N terminus are required for ΔNp73β to suppress cell growth. We also found that, among the 13 residues, residues 6 to 10 are critical to ΔNp73β function. Furthermore, we found that ΔNp73β is capable of inducing some p53 target genes, albeit to a lesser extent than does p73β. Finally, we found that the 13 unique residues, together with the N-terminal PXXP motifs, constitute a novel activation domain. Like ΔNp73β, ΔNp73γ is active in transactivation. However, unlike ΔNp73β, ΔNp73α is inactive in suppressing cell growth. Our data, together with others' previous findings, suggest that ΔNp73β may have distinct functions under certain cellular circumstances.
p73, along with p53 and p63, constitutes the p53 family. p73 shares 63% identity in amino acids with p53 in the DNA-binding domain, including all the DNA contact residues, 38% identity in the tetramerization domain, and 29% identity in the transactivation domain (31, 37, 55). In contrast to the human p53 gene, which is found to only encode one protein, human TP73 produces at least seven alternatively spliced isoforms with different carboxyl termini (p73α-η), termed the TA variant (10, 28, 38, 53). For example, p73α is the longest form of the p73 protein, which contains a sterile α motif (SAM domain) and an extreme C-terminal region, whereas p73β is a smaller polypeptide, missing the extreme C-terminal region and most of the SAM domain in p73α (8, 29, 31, 50). In addition to the alternative splicing in the C terminus, TP73 is also transcribed from a cryptic promoter located in intron 3, which gives rise to at least another seven isoforms (ΔNp73α-η), termed the ΔN variant (28, 55, 56, 58). The ΔN variant does not contain the activation domain in p73 due to lack of sequences encoded by exon 2 (45, 56). However, the ΔN variant acquires 13 unique residues at the N terminus compared with the TA variant (45, 56). Similar to TP73, TP63 encodes both TA and ΔN variants (53-55).
In addition to the significant sequence homology, p53 and p73 share a lot of functional similarities (2, 18, 26, 37, 38, 43, 53, 55). Previous studies showed that p73 can recognize and bind to p53-responsive elements (29). p73 is also able to activate several p53 target genes' promoters in a luciferase assay (11, 22). Overexpression of p73 in both p53+/+ and p53−/− cells promotes cell cycle arrest, apoptosis, and differentiation, as does p53 (1, 18, 29-31, 59). Despite the overlapping function in suppressing cell growth, p73 was found to differentially regulate some putative p53 target genes, which indicates that these proteins maintain separate and unique functions (4, 59). Moreover, p73 can be activated by various stress signals through pathways that are different from the ones that activate p53. For example, p73 can be activated by cisplatin and ionizing radiation in a manner that depends on the nonreceptor tyrosine kinase c-abl (1, 22, 57). Doxorubicin (DOX) stabilizes p73 by induction of p73 acetylation (9). We, and others later, have shown that p73 can be induced transcriptionally by p53, p73, and DNA damage (7, 27). Similarly, E2F1 can directly activate the transcription of the p73 gene via an E2F1-responsive element in the p73 promoter, which is responsible for the E2F1-induced p53-independent apoptosis (25, 49). p73 is also specifically regulated by the transcription repressor ZEB (20). Additionally, viral oncoproteins, such as simian virus 40 large T antigen, human papillomavirus E6, and adenovirus E1B, which efficiently inhibit p53 function, are unable to inactivate p73 (36). MDM2, an important regulator determining the half-life of p53, can bind to p73 and suppress its transcriptional activity but is incapable of targeting p73 for ubiquitination (24). These data suggest that p53 and p73 can be differentially activated and utilized in response to intracellular and extracellular stimuli.
Although p73 shows a significant functional resemblance to p53, it is still not certain whether p73 is a tumor suppressor. Present evidence indicates that p73 does not function as a classic Knudson-type tumor suppressor (53). For example, p73 mutations are extremely rare in human tumors (40, 41, 51). Furthermore, in contrast to p53 knockout mice, p73 knockout mice do not show an increased susceptibility to spontaneous tumors. Instead, these mice exhibit severe neurological defects, including hydrocephalus, hippocampal dysgenesis, and abnormalities in the pheromone sensory pathway (56). However, even though these data argue against the role of p73 as a tumor suppressor, recent evidence showed that p73 is indeed required for p53-induced apoptosis (19).
ΔNp73, which does not contain the activation domain in p73, is presently thought to be transcriptionally inactive (23, 39). Indeed, it has been demonstrated in some experimental systems that ΔNp73 is dominant negative over p53 and/or p73. For example, ΔNp73 prevents p53 or p73 from activating several promoters of p53 target genes in a luciferase assay. In addition, cotransfection of ΔNp73 with p53 or p73 decreases the ability of the latter to induce some endogenous p53 target genes (3, 23, 28, 32, 39, 52, 58). Furthermore, ΔNp73, which is the predominant form in the developing mouse brain, can prevent and rescue the neuronal cells from death, likely mediated by p53, upon withdrawal of the obligate survival factor, nerve growth factor (45, 56). These findings suggest that ΔNp73 is dominant negative. However, many concerns remain. For example, does ΔNp73 have the same role in different cellular contexts and in response to various stress signals? Furthermore, we, and others later, have shown that ΔNp63α, which was regarded as dominant negative, is active in inducing cell cycle arrest and apoptosis and is capable of inducing some p53 family target genes (13-15, 21).
In the process of delineating p73β functional domains (42), we surprisingly found that ΔNp73β is able to induce both cell cycle arrest and apoptosis when stably expressed in cancer cells. Importantly, we found that, when ΔNp73β is expressed at a physiologically relevant level, it is still capable of suppressing cell growth. To further analyze ΔNp73β, we generated stable cell lines that can inducibly express various ΔNp73β mutants. We found that the 13 unique residues at the N terminus are required for ΔNp73β to suppress cell growth. We also found that, among the 13 residues, residues 6 to 10 are critical to ΔNp73β function. Furthermore, we found that ΔNp73β is capable of inducing some p53 target genes, albeit to a lesser extent than p73β. More interestingly, we found that the 13 unique residues, together with the N-terminal PXXP motifs, form a novel activation domain. Like ΔNp73β, ΔNp73γ is active in transactivation. However, ΔNp73α is inactive in suppressing cell growth.
MATERIALS AND METHODS
Plasmids and mutagenesis.
ΔNp73β and various ΔNp73β mutants were generated by PCR with the full-length hemagglutinin (HA)-tagged p73β (HA-p73β) cDNA, and HA-ΔNp73β and ΔNp73β cDNAs afterward, as templates. To generate HA-ΔNp73β, a cDNA fragment encoding amino acids 1 to 201 in ΔNp73 was amplified by using 5′ end primer HA-ΔNp73β-5 (5′ AAG GAT CCA CCA TGT ACC CAT ACG ATG TTC CAG ATT ACG CTC TGT ACG TCG GTG ACC CC 3′) and 3′ end primer HA-ΔNp73β-3 (5′ GAA TTC CGT CCC CAC CTG TG 3′). This fragment was used to replace the region encoding residues 1 to 257 in HA-p73β through an internal EcoRI site. To generate ΔNp73β, a cDNA fragment encoding amino acids 1 to 201 in ΔNp73 was generated by using 5′ end primer ΔNp73β-5 (5′ AAG GAT CCA CCA TGC TGT ACG TCG GTG ACC CCG 3′) and 3′ end primer HA-ΔNp73β-3. This fragment was used to replace the region encoding residues 1 to 257 in HA-p73β through an internal EcoRI site. To generate ΔNp73β(Δ2-5), a cDNA fragment encoding amino acids 1 to 201 but lacking 2-5 in ΔNp73β was generated by using 5′ end primer ΔNp73β(Δ2-5)-5 (5′ GGA ATT CAC CAT GGA CCC CGC ACG GCA C 3′) and 3′ end primer HA-ΔNp73β-3. This fragment was used to replace the region encoding residues 1 to 257 in HA-p73β through an internal EcoRI site. To generate ΔNp73β(Δ2-13), a cDNA fragment encoding amino acids 1 to 201 but lacking 2-13 in ΔNp73β was generated by using 5′ end primer ΔNp73β(Δ2-13)-5 (5′ GGA ATT CAC CAT GGC CCA AGT TCA ATC TG 3′) and 3′ end primer HA-ΔNp73β-3. This fragment was used to replace the region encoding residues 1 to 257 in HA-p73β through an internal EcoRI site. To generate ΔNp73β(Δ6-11), a cDNA fragment encoding amino acids 1 to 201 but lacking 6-11 in ΔNp73β was generated by using 5′ end primer ΔNp73β(Δ6-11)-5 (5′ GGA ATT CAC CAT GCT GTA CGT CGG TGC CAC GGC C 3′) and 3′ end primer HA-ΔNp73β-3. This fragment was used to replace the region encoding residues 1 to 257 in HA-p73β through an internal EcoRI site. To generate ΔNp73β(Neu), a cDNA fragment encoding amino acids 1 to 201 with altered residues 6 and 9 and 10 in ΔNp73β was generated by using 5′ end primer ΔNp73β(Neu)-5 (5′ GGA ATT CAC CAT GCT GTA CGT CGG TGC CCC CGC AGC GGC CCT C 3′) and 3′ end primer HA-ΔNp73β-3. This fragment was used to replace the region encoding residues 1 to 257 in HA-p73β through an internal EcoRI site. To generate HA-ΔNp73α, a cDNA fragment encoding amino acids 248 to 636 in p73α was generated by using 5′ end primer ΔNp73α-5 (5′ ACG GAA TTC ACC ACC ATC CTG 3′) and 3′ end primer ΔNp73α-3 (5′ GGA TCC TCA GTG GAT CTC GGC CTC CGT 3′). This fragment was used to replace the region encoding residues 208 to 459 in HA-ΔNp73β through an EcoRI site and a BamHI site. All the mutations and deletions were confirmed by sequencing. cDNAs were cloned separately into a tetracycline-regulated expression vector, pUHD10-3, and the resulting constructs were used to generate stable cell lines.
