Abstract
The secondary structure and topology of membrane proteins can be described by inspection of two-dimensional 1H-15N dipolar coupling/15N chemical shift polarization inversion spin exchange at the magic angle spectra obtained from uniformly 15N-labeled samples in oriented bilayers. The characteristic wheel-like patterns of resonances observed in these spectra reflect helical wheel projections of residues in both transmembrane and in-plane helices and hence provide direct indices of the secondary structure and topology of membrane proteins in phospholipid bilayers. We refer to these patterns as PISA (polarity index slant angle) wheels. The transmembrane helix of the M2 peptide corresponding to the pore-lining segment of the acetylcholine receptor and the membrane surface helix of the antibiotic peptide magainin are used as examples.
Keywords: solid-state NMR, PISEMA, helical wheels, structure determination, PISA wheels
INTRODUCTION
Membrane proteins adopt unique three-dimensional structures in their functional environment of phospholipid bilayers. Nearly all of the residues in the polypeptide chains are immobile on time scales longer than milliseconds; therefore the relevant dipolar coupling and chemical shift interactions present in backbone sites are not motionally averaged, and a wide variety of solid-state NMR experiments are highly effective and can be used for structure determination (1, 2). In magic angle sample spinning NMR experiments, the resonances are narrowed through averaging of orientational parameters, enabling the measurement of internuclear distances by several different methods (2–4). In contrast, in solid-state NMR spectroscopy of stationary, oriented samples, the uniaxial orientation provides a mechanism for line-narrowing that retains both the distance and the angular information inherent in the anisotropic spin interactions. Although variations of the method have the potential for extending it to a wide range of systems (5, 6), nearly all applications of solid-state NMR spectroscopy to peptides and proteins in oriented lipid bilayer samples (7, 8) have utilized spin interactions between directly bonded nuclei. The bond length is assumed to have a fixed value, enabling the angular constraints to be measured and interpreted directly. Three-dimensional structure determination is feasible when multiple orientationally dependent frequencies are measured for nuclei at each residue, especially 15N-labeled amide sites (9, 10). The complete resolution and assignment of resonances from uniformly labeled samples in multidimensional solid-state NMR spectra is essential (11, 12) for determining the three-dimensional structures of membrane proteins.
In this Communication we demonstrate that, on the path toward three-dimensional structure determination, the secondary structure and topology of membrane proteins can be described by inspection of the two-dimensional 1H-15N dipolar coupling/15N chemical shift PISEMA (polarization inversion with spin exchange at the magic angle) spectra (14) of uniformly 15N-labeled samples in oriented bilayers. The characteristic “wheel-like” patterns of resonances observed in these spectra reflect helical wheel (15) projections of residues in both transmembrane and in-plane helices and hence provide direct indices of secondary structure and topology of membrane proteins in phospholipid bilayers. We refer to these patterns as PISA (polarity index slant angle) wheels. The transmembrane helix of the M2 peptide corresponding to the pore-lining segment of the acetylcholine receptor (AchR) and the membrane surface helix of the antibiotic peptide magainin are used as examples. This approach bears some similarity to the use of isotropic chemical shift indexes for assigning resonances and characterizing the secondary structure of proteins in solution NMR studies prior to the calculation of three-dimensional structures (16, 17). In a parallel and complementary study, Cross and co-workers have independently identified these same characteristics in PISEMA spectra of membrane proteins and present their analysis in an accompanying paper (18).
RESULTS AND DISCUSSION
Helices in Phospholipid Bilayers
The resonance frequencies in PISEMA spectra of oriented samples of membrane proteins depend on helix orientation, as well as on the backbone dihedral angles, the magnitudes and orientations of the principal elements of the amide 15N chemical shift tensor, and the NH bond length. Because the chemical shift tensor and bond length are reasonably well characterized for 15N-labeled amide sites (19), it is possible to calculate solid-state NMR spectra for specific structural models of proteins.
