Abstract
Transforming growth factor beta (TGF-β)/bone morphogenetic protein (BMP) family ligands interact with specific membrane receptor complexes that have serine/threonine kinase activities. The receptor phosphorylation and activation induced by the ligands leads to phosphorylation of the Smad proteins, which translocate to the nucleus, controlling gene expression. Thus, regulation of Smad proteins is a key step in TGF-β/BMP-induced signal transduction. Here we report a novel mechanism of the regulation of SMAD-mediated signaling, by which the Smad1 protein level is controlled through expression of the CHIP protein. CHIP is a U-box-dependent E3 ubiquitin ligase, previously identified as a cochaperon protein. However, we have isolated CHIP as a Smad-interacting protein in a yeast two-hybrid screen using Smad1 as bait. Furthermore we have shown CHIP-Smad interaction using the 35S-labeled CHIP protein, which can interact with glutathione S-transferase (GST)-Smad1 and GST-Smad4 in an in vitro protein-binding assay. The CHIP-Smad interaction has been confirmed in vivo in mammalian cells through coimmunoprecipitation. Interestingly, we demonstrate that the coexpression of Smad1 and Smad4 with the CHIP protein results in the degradation of the Smad proteins through a ubiquitin-mediated process. Consistent with the observation that CHIP induces Smad1 degradation, we further show that the expression of CHIP can inhibit the transcriptional activities of the Smad1/Smad4 complex induced by BMP signals. Intriguingly, pBS/U6/CHIPi, which diminishes CHIP expression, significantly enhanced Smad1/Smad4- or BMPRIB(QD)-induced gene transcription. These results suggest that CHIP can interact with the Smad1/Smad4 proteins and block BMP signal transduction through the ubiquitin-mediated degradation of Smad proteins.
Transforming growth factor beta (TGF-β) superfamily members, including bone morphogenetic proteins (BMPs), TGF-βs, activins, and inhibins, regulate multiple processes in cell proliferation, differentiation, and development (17). TGF-β signals through the activation of the Smad proteins, which are conserved from Drosophila to human (5, 20, 29). TGF-β binds to the type 2 receptor and initiates the assembly of heteromeric complexes of type 2 and type 1 receptors. The type 2 receptor kinase then phosphorylates the type 1 receptor kinase, which further phosphorylates the Smad proteins. Three groups of the Smads have been identified: the receptor-regulated Smads (R-Smad), including Smads 1, 2, 3, 5, and 8, which are phosphorylated by receptor 1 kinase; the common Smad (i.e., Smad4); and the inhibitory Smads (I-Smad) (29). After phosphorylation, R-Smads interact with the Co-Smad, forming an active complex, such as Smad1/Smad4 or Smad2/Smad4. Then, the complex translocates from the cytoplasm into the nucleus to regulate transcription of the target genes. The TGF-β/BMP signal pathways could be inhibited by I-Smads, which include Smads 6 and 7 (22). Recent studies have further shown that a number of partner proteins interact with the Smad complexes to regulate TGF-β/BMP signaling. Some partner proteins, such as FAST-1 and CBP/p300, can interact with Smads to regulate the transactivation of target genes induced by TGF-β (4, 7, 14, 23), whereas several other partner proteins, such as SIP1, Ski, SNIP1, Evi-1, and TGIF, were identified as corepressors of Smads (11, 12, 16, 25, 28). These observations indicate that the function of Smad proteins can be regulated by selective partners.
TGF-β/BMP signal pathways are also regulated at the level of Smad protein turnover. Two novel proteins, Smurf1 and Smurf2, have been identified that interact with R-Smads during BMP signal transduction, leading to Smad ubiquitination and degradation (3, 13, 15, 30, 31). It has also been reported that both Smurf1 and Smurf2 can interact with the TGF-β type 1 receptor through Smad7 and induce receptor degradation (6, 10, 20). During the process of the degradation induced by E3 ubiquitin ligase, other cofactors could also be involved. For example, Smad2 mediates the association of Smurf2 with the transcriptional corepressor SnoN, targeting SnoN for ubiquitin-mediated degradation by the proteasome (3).