To generate pcDNA3 constructs expressing HA-ΔNp73α or HA-ΔNp73β, a full-length cDNA fragment was obtained by excision of pUHD10-3-HA-ΔNp73α or pUHD10-3-HA-ΔNp73β with BamHI. The cDNA fragments were then cloned into pcDNA3 at the BamHI site, respectively. To generate a pcDNA3 construct expressing HA-ΔNp73γ, a cDNA fragment encoding residues 248 to 399 in p73α was generated by using a 5′ end primer (5′ GAA TTC ACC ACC ATC CTG TAC AAC 3′) and a 3′ end primer (5′ ATC CCG GGG CGG CCT CTG TAG GAG CTG CTG CTG CTG 3′); a cDNA fragment encoding residues 450 to 525 in p73α was generated by using a 5′ end primer (5′ CCC GGG CCC CAC TTT GAG GTC ACT TTC CAG CAG 3′) and a 3′ end primer (5′ GGA TCC TCA GTG GAT CTC GGC CTC C 3′). The two cDNA fragments were ligated through an XmaI site and were used to replace the region encoding residues 208 to 587 in HA-ΔNp73α. The resulting constructs were designated pcDNA3-HA-ΔNp73α, pcDNA3-HA-ΔNp73β, and pcDNA3-HA-ΔNp73γ, respectively.
To generate constructs expressing various Gal4 chimeric proteins, cDNA fragments encoding the corresponding residues in ΔNp73 or p73 were either synthesized or were PCR amplified by using ΔNp73β or p73β as a template. The sequences of oligonucleotides for the cDNA fragment encoding ΔNp73(1-13) were sense, 5′ AAT TCA TGC TGT ACG TCG GTG ACC CCG CAC GGC ACC TCG CCA CGC TCG AGT GAT 3′; and antisense, 5′ CTA GAT CAC TCG AGC GTC GCG AGG TGC CGT GCG GGG TCA CCG ACG TAC AGC ATG 3′. The two single-strand DNA fragments were annealed to form a double-strand cDNA fragment. The primers used to amplify ΔNp73(1-35) were 5′ end primer, 5′ GAA TTC ATG CTG TAC GTC GGT GAC CCC 3′; and 3′ end primer, 5′ TCT AGA TCA CTC GAG GCT GGC CGA GGC CGC GCG GCT GCT CAT 3′. ΔNp73(1-67) was generated by the 5′ end primer used for ΔNp73(1-35) generation, and the 3′ end primer (5′ TCT AGA TCA CTC GAG GGG GAT GAC AGG CGC CGC CGA CAT GGT 3′). ΔNp73(4-67) was generated by using a 5′ end primer (5′ GAA TTC ATG GCC CAG TTC AAT CTG CTG AGC AGC 3′) and the 3′ end primer employed for ΔNp73(1-67) generation. p73(1-54) was generated by using a 5′ end primer (5′ GAA TTC ATG GCC CAG TCC ACC GCC ACC TCC 3′) and a 3′ end primer (5′ TCT AGA TCA CTC GAG CAG GTG GAA GAC GTC CAT GCT GGA ATC CGT 3′). p73(1-116) was generated with the 5′ end primer used for p73(1-54) generation and the 3′ end primer used for ΔNp73(1-67) generation. The identity of these cDNA fragments was confirmed by sequencing. Each cDNA was cloned into the EcoRI and XbaI sites of the pcDNA3.1-Gal4 DNA-binding domain. The resulting constructs were designated pcDNA3.1-Gal4-ΔNp73(1-13), pcDNA3.1-Gal4-ΔNp73(1-35), pcDNA3.1-Gal4-ΔNp73(1-67), pcDNA3.1-Gal4-ΔNp73(4-67), pcDNA3.1-Gal4-p73(1-54), and pcDNA3.1-Gal4-p73(1-116), respectively.
Cell lines.
The culture, transfection, and generation of H1299 and MCF-7 cell lines were performed as described previously (34). Individual clones were screened for inducible expression of ΔNp73β, various ΔNp73β mutants, and ΔNp73α protein by Western blot analysis with monoclonal antibodies against the HA epitope or p73. LS174T, DLD-1, SW480, and U251 cells were purchased from the American Type Culture Collection and were treated with camptothecin (CPT) (1 μM), DOX (1 μg/ml), or 5-fluorouracil (5-FU) (400 μM) before the cell extracts were collected for Western blot analysis.
Western blot analysis.
Cells were collected from plates in phosphate-buffered saline (PBS), resuspended with 2× sodium dodecyl sulfate sample buffer, and boiled for 5 min. Western blot analysis was performed as described previously (13, 35). The affinity-purified monoclonal antibodies against p73 (Ab-2 and Ab-3) were purchased from Oncogene Science (Cambridge, Mass.). Affinity-purified anti-p21 polyclonal antibodies (c-19) and anti-HA monoclonal antibody (Y-11) were purchased from Santa Cruz (Santa Cruz, Calif.). The affinity-purified antiactin polyclonal antibodies were purchased from Sigma (St. Louis, Mo.).
Growth rate analysis.
To determine the rate of cell growth, approximately 6 × 104 H1299 cells or 1 × 105 MCF-7 cells were seeded in 60-mm-diameter plates in the presence or absence of tetracycline (2 μg/ml) to regulate the expression of the protein of interest. To determine the effect of ΔNp73β at a physiological level on the rate of cell growth, 2 × 104 H1299 cells were plated in 35-mm dishes in the absence or presence of 2, 0.2, or 0.02 μg of tetracycline/ml to regulate the expression level of ΔNp73β. The media were replaced every 72 h. At time points indicated, two plates were rinsed twice with PBS to remove dead cells and debris. Live cells on the plates were trypsinized and were collected separately. Cells from each plate were counted four times with the Coulter cell counter. The average number of cells from two plates was used for growth rate determination.
Trypan blue dye exclusion assay.
Cells were seeded at approximately n = 6 × 104 for H1299 cells or 1 × 105 for MCF-7 cells in 60-mm-diameter plates in the presence or absence of tetracycline (2 μg/ml) for 3 days. At 72 h after plating, both floating cells in the media and live cells on the plates were collected and concentrated by centrifugation. After being stained with trypan blue dye (Sigma) for 15 min, both live (unstained) and dead (stained) cells were counted twice in a hemocytometer. The percentage of dead cells was calculated by using the number of dead cells divided by the total cells counted.
DNA histogram analysis.
Cells were seeded at 2 × 105 per 90-mm-diameter plate in the presence or absence of tetracycline (2 μg/ml). At 72 h after plating, both floating and dead cells in the media and live cells on the plates were collected and fixed with 1 ml of 100% ethanol for overnight and were then centrifuged and resuspended in 300 μl of PBS solution containing 50 μg each of RNase A (Sigma) and propidium iodide (Sigma) per ml. The stained cells were analyzed in a fluorescence-activated cell sorter (FACSCalibur; Becton Dickinson) within 4 h. The percentage of cells in the sub-G1, G0 and G1, S, and G2 to M phases was determined by using the Cell Quest program. The percentage of cells in the G0 and G1, S, and G2 to M phases was recalculated after subtracting the number of cells in the sub-G1 phase from the total population. The percentage of cells in the sub-G1 phase was used as an index for the degree of apoptosis.
Luciferase assay.
To determine the transcriptional activity of ΔNp73α, ΔNp73β, or ΔNp73γ, 0.5 μg of pcDNA3-HA-ΔNp73α, pcDNA3-HA-ΔNp73β, or pcDNA3-HA-ΔNp73γ was cotransfected into H1299 cells with 0.25 μg of a luciferase reporter, pGL2-GADD45, which is under the control of a p53-responsive element in intron 3 of the GADD45 gene and a c-fos minimum promoter. To determine the transcriptional activity of various Gal4 chimeric proteins, 0.5 μg of pcDNA3.1, pcDNA3.1-Gal4, pcDNA3.1-Gal4-ΔNp73(1-13), pcDNA3.1-Gal4-ΔNp73(1-35), pcDNA3.1-Gal4-ΔNp73(1-67), pcDNA3.1-Gal4-ΔNp73(4-67), pcDNA3.1-Gal4-p73(1-54), or pcDNA3.1-Gal4-p73(1-116) was cotransfected into H1299 cells with 0.25 μg of a luciferase reporter, pGL2-5Gal, which is under the control of a promoter containing five consecutive Gal4 responsive elements. As an internal control, 5 ng of Renilla luciferase assay vector, pRL-CMV (Promega, Madison, Wis.), was also cotransfected. The dual luciferase assay was performed according to the manufacturer's instructions (Promega). The increase (n-fold) in relative luciferase activity is a product of the luciferase activity induced by various ΔNp73 isoforms divided by that induced by pcDNA3 or the luciferase activity induced by various Gal4 chimeric proteins divided by that induced by pcDNA3.1.
RNA isolation and Northern blot analysis.