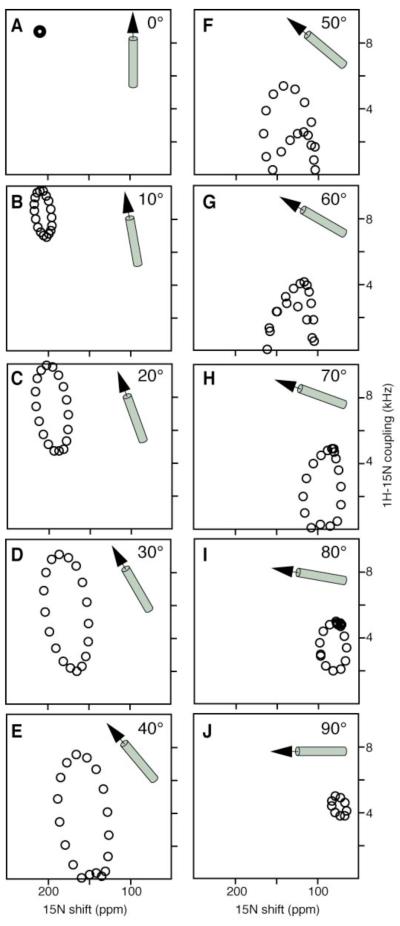
The PISEMA spectra calculated for the full range of possible orientations of an ideal 19-residue α-helix, with 3.6 residues per turn and identical backbone dihedral angles for all residues (ϕ = −65°, ψ = -40°), are shown in Fig. 1. When the helix axis is parallel to the bilayer normal all of the amide sites have an identical orientation relative to the direction of the applied magnetic field, and therefore all of the resonances overlap with the same 1H-15N dipolar coupling and 15N chemical shift frequencies. Since the amide backbone NH bonds in an α-helix are nearly, but not exactly, parallel to the helix axis, the resonance frequencies approach, but do not reach, their maximum values.
FIG. 1.
PISEMA spectra calculated for a 19-residue α-helix with 3.6 residues per turn and uniform dihedral angles at various helix tilt angles relative to the bilayer normal. A. 0°. B. 10°. C. 20°. D. 30°. E. 40°. F. 50°. G. 60°. H. 70°. I. 80°. J. 90°. Spectra were calculated on a Silicon Graphics O2 computer (Mountain View, CA), using the FORTRAN program FINGERPRINT (21, 24). The principal values and molecular orientation of the 15N chemical shift tensor (σ11 = 64 ppm; σ22 = 77 ppm; σ33 = 217 ppm; σ33NH = 17°) and the NH bond distance (1.07 Å) were as previously determined (19).
Tilting the helix away from the membrane normal breaks the symmetry, introducing variations in the orientations of the amide NH bond vectors relative to the field direction. This is manifest in the spectra as dispersions of both the 1H-15N dipolar coupling and the 15N chemical shift frequencies. Since a modest helix tilt of about 10° aligns the NH bond from one amide site and the σ33 amide 15N chemical shift tensor element of another amide site with the magnetic field, the maximum values of both of these frequencies are observed in the spectrum, albeit from different amide resonances because σ33 of the shift tensor is rotated approximately 17° from the NH bond vector (19). Nearly all transmembrane helices are tilted with respect to the bilayer normal (20). It is the combination of the tilt and the 17° difference between the direction of σ33 and the NH bond vector that makes it possible to resolve many resonances from residues in otherwise uniform helices and is responsible for the characteristic wheel-like pattern observed in the PISEMA spectra of uniformly 15N-labeled membrane peptides and proteins (10, 11, 21, 22).