To search further for Smad-interacting proteins, we have carried out yeast two-hybrid screens using Smad1 as bait. In this report we present the evidence that CHIP (carboxyl terminus of Hsc70-interacting protein), which was previously shown as a cochaperon protein and a U-box-dependent E3 ligase (2, 9, 21, 27), interacts with Smad1 and Smad4, and expression of the CHIP protein suppresses the activation of the Smad-mediated signaling pathways, suggesting a novel mechanism of regulating TGF-β/BMP signal transduction through CHIP-mediated Smad degradation.
MATERIALS AND METHODS
Yeast two-hybrid assay.
Yeast two-hybrid screens were performed with the MATCHMAKER two-hybrid system (Clontech). Positive and negative controls were pCL1 and pBGT9, respectively. Full-length human Smad1 (hSmad1), kindly provided by Xiao-Fan Wang at Duke University, was cloned as a fusion protein with the GAL4 DNA-binding domain in plasmid pGBT9 and used as the bait. A human brain library in pACT2 was screened for potential interacting proteins in the yeast strain Y190. Plasmids were transformed into yeast, and protein interactions were assayed by scoring β-galactosidase (β-Gal) activity, according to the protocol provided by Clontech.
GST fusion proteins, in vitro protein binding, and pull-down assays.
The full-length cDNA of hSmads (Smad1/2/3/4) and the full-length cDNA of human CHIP were inserted in frame into pGEX-5x-3 and pGEX-5x-1. Recombinant proteins were expressed in Escherichia coli BL21. The cells were grown for 4 h at 37°C, and the protein was induced with 0.5 mM isopropyl-I-thio-β-galactopyranoside at 30°C for 5 h. E. coli cells were harvested and then lysed by sonication. Glutathione S-transferase (GST) fusion proteins were purified by affinity chromatography on glutathione-Sepharose 4B (Amersham Pharmacia Biotech) according to the manufacturer's instructions.
For in vitro protein translation, full-length human CHIP and hSmad1 cDNA with various deletions were inserted into pcDNA6 (pcDNA6-CHIP), which contained a T7 promoter. 35S-labeled in vitro-translated proteins were produced using the rabbit reticulocyte lysate system (TNT kit; Promega) according to the manufacturer's instructions. For binding assays, 35S-labeled proteins and various GST fusion proteins were mixed and incubated at 4°C for 4 h. Then, the complexes were isolated by binding to glutathione-Sepharose beads (Amersham Pharmacia Biotech). The bound proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Dried gels were visualized with Molecular Dynamics Storm PhosphorImager model 860 (Amersham Pharmacia Biotech).
For GST pull-down assays, full-length Flag-tagged Smads (pRK5-Smad1, Smad2, Smad3, and Smad4) were transfected into 239T cells using Tfx-20 (Promega) according to the manufacturer's instructions. The cells were lysed, and the cell lysates were incubated with the GST-CHIP fusion protein at 4°C for 4 h. The complexes were isolated by binding to glutathione-Sepharose beads, subjected to SDS-polyacrylamide gel electrophoresis, and immunoblotted with an anti-Flag antibody.
Western blot analysis.
Proteins were separated on a 10% SDS-polyacrylamide gel and transferred onto hydrophobic polyvinylidene difluoride membranes (Amersham Pharmacia Biotech). To visualize the transferring efficiency, membranes were stained with Ponceau S (Sigma), destained with Milli-Q water, and blocked in 10% dry milk-TBST (20 mM Tris-HCl [pH 7.6], 127 mM NaCl, 0.1% Tween 20) for 1 h at 37°C. Blots were washed briefly and incubated with the appropriate specific primary antibody in TBST for 1 h at 37°C. Following three washes, membranes were then incubated with secondary antibody (fluorescein-linked anti-mouse immunoglobulin G [IgG]) for 40 min at 37°C in TBST. After another three washes in TBST, membranes were incubated in the third antibody (anti-fluorescein alkaline phosphatase conjugated) at 37°C for 40 min again. The ECF detection system (Amersham Pharmacia Biotech) was used to visualize protein bands. Sources of the purchased antibodies were the following: anti-Flag M2 monoclonal antibody, Sigma; antihemagglutinin (anti-HA) probe mouse monoclonal antibody, Santa Cruz; anti-Myc mouse monoclonal antibody, Santa Cruz; anti-His monoclonal antibody, Qiagen; anti-green fluorescent protein (anti-GFP) rabbit polyclonal antibody, Santa Cruz; anti-Smad1 mouse monoclonal antibody, Santa Cruz. The secondary and the third antibodies were from Amersham Pharmacia Biotechnology.