Total RNA was isolated by using Trizol (GIBCO-BRL) from H1299 and MCF-7 cells, which were uninduced or induced to express p73β or ΔNp73β. Northern blots were prepared by using 10 μg of total RNA. The p21, GADD45, 14-3-3-σ, MDM2, and GAPDH probes were prepared as previously described (59).
RESULTS
ΔNp73β induces cell cycle arrest and apoptosis.
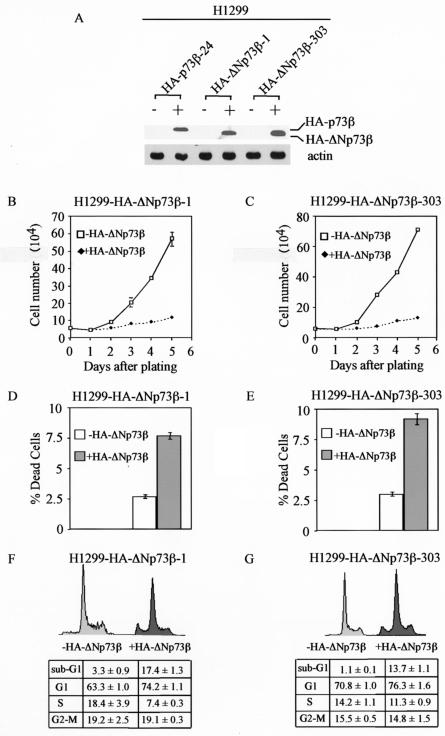
ΔNp73, which lacks the N-terminal activation domain in p73, is thought to be transcriptionally inactive and dominant negative over wild-type p53 and/or p73 (3, 23, 28, 32, 39, 52, 55, 58). However, we, and others later, showed that ΔNp63α, which was once considered functionally inert, is capable of regulating gene expression and suppressing cell growth (13-15, 21). To determine whether various ΔNp73 isoforms are functional, we chose to analyze ΔNp73β. We generated H1299 stable cell lines that inducibly express HA-tagged ΔNp73β (HA-ΔNp73β) under the control of the tetracycline-regulated system. Two representative cell lines, HA-ΔNp73β-1 and HA-ΔNp73β-303, were shown in Fig. 1A. Western blot analysis showed that HA-ΔNp73β was expressed at a level comparable to that of HA-p73β in HA-p73β-24 cells (Fig. 1A). We showed recently that the level of HA-p73β in HA-p73β-24 cells is sufficient to induce cell cycle arrest and apoptosis (42).
FIG. 1.
HA-ΔNp73β induces cell cycle arrest and apoptosis in H1299 cells. (A) Levels of induced expression of HA-p73β and HA-ΔNp73β in H1299 stable cell lines are comparable. Western blot analysis was performed by using cell extracts from uninduced cells (−) and cells that were induced (+) to express HA-p73β or HA-ΔNp73β for 24 h. HA-p73β and HA-ΔNp73β were detected with anti-HA polyclonal antibodies (Y-11). Actin was detected with antiactin polyclonal antibodies and was used as an equal loading control. (B and C) HA-ΔNp73β suppresses cell growth in H1299 cells. Approximately 6 × 104 HA-ΔNp73β-1 cells (B) and HA-ΔNp73β-303 cells (C) were seeded in 60-mm-diameter plates in the presence or absence of tetracycline (2 μg/ml) to regulate the expression of HA-ΔNp73β. At time points indicated, live cells on the plates were trypsinized and were collected separately. Cells from each plate were counted four times by using the Coulter cell counter. The average number of cells from two plates was used for growth rate determination. (D and E) HA-ΔNp73β induces cell death in H1299 cells. H1299 cells were seeded at approximately 6 × 104 in 60-mm-diameter plates in the presence or absence of tetracycline (2 μg/ml) for 3 days. At 72 h after plating, both floating cells in the media and live cells on the plates were collected and stained with trypan blue dye for 15 min. Both live (unstained) and dead (stained) cells were counted two times in a hemocytometer. The percentage of dead cells was calculated by using the number of dead cells dividedby the total cells counted. (F and G) HA-ΔNp73β induces cell cycle arrest and apoptosis in H1299 cells. H1299 cells were seeded at 2 × 105 per 90-mm-diameter plate in the presence or absence of tetracycline (2 μg/ml). At 72 h after plating, both floating and dead cells in the media and live cells on the plates were collected and were fixed with 1 ml of 100% ethanol overnight and were then centrifuged and resuspended in 300 μl of PBS solution containing 50 μg each of RNase A and propidium iodide per ml. The stained cells were measured by fluorescence-activated cell sorter analysis. The percentage of cells in the sub-G1, G0, and G1, S, and G2 to M phases was determined by using the Cell Quest program. The percentage of cells in the G0 and G1, S, and G2 to M phases was recalculated after subtracting the number of cells in the sub-G1 phase from the total population. The percentage of cells in the sub-G1 phase was used as an index for the degree of apoptosis.
To examine whether ΔNp73β is capable of inhibiting cell growth, we performed growth rate analysis and found that cells almost completely failed to proliferate when HA-ΔNp73β was expressed (Fig. 1B and C). This suggests that HA-ΔNp73β is indeed functional. To confirm this, we performed the trypan blue dye exclusion assay and found that the number of dead cells increased from 2.7 to 7.8% in HA-ΔNp73β-1 cells and from 3.0 to 9.2% in HA-ΔNp73β-303 cells when HA-ΔNp73β was induced (Fig. 1D and E). This indicates that HA-ΔNp73β can induce cell death. Additionally, we performed DNA histogram analysis and found that the number of apoptotic cells increased from 3.3 to 17.4% in HA-ΔNp73β-1 cells and from 1.1 to 13.7% in HA-ΔNp73β-303 cells when HA-ΔNp73β was expressed (Fig. 1F and G). Furthermore, cells in the G1 phase increased from 63.3 to 74.2% in HA-ΔNp73β-1 cells and from 70.8 to 76.3% in HA-ΔNp73β-303 cells when HA-ΔNp73β was induced (Fig. 1F and G). Taken together, these data indicate that HA-ΔNp73β can induce both cell cycle arrest and apoptosis.
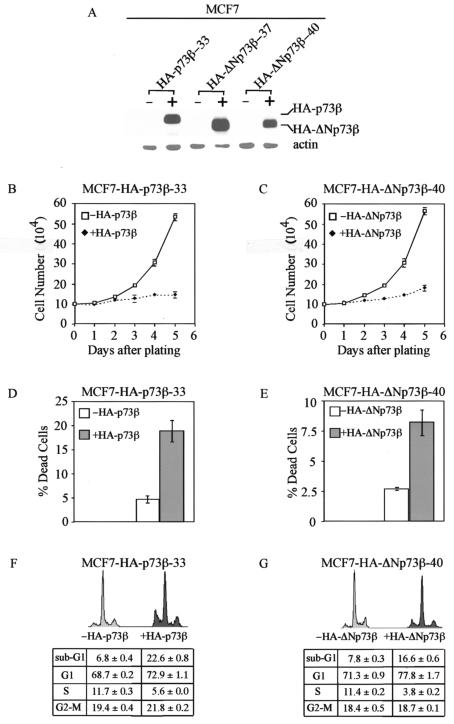
To rule out the possibility that the activity of HA-ΔNp73β observed in H1299 cells is cell type dependent, we generated MCF-7 cell lines that can inducibly express HA-ΔNp73β. Two representative clones were shown in Fig. 2A. We also generated MCF-7 cell lines inducibly expressing HA-p73β, and one representative cell line was shown in Fig. 2A. Western blot analysis revealed that the level of HA-ΔNp73β expressed in these two lines was comparable to that of HA-p73β (Fig. 2A). Next, we performed growth rate analysis and found that both HA-p73β and HA-ΔNp73β were able to efficiently inhibit cell growth in MCF-7 cells (Fig. 2B and C). The trypan blue dye exclusion assay showed that, like HA-p73β, HA-ΔNp73β was capable of inducing cell death in MCF-7 cells (Fig. 2D and E). We then performed DNA histogram analysis and found that the number of apoptotic cells increased from 7.8 to 22.6% and from 7.8 to 16.6% when HA-p73β and HA-ΔNp73β were induced, respectively (Fig. 2F and G). Similarly, the percentage of cells in G1 was increased by HA-p73β (from 68.7 to 72.9%) and HA-ΔNp73β (from 71.3 to 77.8%) (Fig. 2F and G). These results indicate that HA-ΔNp73β is capable of inducing cell cycle arrest and apoptosis both in H1299 and MCF-7 cells.
FIG. 2.
Both HA-p73β and HA-ΔNp73β induce cell cycle arrest and apoptosis in MCF-7 cells. (A) Levels of induced expression of HA-p73β and HA-ΔNp73β in MCF-7 stable cell lines are comparable. (B and C) Both HA-p73β and HA-ΔNp73β suppress cell growth in MCF-7 cells. Approximately 105 MCF-7 cells were seeded in 60-mm plates in the presence or absence of tetracycline (2 μg/ml) to regulate the expression of HA-p73β or HA-ΔNp73β. (D and E) Both HA-p73β and HA-ΔNp73β induce cell death in MCF-7 cells. (F and G) Both HA-p73β and HA-ΔNp73β induce cell cycle arrest and apoptosis in MCF-7 cells.