For all helix orientations other than parallel to the field direction (0°), the PISEMA spectra shown in Fig. 1 have characteristic wheel-like patterns whose frequency breadths reflect the extent of the helix tilt. For helices with tilts equal to or greater than 40° some amide NH bonds adopt orientations relative to the magnetic field equal to the magic angle (54.7°) and have corresponding resonances with 1H-15N dipolar coupling frequencies of 0 kHz (Figs. 1E–1H). Other amide NH bonds are oriented with angles greater than 54.7° relative to the magnetic field resulting in resonances with 1H-15N dipolar coupling frequencies whose sign is inverted. Since PISEMA spectra are symmetric and do not distinguish between positive and negative dipolar coupling frequencies, helix tilts greater than about 40° result in a portion of the wheel-like PISEMA spectrum, which appears to be reflected through the 0-kHz axis. The sign of the dipolar coupling would be a valuable additional orientational constraint in samples that are aligned in a unique direction relative to the magnetic field. Finally, helices oriented parallel to the membrane surface have their amide NH bonds and σ33 of the 15N chemical shift tensor nearly orthogonal to the magnetic field and give highly overlapped PISEMA spectra with all of the resonance frequencies around 5 kHz and 75 ppm.
Figure 1 demonstrates that the wheel-like patterns in the PISEMA spectra of oriented samples are sensitive indicators of protein secondary structure and topology. Even without sequential resonance assignments, the 1H-15N dipolar coupling and 15N chemical shift frequencies of the resonances in the characteristic wheel-like patterns provide strong evidence for the presence of helical secondary structure as well as a direct measure of helix tilt relative to the bilayer normal (21). Variations in the backbone dihedral angles of ±10° lead to excursions as large as ±15 ppm in the 15N chemical shift, but less than ±1 kHz in the 1H-15N dipolar coupling frequencies. Variations in the magnitudes of the principal elements of the 15N chemical shift tensor of ±20 ppm have been observed (23, 24) and lead to excursions as large as ±15 ppm in the calculated 15N chemical shift frequency. Little variability has been observed in the tensor orientations. Finally, variations in the NH bond length as large as ±0.02 Å correspond to excursions of only ±0.6 kHz in the calculated 1H-15N dipolar coupling frequency. This makes the 1H-15N dipolar coupling the more reliable parameter for approximating the tilt of a transmembrane helix.
The experimental PISEMA spectrum of the AchR M2 peptide shown in Fig. 2B is well resolved. This made it possible to measure the 1H-15N dipolar coupling and 15N chemical shift frequencies for each correlation peak assigned to an amide site in the peptide backbone and to determine the structure of the peptide in the membrane (10). Three-dimensional correlation spectra (25) are generally not needed to resolve the resonances of transmembrane helices, even when two or more are present in the protein (22); this is in contrast to the situation for surface bound helices that are orthogonal to the membrane normal, as discussed below and in an accompanying paper (26). Nonetheless, three-dimensional correlation spectra are essential for the measurement of 1H chemical shift frequencies for all resonances, which provide valuable orientational constraints (27).
FIG. 2.

Helical wheel projection and two-dimensional PISEMA spectra of the uniformly 15N-labeled polypeptides in oriented lipid bilayers. A, B, and C. AChR M2. D, E, and F. Magainin II. A. Helical wheel projection of the amide N atoms of AChR M2. B. Experimental PISEMA spectrum of AChR M2, which provided most of the orientational constraints used for structure determination (10). C. Spectrum calculated for an α-helix with 3.6 residues per turn, identical dihedral angles, and a tilt of 12° relative to the membrane normal. D. Helical wheel view of the CA atoms of the magainin II helix with the N-terminus in front. The three-dimensional orientation of the peptide was determined from the angular constraints for Val17 derived from the three-dimensional solid-state NMR spectrum (24). The dashed line marks the boundary between polar (white) and hydrophobic (gray) residues in the amphipathic helix. E. Experimental PISEMA spectrum of magainin II. F. Spectrum calculated from the solution NMR structure of magainin in lipid micelles (PDB file 2MAG). Conditions for sample preparation and for solid-state NMR experiments of AChR M2 (10) and magainin (28) have been described. Spectra were calculated as described in Fig. 1.