Immunoprecipitation.
HEK 293T or COS7 cells were transiently transfected with Flag-tagged Smads (pRK5-Smad1 and Smad4) and Myc-tagged CHIP (pRK1 M-CHIP) or HA-tagged CHIP (pcDNA6-CHIP) using Tfx-20 (Promega) in a 100-mm-diameter dish. Empty vector was added to balance the total DNA amounts. After transfection (36 to 48 h), cells were lysed in 1 ml of the whole-cell extract buffer A (400 mM KCl, 10 mM Na2HPO4, 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 0.5% NP-40 with proteasome inhibitors). Two-hundred-microliter cell lysates were used in immunoprecipitation with anti-Flag M2 monoclonal antibody and immunoblotted with anti-HA monoclonal antibodies. The endogenous CHIP protein was recognized and immunoprecipitated using a rabbit polyclonal antiserum generated in our laboratory. For inhibition of proteasome degradation, cells were incubated with 50 μM MG132 (Sigma) for 4 h.
Metabolic labeling and immunoprecipitation.
HepG2 or Mv1Lu cells were grown in six-well plates and transfected with DNA plasmids as indicated in the text. Twenty-four hours after transfection, cells were starved in methionine-free Dulbecco's modified Eagle medium (Life Technologies) for 1 h and metabolically labeled with [35S]methionine (100 μCi per well) for 4 h at 37°C. Cells were then washed with ice-cold phosphate-buffered saline twice and lysed under nondenaturing conditions with 300 μl of buffer A. All lysates were precleared with 0.5 μg of IgG with the same procedure as for immunoprecipitation. Briefly, reactions were carried out at 4°C for 30 min followed by centrifugation at 500 × g for 5 min. The supernatant was used to carry out the immunoprecipitation. Anti-Smad1 antibody or anti-CHIP serum was added to the supernatant as appropriate and rotated at 4°C for 1 h, and then protein G-agarose was added, with rotation for another 1 h. The pellet was washed with buffer A once, and the immunoprecipitates were eluted with 40 μl of SDS-PAGE sample buffer after incubation for 20 min at 50°C. Then, 35 μl of the aliquot was eluted with 750 μl of buffer B (0.1% Triton, 0.1% SDS in phosphate-buffered saline) containing 0.5% bovine serum albumin and incubated at 50°C for 10 min. To conduct reimmunoprecipitation, anti-Myc mouse monoclonal antibody or anti-CHIP serum was added, followed by incubation for 45 min at room temperature. Thirty microliters of protein G-agarose was then added, with incubation for an additional 45 min. The pellet was washed with buffer B twice, and then 40 μl of 1× SDS-PAGE loading buffer was added and incubated for 20 min at 50°C to elute the reimmunoprecipitated material. The elutant was then analyzed by SDS-PAGE and visualized by autoradiography. In all of the immunoprecipitation assays, 50 μM MG132 was treated for 4 h before harvest.
Ubiquitination assay.
Basically, this assay was performed as previously described by Bonni et al. (3). HA-tagged ubiquitin was cotransfected with Smad1 and CHIP or Smurf2 in the presence of 50 μM MG132 for 4 h before harvest. The cell lysates were precipitated with Ni-nitrilotriacetic acid (NTA) beads. The beads were washed four times for 30 min with washing buffer (20 mM imidazole, 0.05% Tween 20, 50 mM NaH2PO4, 300 mM NaCl [pH. 8.0]). The complexes were eluted with loading buffer and subjected to SDS-PAGE. Anti-HA antibody was used for Western blotting.
Luciferase assay.
Luciferase assays with various expression constructs were carried out using 293T, HepG2, or Mv1Lu cells. Cells at 50 to 60% confluence were transiently transfected with an appropriate combination of the constructs, Smads, and CHIP expression vectors, and the empty vector was used to equalize the total DNA amount. Cells were harvested after 24 h. The luciferase activity was determined by Top Count (Packard) using a kit (Promega). The internal control plasmids were supplied by the company (Promega). The data were normalized with the internal control. The experiments were done in triplicate, and the standard error and mean were calculated. In the experiment using BMP conditioned medium, the expression vector of BMP2 (pAD/hBMP2) was transfected into CHO cells and the medium was collected after 48 h of transfection.