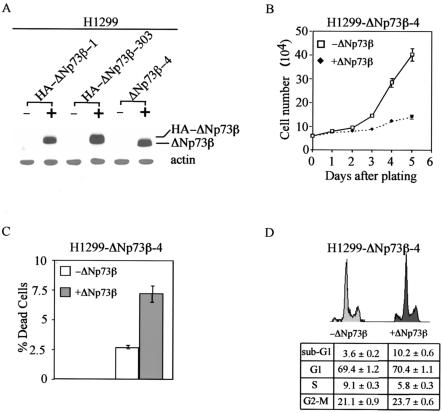
In order to compare the expression level of p73β, ΔNp73β, and their mutants, p73 was tagged with an HA epitope at its N terminus. Even though no report showed to date that the HA epitope can endow a protein with new function, we wanted to exclude such a possibility by generating H1299 cell lines expressing ΔNp73β with no HA epitope. One representative line was shown in Fig. 3A. Western blot analysis showed that the level of ΔNp73β was comparable to that of HA-ΔNp73β when anti-p73 antibody was used (Fig. 3A). Next, we performed growth rate analysis and showed that ΔNp73β was able to suppress H1299 cell proliferation (Fig. 3B). We also performed the trypan blue dye exclusion assay and found that ΔNp73β was potent in inducing cell death (Fig. 3C), which was confirmed by DNA histogram analysis (Fig. 3D). Taken together, we conclude that ΔNp73β is capable of suppressing cell growth and active in inducing cell cycle arrest and apoptosis.
FIG. 3.
ΔNp73β induces cell cycle arrest and apoptosis in H1299 cells. (A) Levels of induced expression of HA-ΔNp73β and ΔNp73β in H1299 stable cell lines are comparable. (B to D) ΔNp73β induces growth suppression (B), cell death (C), and cell cycle arrest and apoptosis (D) in H1299 cells.
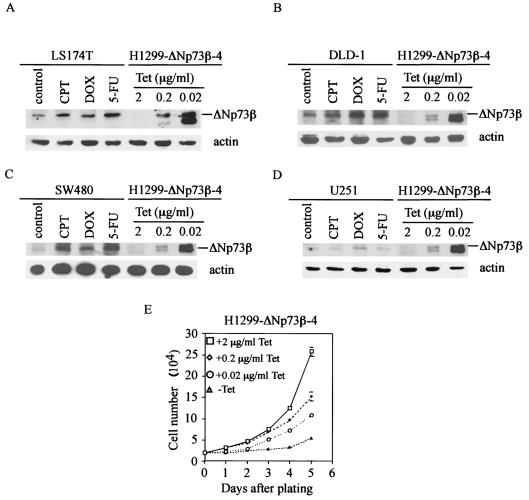
Furthermore, in order to eliminate the concern that the activity of ΔNp73β detected above might be artificial due to its overexpression in inducible cell lines, we titrated the concentration of tetracycline in the medium to regulate the expression level of ΔNp73β. We found that, in the presence of 0.2 or 0.02 μg of tetracycline/ml, the level of exogenous ΔNp73β in H1299 cells was comparable to the physiological level of endogenous ΔNp73β in LS174T, DLD-1, SW480, and U251 cells induced by CPT, DOX, or 5-FU (Fig. 4A to D). Growth rate analysis showed that ΔNp73β at the physiological level was capable of suppressing cell growth, though to a lesser extent than that expressed in the absence of tetracycline (Fig. 4E). This suggests that the inhibitory effect of endogenous ΔNp73β in these cells induced by DNA damage agents, although relatively small, may partially contribute to DNA damage-induced growth suppression.
FIG. 4.
ΔNp73β expressed at a physiological level is capable of suppressing cell growth. (A to D) The ΔNp73β expression level in the presence of 0.2 or 0.02 μg of tetracycline/ml is comparable to a physiological level of endogenous ΔNp73β in LS174T, DLD-1, SW480, and U251 cells. H1299-ΔNp73β-4 cells were seeded on 10-cm-diameter dishes in the presence of 0.02, 0.2, or 2 μg of tetracycline/ml to regulate ΔNp73β expression. LS174T, DLD-1, SW480, and U251 cells were treated with CPT (1 μM), DOX (1 μg/ml), or 5-FU (400 μM) for 24 h. Cell extracts were collected for Western blot analysis. ΔNp73β was detected by using anti-p73 (Ab2 and Ab3) monoclonal antibodies. (E) ΔNp73β expressed at a physiological level is capable of suppressing cell growth. Approximately 2 × 104 H1299-ΔNp73β-4 cells were seeded in the absence or presence of 0.02, 0.2, or 2 μg of tetracycline/ml to regulate ΔNp73β expression.
The 13 unique residues at the N terminus are necessary for ΔNp73β to suppress cell growth.
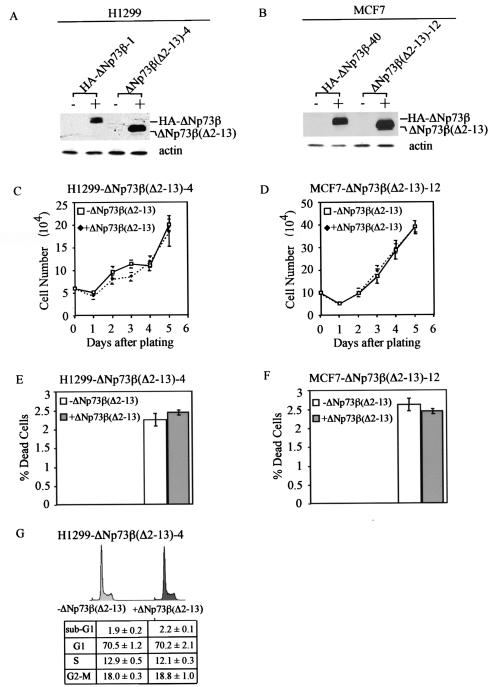
ΔNp73β possesses 13 unique residues at its N terminus but lacks the first 62 residues in p73β, which includes the N-terminal activation domain within residues 1 to 54 (6, 55). We have shown recently that p73β(Δ1-54), which lacks the N-terminal 54 residues, is functionally inert (42). This prompts us to ask whether the function of ΔNp73β is endowed by the acquisition of these 13 unique residues. To address this, we generated H1299 cell lines that can inducibly express a ΔNp73β mutant lacking residues 2-13. A representative clone, ΔNp73β(Δ2-13)-4, was shown in Fig. 5A. Western blot analysis showed that the level of this mutant expressed in ΔNp73β(Δ2-13)-4 cells was higher than that of ΔNp73β in HA-ΔNp73β-1 cells (Fig. 5A). Interestingly, we found that ΔNp73β(Δ2-13) was incapable of suppressing cell growth (Fig. 5C). Additionally, we performed the trypan blue dye exclusion assay and found that ΔNp73β(Δ2-13) was unable to induce cell death (Fig. 5E). Furthermore, DNA histogram analysis showed no significant changes in any phase of the cell cycle (Fig. 5G). These data indicate that the 13 unique residues confer ΔNp73β with the ability to suppress cell growth. To confirm the data obtained in H1299 cells, we generated MCF-7 cell lines that can inducibly express ΔNp73β(Δ2-13) (Fig. 5B). Both growth rate analysis and the trypan blue dye exclusion assay showed results similar to those observed in H1299 cells (compare Fig. 5C and D and 5E and F). Taken together, we conclude that the 13 unique residues are necessary for the function of ΔNp73β.
FIG. 5.
The 13 unique residues at the N terminus are necessary for ΔNp73β to suppress cell growth. (A and B) Levels of induced expression of HA-ΔNp73β and ΔNp73β(Δ2-13) in H1299 (A) or MCF-7 (B) stable cell lines are comparable. Western blot analysis was performed by using anti-p73 (Ab-3) monoclonal antibody. (C and D) ΔNp73β(Δ2-13) is incapable of suppressing cell growth in H1299 cells (C) and in MCF-7 cells (D). (E and F) ΔNp73β(Δ2-13) is incapable of inducing cell death in H-1299 cells (E) and in MCF-7 cells (F). (G) ΔNp73β(Δ2-13) is incapable of inducing cell cycle arrest and apoptosis in H1299 cells.
Residues 6-11 are critical to the activity of ΔNp73β.