The resonances in the assigned experimental PISEMA spectrum of uniformly 15N-labeled AchR M2 (Fig. 2B) trace out a wheel-like pattern similar to those described in Fig. 1. The spectrum calculated for an ideal transmembrane α-helix tilted by 12° with 3.6 residues per turn and identical dihedral angles (ϕ = −65°, ψ = -40°) is shown in Fig. 2C. Differences between the experimental spectrum of AchR M2, whose helix was found to tilt by about 12° with respect to the membrane normal through structure determination (10), and the calculated spectrum are largely due to deviations in the experimentally determined backbone dihedral angles of AChR M2 (PDB file 1cek) from those of an ideal α-helix. Thus, even without resonance assignments, comparison of the experimental and calculated spectra leads to the conclusion that the AChR M2 helix is transmembrane with a tilt angle in the range of 10 to 20° from the bilayer normal. A similar comparison was used to determine that the single transmembrane helix in Vpu, a membrane bound accessory protein from HIV-1, is tilted by about 15° (21).
In the experimental PISEMA spectrum of AchR M2, the wheel-like pattern of assigned resonances is identical to the helical wheel projection of the peptide shown in Fig. 2A, with the exceptions of Val15 and Ile7. The helical wheel in Fig. 2A is arranged so that the residues align with their corresponding amide resonances in the assigned experimental PISEMA spectrum (Fig. 2B). This is exactly as predicted by the assigned resonances in the calculated spectrum (Fig. 2C) and reinforces the point that the solid-state NMR spectra of oriented proteins contain all of the structural information necessary for structure determination. Indeed, the polarity of the wheel-like pattern in a PISEMA spectrum provides a direct measure of the angle of helix rotation about its long axis within the membrane.
The correspondence among the polarity of the helical wheel, the helix rotation and tilt, and the polarity of the wheel-like pattern in the resulting PISEMA spectrum is illustrated in Fig. 3. In a mutually orthogonal laboratory frame of reference (x, y, z) with the z axis parallel to the lipid bilayer normal (n) and to the direction of the applied magnetic field (B°), we define the angle χ = 0° for a helix axis (HA) parallel to z and the amide nitrogen of residue 2 aligned with the x axis. The helix polarity is defined by a right-handed rotation of the helical wheel about the z axis through the angle χ (Figs. 3A and 3D). The helix tilt from the bilayer normal is prescribed by a right-handed rotation of the helix axis about y, through an angle δ (Figs. 3B and 3E). Therefore, δ is the angle between HA and the direction of B°, and χ is the polar angle between the projection of B° on the helical wheel and a vector drawn between residue 2 and the center of the wheel. These angles (χ, δ) are sufficient to fully define the orientation of a helix in a lipid bilayer membrane.
FIG. 3.
Correspondence between membrane protein helix tilt and polarity, and the resulting PISEMA spectra for uniformly 15N-labeled protein in oriented bilayers. A, D, G, and J. Helical wheels rotated by various values of the polar angle χ. B, E, H, and K. Helices rotated through various values of χ about their long axes (HA) and tilted by δ = 12° (B, E) and δ = 90° (H, K) away from the membrane normal (n). The y axis of the laboratory frame points out of the page. C, F, I, and L. Calculated PISEMA spectra for the various helix rotations and tilts. The NH bond vectors of the polar opposite residues 2 and 11 in the helical wheels are highlighted. The light gray areas in B, E, H, and K represent the lipid bilayer.