SiRNA construction.
The pBS/U6/CHIPi was constructed according to a previously published protocol (24). Briefly, a fragment starting with GGG in the position of 233 to 251 bp from ATG in the CHIP cDNA was synthesized, and the hairpin structure was generated by a GGGCCC palindrome. The insert was constructed in the pBS/U6 vector. The vector and pBS/U6/EGFPi were kindly provided by Yang Shi of Harvard University.
RESULTS
CHIP was isolated as a Smad-interacting protein.
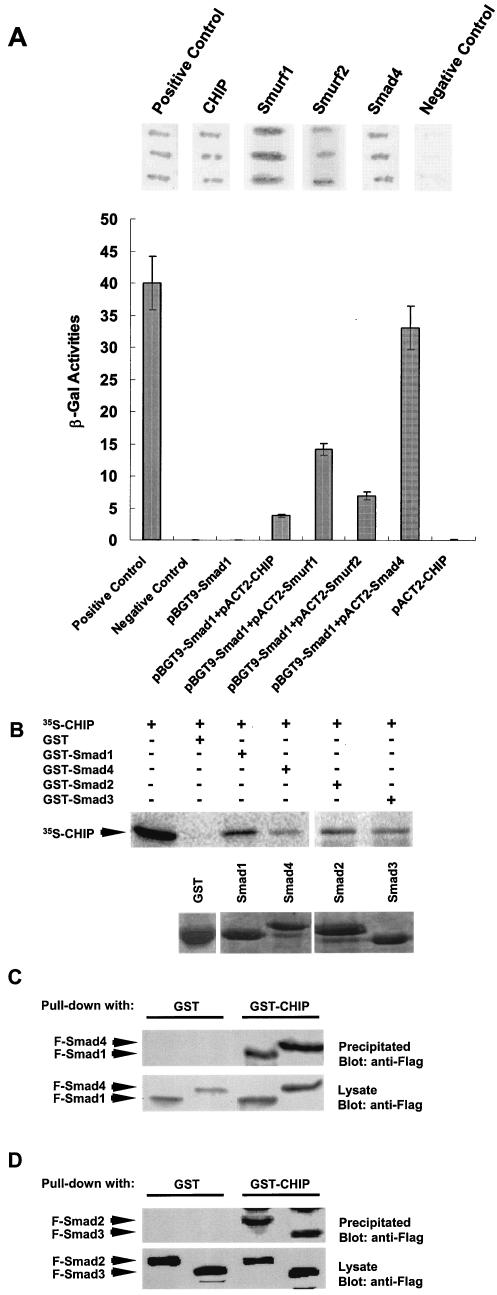
To identify possible additional partner protein(s) interacting with Smads, we performed a yeast two-hybrid screen using Smad1 as bait to search for interacting proteins in a human brain cDNA library. We identified 20 positive clones and later confirmed the interaction by a liquid β-Gal assay. After sequencing all of the 20 positive clones, we found those candidate clones encoding known Smad1-interacting proteins, including Smad4, Smurf1, and Smurf2, indicating that our screen was successful. Interestingly, we have identified two positive clones coding for the CHIP protein (Fig. 1A), which was previously characterized as a cochaperon protein (2). To confirm the yeast two-hybrid results, we made prey vectors with full-length CHIP, Smad4, Smurf1, and Smurf2 to carry out a filter assay (Fig. 1A, top) and liquid β-Gal assay (Fig. 1A, bottom). The data clearly showed that full-length CHIP could interact with Smad1 in a yeast two-hybrid screening. The interaction affinities of Smad1 with Smad4, Smurf1, Smurf2, and CHIP could vary due to unknown factors, but the specific interactions obviously exist, and the controls were negative.
FIG. 1.