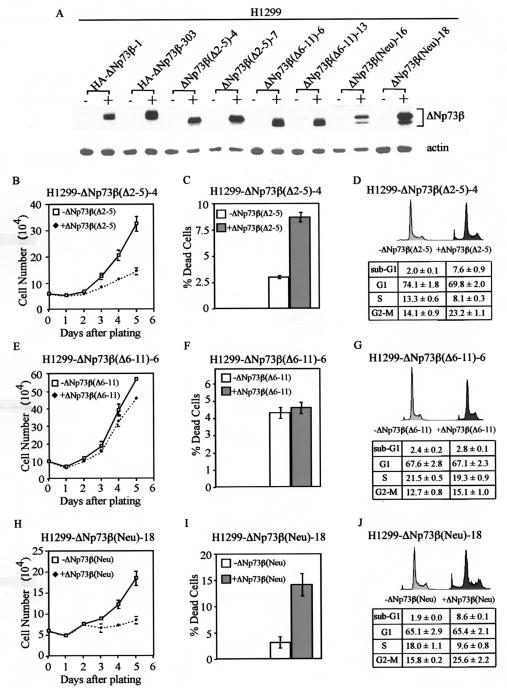
To further determine which part of these 13 residues is critical to the activity of ΔNp73β, we generated H1299 cell lines that can inducibly express a ΔNp73β mutant lacking residues 2-5, designated ΔNp73β(Δ2-5) (Fig. 6A). We chose the cell line ΔNp73β(Δ2-5)-4 for further studies because the level of the protein expressed was comparable to that expressed in HA-ΔNp73β-1 cells (Fig. 6A). We performed growth rate analysis, the trypan blue dye exclusion assay, and DNA histogram analysis and found that ΔNp73β(Δ2-5) was still active in inhibiting cell growth and in inducing apoptosis (Fig. 6B to D), albeit to a lesser extent than ΔNp73β (Fig. 1). This suggests that residues 2-5 are not required but that other residues might be critical for the activity of ΔNp73β. To address this, we generated H1299 cell lines that can inducibly express a ΔNp73β mutant lacking residues 6 to 11, designated ΔNp73β(Δ6-11). We then chose ΔNp73β(Δ6-11)-6 for growth rate analysis, the trypan blue dye exclusion assay, and DNA histogram analysis. We found no significant growth suppression and cell death induced by ΔNp73β(Δ6-11) (Fig. 6E to G). This suggests that residues 6-11 are critical to the function of ΔNp73β. Within this region, there are three residues with a charged polar side chain, namely, residue 6 (Asp), residue 9 (Arg), and residue 10 (His). To examine whether these residues are necessary for the activity of ΔNp73β, we generated a ΔNp73β mutant [ΔNp73β(Neu)], in which these three amino acids were replaced with alanine. Alanine has a nonpolar side chain. We then generated H1299 cell lines expressing this mutant. We found that ΔNp73β(Neu) was still capable of inhibiting cell growth and inducing apoptosis (Fig. 6H to J). This suggests that either mutation of these amino acids does not significantly alter the conformation of the ΔNp73β at the N terminus or that the altered conformation is still competent. In order to confirm the results obtained in H1299 cells, we also generated MCF-7 cell lines that can inducibly express ΔNp73β(Δ2-5), ΔNp73β(Δ6-11), and ΔNp73β(Neu). We found results similar to those detected in H1299 cells (data not shown).
FIG. 6.
Residues 6 to 11 are critical to the activity of ΔNp73β. (A) Levels of induced expression of HA-ΔNp73β, ΔNp73β(Δ2-5), ΔNp73β(Δ6-11), and ΔNp73β(Neu) in H1299 stable cell lines are comparable. Western blot analysis was performed by using anti-p73 (Ab-3) monoclonal antibody. (B and H) ΔNp73β(Δ2-5) (B) and ΔNp73β(Neu) (H) are able to suppress cell growth in H1299 cells. (C and I) ΔNp73β(Δ2-5) (C) and ΔNp73β(Neu) (I) are able to induce cell death in H1299 cells. (D and J) ΔNp73β(Δ2-5) (D) and ΔNp73β(Neu) (J) are able to induce cell cycle arrest and apoptosis in H1299 cells. (E to G) ΔNp73β(Δ6-11) is incapable of inducing growth suppression (E), cell death (F), and cell cycle arrest and apoptosis (G) in H1299 cells.
DNp73α is inactive in suppressing cell growth.
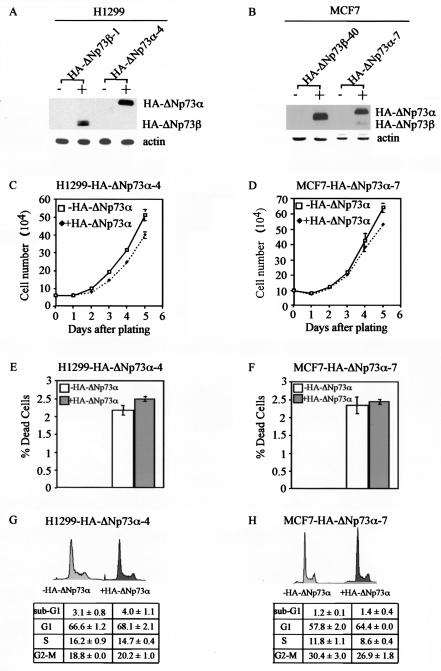
In contrast to p53, p73 has at least seven alternatively spliced isoforms with different C termini. p73α is the longest form of the p73 protein, which contains a SAM domain and an extreme C-terminal region. p73β lacks the extreme C-terminal region and most of the SAM domain (8, 29, 31, 50). Previous studies showed that p73β is more potent than p73α in inducing some p53 target genes and in growth suppression (12, 32, 46). This suggests that the C-terminal region in p73α may suppress its own function. Indeed, a recent report has provided evidence that the C-terminal region of p73α in a fusion protein represses transcription to an extent similar to that of the repressor protein E1B-55kDa from adenovirus (32). Moreover, the extreme C-terminal region in p63α, which shares significant sequence identity with that in p73α, is capable of interacting with its own activation domain at the N terminus and repressing its own transcriptional activity (48). All this evidence points to the possibility that the C-terminal region in p73α is likely to repress p73 function. Since we have shown that ΔNp73β is functional in inhibiting cell growth, it is important to determine whether ΔNp73α is also able to suppress cell growth. To address this, we generated H1299 stable cell lines that can inducibly express ΔNp73α (Fig. 7A). We note that the level of ΔNp73α protein was higher than that of ΔNp73β protein (Fig. 7A). We then performed growth rate analysis, the trypan blue dye exclusion assay, and DNA histogram analysis. We found that, unlike ΔNp73β, ΔNp73α was unable to inhibit cell growth and induce apoptosis (Fig. 7C, E, and G). To confirm our observation in H1299 cells, we generated MCF-7 cell lines inducibly expressing ΔNp73α. Similar results were obtained (Fig. 7B, D, F, and H). This suggests that the C-terminal region is capable of inhibiting ΔNp73α activity.
FIG. 7.
ΔNp73α is inactive in suppressing cell growth. (A and B) Levels of induced expression of HA-ΔNp73β and HA-ΔNp73α in H1299 (A) or MCF-7 (B) stable cell lines are comparable. (C and D) HA-ΔNp73α is unable to inhibit cell growth in H1299 cells (C) and in MCF-7 cells (D). (E and F) HA-ΔNp73α is unable to induce cell death in H1299 cells (C) and in MCF-7 cells (D). (G and H) HA-ΔNp73α is unable to induce cell cycle arrest and apoptosis in H1299 cells (G) and in MCF-7 cells (H).
ΔNp73β is active in transactivation.
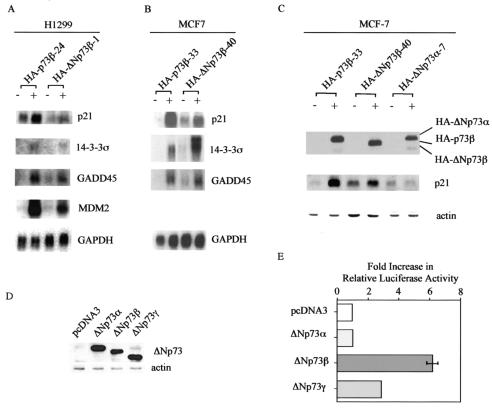
p73β induces both cell cycle arrest and apoptosis mainly through regulating some p53 target genes (6, 59). Since ΔNp73β is also capable of inducing both cell cycle arrest and apoptosis, we speculate that ΔNp73β is transcriptionally active. To address this, we performed Northern blot analysis and found that ΔNp73β was capable of inducing p21, 14-3-3σ, GADD45, and MDM2 in H1299 cells, albeit to a lesser extent than that by p73β (Fig. 8A). Similarly, we found that p21, 14-3-3σ, and GADD45 were induced by ΔNp73β in MCF-7 cells (Fig. 8B). It should be noted that ΔNp73β was more active in inducing 14-3-3σ than was p73β in MCF-7 cells (Fig. 8B). In addition, Western blot analysis showed that ΔNp73β was able to up-regulate p21 expression (Fig. 8C). In contrast, ΔNp73α was unable to induce the expression of p21 (Fig. 8C), consistent with the above data showing that ΔNp73α was incapable of inducing growth suppression. Like ΔNp73β, ΔNp73γ does not possess the SAM domain and the extreme C-terminal region in ΔNp73α, though ΔNp73γ is different from ΔNp73β in its C terminus. Thus, it is possible that ΔNp73γ is also active in transactivation. To address this, we performed a luciferase assay with a reporter containing a p53-responsive element in intron 3 of the GADD45 gene. We first examined the expression of ΔNp73α, ΔNp73β, and ΔNp73γ in the luciferase assay and found that their expression levels were comparable (Fig. 8D). We then found that both ΔNp73β and ΔNp73γ were able to activate the reporter (Fig. 8E). However, ΔNp73α was incapable of inducing the luciferase activity (Fig. 8E). These results further support the notion that the C-terminal region in the α isoforms has inhibitory activity.
FIG. 8.
ΔNp73β is active in transactivation. (A) ΔNp73β is capable of inducing some p53 target genes in H1299 cells. Northern blots were prepared by using 10 μg of total RNA isolated from uninduced cells (−) or cells that were induced (+) to express HA-p73β or HA-ΔNp73β. The blots were probed with 14-3-3σ, GADD45, p21, MDM2, and GAPDH, respectively. (B) ΔNp73β is capable of inducing some p53 target genes in MCF-7 cells. Northern blots were prepared as described for panel A. The blots were probed with 14-3-3σ, GADD45, p21, and GAPDH, respectively. (C) ΔNp73β, but not ΔNp73α, induces p21 expression. Levels of p73, p21, and actin in MCF-7 cells grown in the absence (−) or presence (+) of HA-p73β, HA-ΔNp73β, or HA-ΔNp73α for 24 h were assayed by Western blot analysis. (D) The levels of various ΔNp73 isoforms in the luciferase assay (E) are comparable. ΔNp73 was detected by anti-p73 (Ab-2 and Ab-3) monoclonal antibodies. (E) ΔNp73β and ΔNp73γ, but not ΔNp73α, are able to activate a luciferase reporter, pGL2-GADD45, which is under the control of a p53-responsive element in intron 3 of the GADD45 gene. Half a microgram of a control vector, pcDNA3, or of a vector expressing ΔNp73α, ΔNp73β, or ΔNp73γ was cotransfected into H1299 cells with 0.25 μg of pGL2-GADD45. As an internal control, 5 ng of Renilla luciferase assay vector, pRL-CMV, was also cotransfected. The increase (n-fold) in relative luciferase activity is a product of the luciferase activity induced by various ΔNp73 isoforms divided by that induced by pcDNA3.