In Figs. 3C and 3F, the wheels traced out in the PISEMA spectra for two different helix orientations, with equal values of δ, but different values of χ, are identical because the 1H-15N dipolar coupling and 15N chemical shift frequencies reflect only the angle of tilt of the helix axis relative to the bilayer normal (21). However the spectra differ in their resonance assignments, whose patterns mirror exactly the polarity χ of their corresponding helical wheels. In Fig. 3D, helix rotation through χ aligns a vector drawn between the amide N atoms of residues 2 and 11 with the laboratory y axis. The helix is then tilted through δ around this vector so that the NH bond of residue 11 is aligned with the magnetic field, while the NH bond of residue 2 makes a larger angle with respect to the field. As a result, the corresponding resonances have the highest (residue 11) and lowest (residue 2) 1H-15N dipolar coupling frequencies in the wheel-like pattern, which is similar to that observed for AchR M2 (Fig. 2B).
Importantly, this implies that when a wheel-like PISEMA spectrum is observed no assignments are needed to determine the tilt of a transmembrane helix (21) in the membrane, and a single resonance assignment is sufficient to determine helix rotation (see below). These are important parameters because the tilt and rotation of transmembrane helices determine the supramolecular architectures of membrane protein assemblies. For example, the structure of the AchR M2 determined by solid-state NMR spectroscopy in lipid bilayers is tilted by 12° and rotated about its helix axis so that the hydrophilic residues face the N-terminal side of the membrane. This has important consequences for ion channel pore geometry and conduction as it leads to the assembly of a symmetric, pentameric, funnel-like pore with its wide opening at the N-terminal side of the membrane (10). All of these conclusions about the structure of the peptide in bilayers are immediately apparent from inspection of the assigned PISEMA spectrum in Fig. 2B, prior to complete structure determination.
In-Plane Helices Parallel to the Membrane Surface
Typically membrane surface associated α-helices orient with their axes parallel to the bilayer surface and orthogonal to the direction of the applied magnetic field. As shown in Fig. 1J and described in the accompanying paper (26), this results in PISEMA spectra that are highly overlapped and require the use of three-dimensional correlation spectroscopy (25) in order to separate amide backbone resonances on the basis of 1H chemical shift differences. Tilting the helix from this 90° orientation results in local angular differences, which give dispersions in both the 1H-15N dipolar coupling and the 15N chemical shift frequency dimensions.
Nevertheless, as shown in Figs. 1I and 1J, characteristic wheel-like resonance patterns are also observed in the PISEMA spectra of a-helices aligned parallel to the membrane surface. The experimental PISEMA spectrum of uniformly 15N-labeled magainin, a helical peptide that lies in the plane of the bilayer, is shown in Fig. 2E. The resonance assigned to the amide nitrogen of Val17 is marked. Many features of the experimental solid-state NMR spectrum in Fig. 2E are reproduced in the spectrum in Fig. 2F, which was calculated based on the coordinates of the structure of magainin II determined in lipid micelles by solution NMR spectroscopy (28; PDB file 1MAG) and the orientation of the Val17 peptide plane determined from solid-state NMR data (29). Differences between the experimental and calculated magainin spectra are most likely due to variations in the backbone dihedral angles between the structure in lipid micelles and lipid bilayers, variations in the 15N chemical shift tensors, and in the NH bond lengths.
For an α-helix with 3.6 residues per turn, adjacent amide residues are separated by 100° of the arc of the wheel, resulting in 18 different amide NH bond orientations that can be sampled for any given helix orientation (15). However, this is effectively reduced to 9 orientations when the helix axis is oriented parallel to the membrane surface. This is because, for this helix tilt (δ = 90°), polar opposite residue pairs that are separated by 180° of the arc of the wheel have NH bonds with angles θ or 180 - θ relative to the magnetic field (e.g., residues 11 and 2 in Figs. 3G–3L). These angles are indistinguishable in solid-state NMR spectroscopy of uniaxially oriented samples; therefore, amide sites separated by 8 residues and at opposite poles of the helical wheel have identical 1H-15N dipolar coupling and 15N chemical shift frequencies. In the calculated PISEMA spectrum of magainin (Fig. 2F) the polar opposite residue pairs Val17/Ser8, Ala15/Leu6, and Lys14/Phe5 have overlapping resonances, as expected for a α-helix lying in the plane of the bilayer.