Interactions between CHIP and Smads in vitro. (A) Results of yeast two-hybrid screenings shown by β-Gal filter and liquid assays. Three growing colonies of yeast cells cotransformed with bait plasmid(pGBT9-Smad1), as indicated, and the prey plasmids were spread onto the filter paper or cultured in test tubes. Dark staining indicated the interaction. Bottom columns showed the interaction measured by β-Gal activities. Data were averaged by three clones. The positive (pCL1) and negative (pBGT9 vector) controls were supplied by Clontech. (B) In vitro GST binding assay. GST-Smad1, GST-Smad2, GST-Smad3, or GST-Smad4 was examined for direct interaction with in vitro-translated 35S-CHIP (upper panel). The amounts of Smad proteins used were at similar levels, as indicated by protein staining (lower panel). Input (1/5) of 35S-labeled protein was loaded. (C and D) Pull-down assay. GST-CHIP was used to pull down Smad1, Smad2, Smad3, and Smad4 from 293T cell lysate. All Smads interacted with GST-CHIP, while GST alone did not (upper panel). Expression levels of F-Smads in whole-cell lysates were similar, as shown in the lower panel.
To confirm whether CHIP interacts with Smad1 physically, we synthesized 35S-labeled CHIP in vitro and expressed Smad1 protein in a GST fusion vector. Next, we carried out a GST binding assay. The data clearly show that the GST-Smad1 fusion protein binds directly to the CHIP protein while the GST protein alone does not (Fig. 1B), confirming the result from the yeast two-hybrid screen. Since Smad4 could form a complex with Smad1 in response to BMP signals, we examined further whether Smad4 can interact with CHIP. As shown in Fig. 1B, the GST-Smad4 fusion protein could also pull down the CHIP protein in the same binding assay. The weaker CHIP band in the Smad4 lane was probably due to the use of less Smad4 than Smad1 in the assay (see the lower panel with GST-Smad1 and GST-smad4). Moreover, when the GST-Smad2 or GST-Smad3 fusion protein was used, the CHIP protein was also pulled down (Fig. 1B). These results suggested that CHIP interacts with these Smad proteins in vitro.
Besides using GST-Smad fusion proteins generated from E. coli, we also expressed Smad1 and Smad4 proteins tagged with the Flag epitope in 293T cells and COS-7 cells by transient transfection. Then, we examined whether a GST-CHIP fusion protein could pull down these Smad proteins isolated from these mammalian cells. As showed in Fig. 1C, the purified GST-CHIP pulled down the Flag-tagged Smad1 and Smad4 expressed in both 293T cells (Fig. 1C) and COS-7 cells (data not shown). The varied intensities of Smad1 and Smad4 bands in Fig. 1B and C may be due to loading variations. In addition, purified GST-CHIP also pulled down the Flag-tagged Smad2 and Smad3 in 293T cells (Fig. 1D). Thus, CHIP is a SMAD-interacting protein, as indicated by the above results that Smads could pull down the CHIP protein, and CHIP also could pull down Smad1, Smad2, Smad3, and Smad4 proteins under the in vitro conditions. In the following experiments, we focused on the interaction of CHIP with Smad1 and Smad4, which mediate BMP signal transduction.
CHIP interacts with Smad1 and Smad4 in vivo.