The 13 unique residues and the N-terminal PXXP motifs in ΔNp73 form a novel activation domain.
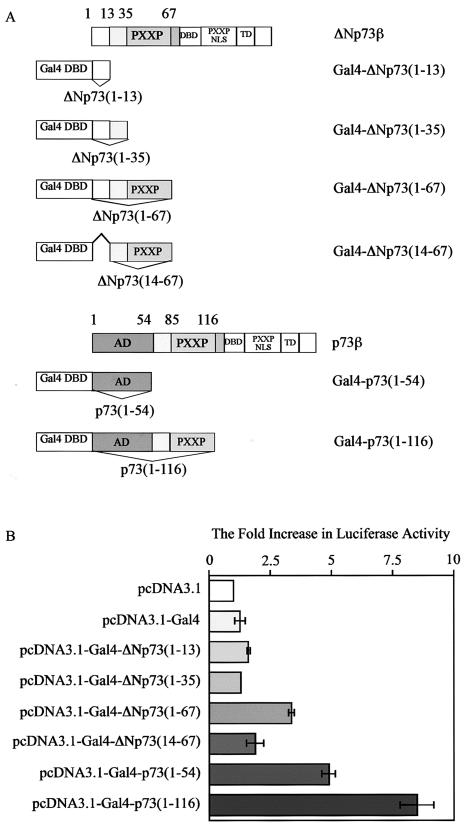
We showed that ΔNp73β, which does not contain the activation domain in TAp73, is still capable of inducing some p53 target genes. However, we have found that p73β(Δ1-54), which lacks the N-terminal activation domain, is inactive in transactivation (42). This suggests that an activation domain must exist in ΔNp73 and that the 13 unique residues at the N terminus are a key component of this activation domain. Previously, we and others have shown that the proline-rich domain in p53, which consists of five PXXP motifs, is required for p53-mediated apoptosis. The proline-rich domain, together with the residues adjacent to the originally defined activation domain in p53, forms a second activation domain (47, 60). p73 contains two N-terminal PXXP motifs between residues 36 and 67 in ΔNp73 or 84 and 117 in TAp73 (6, 55). Thus, to determine whether the 13 unique residues at the N terminus, alone or together with other adjacent residues including the N-terminal PXXP motifs, form an activation domain, we generated constructs expressing various Gal4 chimeric proteins, with the Gal4 DNA-binding domain fused separately with residues 1 to 13, 1 to 35, or 1 to 67 in ΔNp73 (Fig. 9A). To demonstrate the requirement of the 13 unique residues for this potential activation domain, we generated a construct expressing Gal4-ΔNp73(4-67) (Fig. 9A). To compare the transcriptional activity of the potential activation domain in ΔNp73 and the activation domain in TAp73, we generated a construct expressing Gal4-p73(1-54) or Gal4-p73(1-116) (Fig. 9A). We then performed a luciferase assay with a luciferase reporter under the control of five consecutive Gal4 responsive elements. We found that Gal4-ΔNp73(1-13) was unable to activate the luciferase reporter (Fig. 9B), suggesting that the 13 residues alone do not constitute an activation domain. We found that Gal4-ΔNp73(1-35) was also unable to activate the luciferase reporter (Fig. 9B). Similarly, we found that Gal4-ΔNp73(4-67), which lacks the 13 unique residues, was unable to significantly activate the luciferase reporter (Fig. 9B). In contrast, we found that Gal4-ΔNp73(1-67), which contains the 13 unique residues and the N-terminal PXXP motifs, significantly activated the luciferase reporter (∼3- to 4-fold) (Fig. 9B). This implies that the first 67 residues in ΔNp73 form a novel activation domain and that the 13 unique residues are a key component of this activation domain. This is consistent with our data indicating that ΔNp73β(Δ2-13) was incapable of inducing both cell cycle arrest and apoptosis. As a positive control, we found that Gal4-p73(1-54), which contains the N-terminal activation domain in TAp73, significantly induced luciferase activity (Fig. 9B). Moreover, we found that Gal4-p73(1-116), which contains both the N-terminal activation domain and the N-terminal PXXP motifs in p73, was more active in inducing the luciferase activity than was Gal4-p73(1-54) (Fig. 9B). This indicates that the N-terminal PXXP motifs can increase the activity of the activation domain in p73, consistent with the above evidence that the N-terminal PXXP motifs are required for the formation of the activation domain in ΔNp73. Additionally, we found that both Gal4-p73(1-54) and Gal4-p73(1-116) were more active than Gal4-ΔNp73(1-67) in inducing luciferase activity (Fig. 9B), consistent with the above evidence that p73β is more active than ΔNp73β in inducing some p53 target genes (Fig. 8).
FIG. 9.
The 13 unique residues together with the N-terminal PXXP motifs in ΔNp73 form a novel activation domain. (A) Schematic representation of structures of p73β, ΔNp73β, and various Gal4-p73 chimeric proteins. (B) The 13 unique residues together with the N-terminal PXXP motifs in ΔNp73 form an activation domain. Half a microgram of a control vector, pcDNA3.1, or of a vector expressing Gal4 and various Gal4-p73 chimeric proteins as listed on the left was cotransfected into H1299 cells with 0.25 μg of a luciferase reporter, pGL2-5Gal. The increase (n-fold) in relative luciferase activity is a product of the luciferase activity induced by Gal4 or various Gal4-p73 chimeric proteins divided by that induced by pcDNA3.1.
DISCUSSION
ΔNp73β is active in inducing cell cycle arrest and apoptosis.
ΔNp73 was thought at first glance to be transcriptionally inactive due to apparent lack of the N-terminal activation domain in TAp73. In fact, several studies have provided evidence that ΔNp73 is not only transcriptionally inactive but is also dominant negative over wild-type p53 and/or p73 probably via heterotetramerization between ΔNp73 and wild-type p53 or TAp73 (3, 23, 28, 32, 39, 45, 52, 58). However, quite unexpectedly and surprisingly, we found that ΔNp73β is indeed capable of inducing both cell cycle arrest and apoptosis in H1299 stable cell lines. In order to rule out the possibility that ΔNp73β functions in a cell-type-dependent manner, we have generated MCF-7 cell lines that can inducibly express HA-ΔNp73β and demonstrated that ΔNp73β has activities similar to those in H1299 cells. Moreover, to eliminate the concern that the HA epitope may potentially endow such activities to ΔNp73β, even though no such report exists to date, we have even generated a cell line that can inducibly express ΔNp73β with no HA epitope and found that ΔNp73β has the same function as does HA-ΔNp73β. Finally, in order to rule out the possibility that the ΔNp73β functions detected in our system might be artificial due to its overexpression, we titrated the concentration of tetracycline in the medium to achieve an expression comparable to the physiological level of endogenous ΔNp73β in LS174T, DLD-1, SW480, and U251 cells induced by CPT, DOX, or 5-FU. We found that exogenous ΔNp73β expressed at the physiological level is capable of suppressing cell growth. Therefore, our findings firmly establish the idea that ΔNp73β is functional in inducing cell cycle arrest and apoptosis when it is inducibly expressed in stable cell lines.
How do we explain the activity exhibited by ΔNp73β in our systems? Previous studies that examined ΔNp73β were performed primarily by transient-transfection assays or by using cell lines that constitutively express ΔNp73β (3, 23, 28, 32, 39, 52, 58). Because transient transfection is by nature temporary and short-lived, we believe that this method may not have been sensitive enough to detect the activity of ΔNp73β in vivo, especially the long-term effect of ΔNp73β on cell proliferation. On the other hand, when noninducible stable cell lines, which constitutively express ΔNp73, are used, the activity of ΔNp73 would not be uncovered since p73-producing cells, if sensitive to ΔNp73, would not proliferate due to cell cycle arrest and apoptosis. Thus, only those cells that can tolerate ΔNp73 would proliferate, become a cell line, and hence be examined. In our system, ΔNp73β can be inducibly expressed after withdrawal of tetracycline from the culture medium. The uninduced and induced cells are isogenic and ideal for the examination of ΔNp73β function, especially its long-term effects on cell growth.
A novel activation domain in the ΔN variant of p73.