Thus, as observed for transmembrane helices, the PISEMA spectra of in-plane a-helices also reflect the helix tilt and rotation described in terms of the angles δ and χ. The spectra calculated for two helices oriented parallel to the membrane surface (δ = 90°) but with different rotations of χ about their respective helix axes are shown in Figs. 3I and 3L. The spectra trace out identical wheel-like patterns since they reflect only the tilt of the helix. However, the resonance assignments are markedly different and mirror both the helix rotation at the membrane (Figs. 3H and 3K) and the corresponding helical wheel projections (Figs. 3G and 3J). As discussed above, resonances from polar opposite residue pairs overlap. Because the resonances in the PISEMA spectra of in-plane helices have 1H-15N dipolar couplings of opposite sign to those of transmembrane helices and are reflected through 0 kHz, their wheel-like resonance patterns have a handedness contrary to that observed for transmembrane helices.
The sensitivity of assigned PISEMA spectra to the polarity of a helix bound to the membrane surface is particularly useful for determining the mode of binding of an amphipathic helix like magainin II. This peptide lies on the membrane surface exposing its apolar side to the hydrophobic core of the lipid bilayer and with its polar–apolar interface parallel to the bilayer surface (29). In an α-helix, amide NH bonds at sites near the polar–apolar interface make larger angles with respect to the direction of the field (residues 2 and 11 in Fig. 3J) than do sites farther from the interface (residues 2 and 11 in Fig. 3G). This results in PISEMA spectra where the resonances are effectively polarized: amide sites near the polar–apolar boundary have the smallest 1H-15N dipolar couplings and 15N chemical shifts, while amide sites farthest from the boundary have the largest 1H-15N dipolar couplings and 15N chemical shifts. This is clearly observed in the spectrum of magainin (Figs. 2E and 2F). Residues Met21, Phe5, Lys14, and Glu19 are located near the polar–apolar interface (dashed line in Fig. 2D) and have corresponding PISEMA resonances with smaller 1H-15N dipolar coupling and 15N chemical shift frequencies. On the other hand, residues Lys4 and Asn22 in the center of the hydrophilic face of the helix (white atoms in Fig. 2D) and residues Leu6, Gly13, and Ile20 in the center of the hydrophobic face of the helix (gray atoms in Fig. 2D) have PISEMA resonances with larger frequencies.
CONCLUSIONS
Only the resolution and sequential assignment of resonances from all residues, and the subsequent calculation of the three-dimensional structure, can provide the definitive answer about the three-dimensional structure of a membrane protein. However, the helical wheel patterns observed in the PISEMA spectra of uniformly 15N-labeled polypeptides associated with oriented lipid bilayers provide a powerful qualitative measure of membrane protein secondary structure and topology. In solution NMR spectra of proteins the sensitivity of the isotropic chemical shifts to local molecular geometry provides a method for determining secondary structural elements based on simple inspection of resonance frequencies (16, 17). This chemical shift index is so effective that it is routinely used to identify macromolecular structural features and guide structure refinement. Similarly, the PISA wheel patterns observed in PISEMA spectra of uniformly 15N-labeled samples of proteins in oriented bilayers identify the structure and topology of membrane proteins. In addition, the spectral parameters provide constraints for the calculation of the three-dimensional structures of membrane proteins.
ACKNOWLEDGMENTS
We thank T. A. Cross for discussing his results and sending us a copy of his manuscript prior to submission for publication. We also thank M. Montal and M. Zasloff for their participation in the long-term collaborations that led to the expression and uniform 15N-labeling of the recombinant polypeptides used in these experiments. This research was supported by grants from the National Institute of General Medical Sciences (R01 GM29754 and P01 GM56538) to S.J.O. and the W. W. Smith Charitable Trust to F.M.M. It utilized the Resource for Solid-State NMR of Proteins at the University of Pennsylvania, supported by Grant P41 RR09731 from the Biomedical Research Technology Program, National Center for Research Resources, National Institutes of Health.
REFERENCES
- 1.Opella SJ. NMR and membrane proteins. Nat. Struct. Biol. NMR I Suppl. 1997;4:845–848. [PubMed] [Google Scholar]
- 2.Griffin R. Dipolar recoupling in MAS spectra of biological solids. Nat. Struct. Biol. NMR II Suppl. 1998;5:508–512. doi: 10.1038/749. [DOI] [PubMed] [Google Scholar]
- 3.Smith SO, Ascheim K, Groesbeck M. Magic angle spinning NMR spectroscopy of membrane proteins. Q. Rev. Biophys. 1996;29:395–449. doi: 10.1017/s0033583500005898. [DOI] [PubMed] [Google Scholar]
- 4.McDowell LM, Schaefer J. High resolution NMR of biological solids. Curr. Opin. Struct. Biol. 1996;6:624–629. doi: 10.1016/s0959-440x(96)80028-5. [DOI] [PubMed] [Google Scholar]
- 5.Glaubitz C, Watts A. Magic angle-oriented sample spinning (MAOSS): A new approach toward biomembrane studies. J. Magn. Reson. 1998;130:305–316. doi: 10.1006/jmre.1997.1344. [DOI] [PubMed] [Google Scholar]
- 6.Grage SL, Ulrich AS. Structural parameters from 19F homonuclear dipolar couplings, obtained by multipulse solid-state NMR on static and oriented systems. J. Magn. Reson. 1999;138:98–106. doi: 10.1006/jmre.1999.1726. [DOI] [PubMed] [Google Scholar]
- 7.Cross TA, Opella SJ. Solid-state NMR structural studies of peptides and proteins in membranes. Curr. Opin. Struct. Biol. 1994;4:574–581. [Google Scholar]
- 8.Marassi FM, Opella SJ. NMR structural studies of membrane proteins. Curr. Opin. Struct. Biol. 1998;8:640–648. doi: 10.1016/s0959-440x(98)80157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ketchem RR, Hu W, Cross TA. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science. 1993;261:1457–1460. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- 10.Opella SJ, Marassi FM, Gesell JJ, Valente AP, Kim Y, Oblatt-Montal M, Montal M. Three-dimensional structure of the membrane-embedded M2 channel-lining segment from nicotinic acetylcholine receptors and NMDA receptors by NMR spectroscopy. Nat. Struct. Biol. 1999;6:374–379. doi: 10.1038/7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marassi FM, Ramamoorthy A, Opella SJ. Complete resolution of the solid-state NMR spectrum of a uniformly 15N-labeled membrane protein in phospholipid bilayers. Proc. Natl. Acad. Sci. USA. 1997;94:8551–8556. doi: 10.1073/pnas.94.16.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marassi FM, Gesell JJ, Valente AP, Oblatt-Montal M, Montal M, Opella SJ. Dilute spin-exchange assignment of solid-state NMR spectra of oriented proteins: acetylcholine M2 in bilayers. J. Biomol. NMR. 1999;14:141–148. doi: 10.1023/a:1008391823293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan WM, Gu Z, Zeri AC, Opella SJ. Solid-state NMR triple resonance backbone assignments in a protein. J. Biomol. NMR. 1999;13:337–342. doi: 10.1023/a:1008379105545. [DOI] [PubMed] [Google Scholar]
- 14.Wu CH, Ramamoorthy A, Opella SJ. High resolution heteronuclear dipolar solid-state NMR spectroscopy. J. Magn. Reson. A. 1994;109:270–272. [Google Scholar]