ΔNp73 does not contain the activation domain in TAp73, but it gains 13 unique residues at its N terminus (28, 56). We have demonstrated that ΔNp73β is functional in inducing cell cycle arrest and apoptosis, whereas ΔNp73β(Δ2-13) is inactive. This clearly suggests that the N-terminal 13 unique residues, although very short, are required for the function of ΔNp73β. In addition, we have shown that residues 6 to 11 are more critical than residues 2 to 5 to the function of ΔNp73β since deletion of residues 6 to 11 but not of residues 2 to 5 nearly abolishes its function. Considering that ΔNp73β is still transcriptionally active, a novel activation domain must exist in ΔNp73β. Indeed, we have provided strong evidence that the N-terminal 13 unique residues, together with the N-terminal PXXP motifs, constitute a novel activation domain for ΔNp73β. Furthermore, we have found that ΔNp73β is capable of inducing some p53 target genes, though to a lesser extent than p73β. It is well known that p53 family proteins are dependent upon their ability to up-regulate some p53 target genes, which then mediate cell cycle arrest and apoptosis (33). Thus, the ability of ΔNp73β to regulate some p53 target genes is, at least in part, responsible for its function. However, it is unknown at this moment whether ΔNp73β can induce its own unique target genes.
Previous studies have shown that p73β is more potent than p73α in the regulation of some p53 target genes (12, 20, 46). Compared to p73α, p73β lacks the extreme C-terminal region and most of the SAM domain. It has been demonstrated that the extreme C-terminal region in p63α, which shares significant homology with that in p73α, can interact with the activation domain in p63α and repress its own activity (48). Since we have shown that ΔNp73α has no significant activity in inducing cell cycle arrest and apoptosis, it is likely that the extreme C-terminal region in the alpha isoforms represses the activity of the novel activation domain in ΔNp73. This is further suggested by our finding that, like ΔNp73β, ΔNp73γ, which also lacks the SAM domain and the extreme C-terminal region in the alpha isoforms, is active in transactivation.
Τhe physiological significance of ΔNp73β function.
TAp73 and ΔNp73 are transcribed by using two different promoters in the same gene locus. However, it is totally unknown to date how the cells choose the differential usage of the two promoters, namely, when or how to express TAp73, ΔNp73, or both when they are in different development stages or exposed to various extracellular stimuli. It is also unclear which isoform of TAp73 or ΔNp73 variants will be expressed in those situations. Thus, we believe that it is critical to know the function of each individual isoform of TA and ΔNp73 variants when a specific isoform is expressed alone or in combination with other isoforms in cells. We have found that ΔNp73β is active in transactivation and in suppressing cell growth. The function of ΔNp73β is convincing especially when we found that ΔNp73β expressed at a physiological level is capable of inhibiting cell growth, though to a lesser extent than that expressed in the absence of tetracycline. It indicates that the inhibitory effect of ΔNp73β at the physiological level, while relatively small, may partially contribute to DNA damage-induced growth suppression. However, previous studies found that ΔNp73 is the predominant form in developing brain and sympathetic neuronal cells and that its function seems to counteract the p53-induced apoptosis (45, 56). This evidence suggests that endogenous ΔNp73 is dominant negative in some specific circumstance. Nevertheless, we think that it is not necessarily contradictory to our findings that ΔNp73β is capable of inducing cell cycle arrest and apoptosis. The repression of apoptosis induced by p53 is critical for the normal development of the neural system. As we predicted above, some unknown cellular factors probably determine the usage of the ΔNp73 promoter, some of which could abolish its transcriptional activity in the developing stage. This will meet the requirement of normal development of the neural system. Once the neuronal cells pass the specific developing stage, those cellular factors that repress the activity of ΔNp73β may be dislocated or degraded. Thus, ΔNp73β is then active to induce neuronal differentiation through up-regulating some target genes, for example, p21. Actually, previous studies showed that p21 is involved in differentiation of neuronal cells (5, 16, 17, 44).
In summary, we have found that ΔNp73β is active in inducing cell cycle arrest and apoptosis. The N-terminal 13 unique residues, together with the N-terminal PXXP motifs, form a novel activation domain and are required for ΔNp73β function.
Acknowledgments
This work is supported in part by NIH grant 5 R01 CA081237.
REFERENCES
- 1.Agami, R., G. Blandino, M. Oren, and Y. Shaul. 1999. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature 399:809-813. [DOI] [PubMed] [Google Scholar]
- 2.Alarcon-Vargas, D., S. Y. Fuchs, S. Deb, and Z. Ronai. 2000. p73 transcriptional activity increases upon cooperation between its spliced forms. Oncogene 19:831-835. [DOI] [PubMed] [Google Scholar]
- 3.Allart, S., H. Martin, C. Detraves, J. Terrasson, D. Caput, and C. Davrinche. 2002. Human cytomegalovirus induces drug resistance and alteration of programmed cell death by accumulation of deltaN-p73alpha. J. Biol. Chem. 277:29063-29068. [DOI] [PubMed] [Google Scholar]
- 4.Blint, E., A. C. Phillips, S. Kozlov, C. L. Stewart, and K. H. Vousden. 2002. Induction of p57(KIP2) expression by p73beta. Proc. Natl. Acad. Sci. USA 99:3529-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter, B. D., U. Zirrgiebel, and Y. A. Barde. 1995. Differential regulation of p21ras activation in neurons by nerve growth factor and brain-derived neurotrophic factor. J. Biol. Chem. 270:21751-21757. [DOI] [PubMed] [Google Scholar]
- 6.Chen, X. 1999. The p53 family: same response, different signals? Mol. Med. Today 5:387-392. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X., Y. Zheng, J. Zhu, J. Jiang, and J. Wang. 2001. p73 is transcriptionally regulated by DNA damage, p53, and p73. Oncogene 20:769-774. [DOI] [PubMed] [Google Scholar]
- 8.Chi, S. W., A. Ayed, and C. H. Arrowsmith. 1999. Solution structure of a conserved C-terminal domain of p73 with structural homology to the SAM domain. EMBO J. 18:4438-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costanzo, A., P. Merlo, N. Pediconi, M. Fulco, V. Sartorelli, P. A. Cole, G. Fontemaggi, M. Fanciulli, L. Schiltz, G. Blandino, C. Balsano, and M. Levrero. 2002. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol. Cell 9:175-186. [DOI] [PubMed] [Google Scholar]
- 10.De Laurenzi, V., A. Costanzo, D. Barcaroli, A. Terrinoni, M. Falco, M. Annicchiarico-Petruzzelli, M. Levrero, and G. Melino. 1998. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J. Exp. Med. 188:1763-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Laurenzi, V., A. Rossi, A. Terrinoni, D. Barcaroli, M. Levrero, A. Costanzo, R. A. Knight, P. Guerrieri, and G. Melino. 2000. p63 and p73 transactivate differentiation gene promoters in human keratinocytes. Biochem. Biophys. Res. Commun. 273:342-346. [DOI] [PubMed] [Google Scholar]
- 12.Dobbelstein, M., S. Wienzek, C. Konig, and J. Roth. 1999. Inactivation of the p53-homologue p73 by the mdm2-oncoprotein. Oncogene 18:2101-2106. [DOI] [PubMed] [Google Scholar]
- 13.Dohn, M., S. Zhang, and X. Chen. 2001. p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene 20:3193-3205. [DOI] [PubMed] [Google Scholar]
- 14.Duijf, P. H., K. R. Vanmolkot, P. Propping, W. Friedl, E. Krieger, F. McKeon, V. Dotsch, H. G. Brunner, and H. van Bokhoven. 2002. Gain-of-function mutation in ADULT syndrome reveals the presence of a second transactivation domain in p63. Hum. Mol. Genet. 11:799-804. [DOI] [PubMed] [Google Scholar]
- 15.Ellisen, L. W., K. D. Ramsayer, C. M. Johannessen, A. Yang, H. Beppu, K. Minda, J. D. Oliner, F. McKeon, and D. A. Haber. 2002. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol. Cell 10:995-1005. [DOI] [PubMed] [Google Scholar]
- 16.Erhardt, J. A., and R. N. Pittman. 1998. Ectopic p21(WAF1) expression induces differentiation-specific cell cycle changes in PC12 cells characteristic of nerve growth factor treatment. J. Biol. Chem. 273:23517-23523. [DOI] [PubMed] [Google Scholar]
- 17.Erhardt, J. A., and R. N. Pittman. 1998. p21WAF1 induces permanent growth arrest and enhances differentiation, but does not alter apoptosis in PC12 cells. Oncogene 16:443-451. [DOI] [PubMed] [Google Scholar]
- 18.Fang, L., S. W. Lee, and S. A. Aaronson. 1999. Comparative analysis of p73 and p53 regulation and effector functions. J. Cell Biol. 147:823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flores, E. R., K. Y. Tsai, D. Crowley, S. Sengupta, A. Yang, F. McKeon, and T. Jacks. 2002. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416:560-564. [DOI] [PubMed] [Google Scholar]