- 15.Schiffer M, Edmunson AB. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys. J. 1967;7:121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wishart DS, Sykes BD, Richards FM. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 17.Kuszewski J, Qin J, Gronenborn AM, Clore GM. The impact of direct refinement against 13Cg and 13Cb chemical shifts on protein structure determination by NMR. J. Magn. Reson. B. 1995;106:92–96. doi: 10.1006/jmrb.1995.1017. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Denny J, Tian C, Kim S, Mo Y, Kovacs F, Song Z, Nishimura K, Gan Z, Fu R, Quine JR, Cross TA. Imaging membrane protein helical wheels. J. Magn. Reson. 2000;144:162–167. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- 19.Wu C, Ramamoorthy A, Gierasch LM, Opella SJ. Simultaneous characterization of the amide 1H chemical shift, 1H-15N dipolar, and 15N chemical shift interaction tensors in a peptide bond by three-dimensional solid-state NMR spectroscopy. J. Am. Chem. Soc. 1995;117:6148–6149. [Google Scholar]
- 20.Bowie JU. Helix packing in membrane proteins. J. Mol. Biol. 1997;272:780–789. doi: 10.1006/jmbi.1997.1279. [DOI] [PubMed] [Google Scholar]
- 21.Marassi FM, Ma C, Gratkowski H, Straus SK, Strebel K, Oblatt-Montal M, Montal M, Opella SJ. Correlation of the structural and functional domains in the membrane protein Vpu from HIV-1. Proc. Natl. Acad. Sci. USA. 1999;96:14336–14341. doi: 10.1073/pnas.96.25.14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y, Valentine K, Opella SJ, Schendel SL, Cramer WA. Solid-state NMR studies of the membrane-bound closed state of the colicin E1 channel domain in lipid bilayers. Protein Sci. 1998;7:342–348. doi: 10.1002/pro.5560070214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mai W, Hu W, Wang C, Cross TA. Orientational constraints as 3-dimensional structural constraints from chemical-shift anisotropy— The polypeptide backbone of gramicidin-A in a lipid bilayer. Protein Sci. 1993;2:532–542. doi: 10.1002/pro.5560020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fushman D, Cowburn D. Model-independent analysis of 15N chemical shift anisotropy from NMR relaxation data. Ubiquitin as a test example. J. Amer. Chem. Soc. 1998;120:7109–7110. [Google Scholar]
- 25.Ramamoorthy A, Wu CH, Opella SJ. Three-dimensional solid-state NMR experiment that correlates the chemical shift and dipolar coupling frequencies of two heteronuclei. J. Magn. Reson. B. 1995;107:88–90. doi: 10.1006/jmrb.1995.1063. [DOI] [PubMed] [Google Scholar]
- 26.Marassi FM, Ma C, Gesell JJ, Opella SJ. Three-dimensional solid-state NMR spectroscopy is essential for resolution of resonances from in-plane residues in uniformly 15N-labeled helical membrane proteins in oriented lipid bilayers. J. Magn. Reson. 2000;144:156–161. doi: 10.1006/jmre.2000.2036. [DOI] [PubMed] [Google Scholar]
- 27.Marassi FM, Ma C, Gesell JJ, Opella SJ. The roles of homonuclear line narrowing and the 1H amide chemical shift tensor in structure determination of proteins by solid-state NMR spectroscopy. Appl. Magn. Reson. 1999;17:433–447. [Google Scholar]
- 28.Gesell JJ, Zasloff M, Opella SJ. Two-dimensional 1H NMR experiments show that the 23-residue magainin antibiotic peptide is an alpha helix in dodecylphosphocholine micelles, sodium dodecylsulfate micelles, and trifluoroethanol/water solution. J. Biomol. NMR. 1997;9:127–135. doi: 10.1023/a:1018698002314. [DOI] [PubMed] [Google Scholar]
- 29.Ramamoorthy A, Marassi FM, Zasloff M, Opella SJ. Three-dimensional solid-state NMR spectroscopy of a peptide oriented in membrane bilayers. J. Biomol. NMR. 1995;6:329–334. doi: 10.1007/BF00197814. [DOI] [PMC free article] [PubMed] [Google Scholar]