- 20.Fontemaggi, G., A. Gurtner, S. Strano, Y. Higashi, A. Sacchi, G. Piaggio, and G. Blandino. 2001. The transcriptional repressor ZEB regulates p73 expression at the crossroad between proliferation and differentiation. Mol. Cell. Biol. 21:8461-8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghioni, P., F. Bolognese, P. H. Duijf, H. Van Bokhoven, R. Mantovani, and L. Guerrini. 2002. Complex transcriptional effects of p63 isoforms: identification of novel activation and repression domains. Mol. Cell. Biol. 22:8659-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong, J. G., A. Costanzo, H. Q. Yang, G. Melino, W. G. Kaelin, Jr., M. Levrero, and J. Y. Wang. 1999. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 399:806-809. [DOI] [PubMed] [Google Scholar]
- 23.Grob, T. J., U. Novak, C. Maisse, D. Barcaroli, A. U. Luthi, F. Pirnia, B. Hugli, H. U. Graber, V. De Laurenzi, M. F. Fey, G. Melino, and A. Tobler. 2001. Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ. 8:1213-1223. [DOI] [PubMed] [Google Scholar]
- 24.Gu, J., L. Nie, H. Kawai, and Z. M. Yuan. 2001. Subcellular distribution of p53 and p73 are differentially regulated by MDM2. Cancer Res. 61:6703-6707. [PubMed] [Google Scholar]
- 25.Irwin, M., M. C. Marin, A. C. Phillips, R. S. Seelan, D. I. Smith, W. Liu, E. R. Flores, K. Y. Tsai, T. Jacks, K. H. Vousden, and W. G. Kaelin, Jr. 2000. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407:645-648. [DOI] [PubMed] [Google Scholar]
- 26.Irwin, M. S., and W. G. Kaelin. 2001. p53 family update: p73 and p63 develop their own identities. Cell Growth Differ. 12:337-349. [PubMed] [Google Scholar]
- 27.Irwin, M. S., K. Kondo, M. C. Marin, L. S. Cheng, W. C. Hahn, and W. G. Kaelin. 2003. Chemosensitivity linked to p73 function. Cancer Cell 3:403-410. [DOI] [PubMed] [Google Scholar]
- 28.Ishimoto, O., C. Kawahara, K. Enjo, M. Obinata, T. Nukiwa, and S. Ikawa. 2002. Possible oncogenic potential of DeltaNp73: a newly identified isoform of human p73. Cancer Res. 62:636-641. [PubMed] [Google Scholar]
- 29.Jost, C. A., M. C. Marin, and W. G. Kaelin, Jr. 1997. p73 is a human p53-related protein that can induce apoptosis. Nature 389:191-194. [DOI] [PubMed] [Google Scholar]
- 30.Kaelin, W. G., Jr. 1999. The p53 gene family. Oncogene 18:7701-7705. [DOI] [PubMed] [Google Scholar]
- 31.Kaghad, M., H. Bonnet, A. Yang, L. Creancier, J. C. Biscan, A. Valent, A. Minty, P. Chalon, J. M. Lelias, X. Dumont, P. Ferrara, F. McKeon, and D. Caput. 1997. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90:809-819. [DOI] [PubMed] [Google Scholar]
- 32.Kartasheva, N. N., A. Contente, C. Lenz-Stoppler, J. Roth, and M. Dobbelstein. 2002. p53 induces the expression of its antagonist p73 Delta N, establishing an autoregulatory feedback loop. Oncogene 21:4715-4727. [DOI] [PubMed] [Google Scholar]
- 33.Ko, L. J., and C. Prives. 1996. p53: puzzle and paradigm. Genes Dev. 10:1054-1072. [DOI] [PubMed] [Google Scholar]
- 34.Liu, G., and X. Chen. 2002. The ferredoxin reductase gene is regulated by the p53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene 21:7195-7204. [DOI] [PubMed] [Google Scholar]
- 35.Liu, G., T. Xia, and X. Chen. 2003. The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CREB-binding protein. J. Biol. Chem. 278:17557-17565. [DOI] [PubMed] [Google Scholar]
- 36.Marin, M. C., C. A. Jost, M. S. Irwin, J. A. DeCaprio, D. Caput, and W. G. Kaelin, Jr. 1998. Viral oncoproteins discriminate between p53 and the p53 homolog p73. Mol. Cell. Biol. 18:6316-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melino, G., V. De Laurenzi, and K. H. Vousden. 2002. p73: friend or foe in tumorigenesis. Nat. Rev. Cancer 2:605-615. [DOI] [PubMed] [Google Scholar]
- 38.Moll, U. M., S. Erster, and A. Zaika. 2001. p53, p63 and p73—solos, alliances and feuds among family members. Biochim. Biophys. Acta 1552:47-59. [DOI] [PubMed] [Google Scholar]
- 39.Nakagawa, T., M. Takahashi, T. Ozaki, K.-I. Watanabe, S. Todo, H. Mizuguchi, T. Hayakawa, and A. Nakagawara. 2002. Autoinhibitory regulation of p73 by ΔNp73 to modulate cell survival and death through a p73-specific target element within the ΔNp73 promoter. Mol. Cell. Biol. 22:2575-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nimura, Y., M. Mihara, S. Ichimiya, S. Sakiyama, N. Seki, M. Ohira, N. Nomura, M. Fujimori, W. Adachi, J. Amano, M. He, Y. M. Ping, and A. Nakagawara. 1998. p73, a gene related to p53, is not mutated in esophageal carcinomas. Int. J. Cancer 78:437-440. [DOI] [PubMed] [Google Scholar]
- 41.Nomoto, S., N. Haruki, M. Kondo, H. Konishi, and T. Takahashi. 1998. Search for mutations and examination of allelic expression imbalance of the p73 gene at 1p36.33 in human lung cancers. Cancer Res. 58:1380-1383. [PubMed] [Google Scholar]
- 42.Nozell, S., Y. Wu, K. McNaughton, G. Liu, A. Willis, J. C. Paik, and X. Chen. 2003. Characterization of p73 functional domains necessary for transactivation and growth suppression. Oncogene 22:4333-4347. [DOI] [PubMed] [Google Scholar]
- 43.Ohtsuka, T., H. Ryu, Y. A. Minamishima, A. Ryo, and S. W. Lee. 2003. Modulation of p53 and p73 levels by cyclin G: implication of a negative feedback regulation. Oncogene 22:1678-1687. [DOI] [PubMed] [Google Scholar]
- 44.Poluha, W., C. M. Schonhoff, K. S. Harrington, M. B. Lachyankar, N. E. Crosbie, D. A. Bulseco, and A. H. Ross. 1997. A novel, nerve growth factor-activated pathway involving nitric oxide, p53, and p21WAF1 regulates neuronal differentiation of PC12 cells. J. Biol. Chem. 272:24002-24007. [DOI] [PubMed] [Google Scholar]
- 45.Pozniak, C. D., S. Radinovic, A. Yang, F. McKeon, D. R. Kaplan, and F. D. Miller. 2000. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science 289:304-306. [DOI] [PubMed] [Google Scholar]
- 46.Roth, J., and M. Dobbelstein. 1999. Failure of viral oncoproteins to target the p53-homologue p51A. J. Gen. Virol. 80:3251-3255. [DOI] [PubMed] [Google Scholar]
- 47.Sakamuro, D., P. Sabbatini, E. White, and G. C. Prendergast. 1997. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene 15:887-898. [DOI] [PubMed] [Google Scholar]
- 48.Serber, Z., H. C. Lai, A. Yang, H. D. Ou, M. S. Sigal, A. E. Kelly, B. D. Darimont, P. H. Duijf, H. Van Bokhoven, F. McKeon, and V. Dotsch. 2002. A C-terminal inhibitory domain controls the activity of p63 by an intramolecular mechanism. Mol. Cell. Biol. 22:8601-8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stiewe, T., and B. M. Putzer. 2000. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat. Genet. 26:464-469. [DOI] [PubMed] [Google Scholar]
- 50.Thanos, C. D., and J. U. Bowie. 1999. p53 family members p63 and p73 are SAM domain-containing proteins. Protein Sci. 8:1708-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsao, H., X. Zhang, P. Majewski, and F. G. Haluska. 1999. Mutational and expression analysis of the p73 gene in melanoma cell lines. Cancer Res. 59:172-174. [PubMed] [Google Scholar]
- 52.Vossio, S., E. Palescandolo, N. Pediconi, F. Moretti, C. Balsano, M. Levrero, and A. Costanzo. 2002. DN-p73 is activated after DNA damage in a p53-dependent manner to regulate p53-induced cell cycle arrest. Oncogene 21:3796-3803. [DOI] [PubMed] [Google Scholar]
- 53.Yang, A., M. Kaghad, D. Caput, and F. McKeon. 2002. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 18:90-95. [DOI] [PubMed] [Google Scholar]
- 54.Yang, A., M. Kaghad, Y. Wang, E. Gillett, M. D. Fleming, V. Dotsch, N. C. Andrews, D. Caput, and F. McKeon. 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2:305-316. [DOI] [PubMed] [Google Scholar]
- 55.Yang, A., and F. McKeon. 2000. P63 and P73: P53 mimics, menaces and more. Nat. Rev. Mol. Cell Biol. 1:199-207. [DOI] [PubMed] [Google Scholar]
- 56.Yang, A., N. Walker, R. Bronson, M. Kaghad, M. Oosterwegel, J. Bonnin, C. Vagner, H. Bonnet, P. Dikkes, A. Sharpe, F. McKeon, and D. Caput. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404:99-103. [DOI] [PubMed] [Google Scholar]
- 57.Yuan, Z. M., H. Shioya, T. Ishiko, X. Sun, J. Gu, Y. Y. Huang, H. Lu, S. Kharbanda, R. Weichselbaum, and D. Kufe. 1999. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature 399:814-817. [DOI] [PubMed] [Google Scholar]
- 58.Zaika, A. I., N. Slade, S. H. Erster, C. Sansome, T. W. Joseph, M. Pearl, E. Chalas, and U. M. Moll. 2002. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J. Exp. Med. 196:765-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu, J., J. Jiang, W. Zhou, and X. Chen. 1998. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 58:5061-5065. [PubMed] [Google Scholar]
- 60.Zhu, J., J. Jiang, W. Zhou, K. Zhu, and X. Chen. 1999. Differential regulation of cellular target genes by p53 devoid of the PXXP motifs with impaired apoptotic activity. Oncogene 18:2149-2155. [DOI] [PubMed] [Google Scholar]