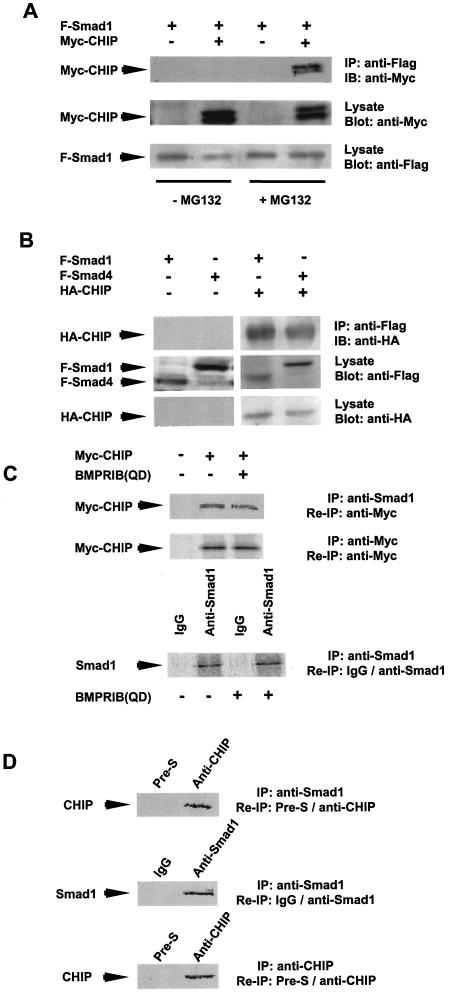
We next investigated whether CHIP could interact with Smad1 or Smad4 when both of these two proteins were expressed in mammalian cells. In the initial test, we could not precipitate Flag-tagged Smad1 coexpressed with Myc-CHIP in mammalian cell lysates (Fig. 2A, left two lanes). Since it was shown that the CHIP protein could mediate protein degradation, we reasoned that the interacting complex of CHIP with Smad1 might turn over too quickly to be precipitated down. To overcome this, cells were pretreated with a proteosome inhibitor (MG132) before making the cell lysates. As expected, the Flag-tagged Smad1 fusion protein (F-Smad1) precipitated the expressed Myc-tagged CHIP fusion protein in the presence of MG132 (Fig. 2A, right lane). In the following experiments, revealing the interaction of CHIP and Smads using mammalian cell lysates, we always added MG132 before harvesting the cells. These results further indicate that CHIP could interact with the Smad proteins expressed in mammalian cells in vitro.
FIG. 2.
Immunoprecipitation assays showing CHIP-Smad interaction in vivo. (A) Overexpressed Flag-tagged Smad1 precipitated Myc-tagged CHIP in the presence of MG132. Myc-tagged CHIP was cooverexpressed with Flag-tagged Smad1 in the absence or presence of 50 μM MG132. The precipitated band was as indicated. (B) CHIP interacts with Smad1 and Smad4 in mammalian cells. 293T cells were transfected with plasmids expressing Flag-tagged full-size Smad1 or Smad4 or HA-tagged full-length CHIP as indicated. The whole-cell lysates were used in immunoprecipitation (IP) with anti-Flag M2 antibody and blotted with anti-HA antibody (IB) (upper panel). The levels of F-Smads in the whole-cell lysates (middle panel) and HA-CHIP (bottom panel) have been shown as indicated. MG132 was added to the transfected cells. (C) Myc-CHIP interacts with endogenous Smad1 in vivo. HepG2 cells were transfected with Myc-tagged CHIP or a constitutively active form of BMPRIB(QD). After 24 h of transfection, cells were metabolically labeled with 35S-methionine and IP was then carried out. SDS-PAGE gel was dried and visualized by autoradiograph. The expression of Myc-CHIP and endogenous Smad1 are shown in the middle and lower panels separately. (D) EndogenousCHIP interacts with endogenous Smad1 in vivo. The HepG2 cells were metabolically labeled with [35S]methionine. The cell lysate was immunoprecipitated with either anti-Smad1 antibody or anti-CHIP serum. The precipitated complex with anti-Smad1 antibody was then reimmunoprecipitated with either anti-Smad1 antibody to show the expression of endogenous Smad1 (middle panel) or anti-CHIP serum to show the interaction of Smad1 and CHIP (top panel). The precipitated complex with anti-CHIP serum was reimmunoprecipitated with anti-CHIP serum to show the endogenous expression of CHIP (bottom panel) in the cells. Mouse IgG and preimmune rabbit serum (Pre-S) were always used as negative controls for the reimmunoprecipitation as indicated.
Furthermore, we coexpressed the CHIP protein fused with HA tag and Smad1 or Smad4 fused with Flag tag in 293T cells by transient transfection. The immunoprecipitation assays were performed 48 h after transfection. As shown in Fig. 2B, the anti-Flag antibody detected Smad1 or Smad4, respectively, from cell lysates (Fig. 2B, middle panel). The CHIP protein was coimmunoprecipitated with anti-Flag antibody, as indicated by an immunoblot with anti-HA (Fig. 2B, right, the upper panel). These results suggest that CHIP forms a complex with either Smad1 or Smad4 in these mammalian cells in vivo.
In order to test whether Myc-CHIP could precipitate the endogenous Smad1, we performed metabolic labeling and immunoprecipitation in HepG2 cells. We specifically precipitated endogenous Smad1 using anti-Smad1 antibody (Fig. 2C, bottom). Interestingly, we successfully reprecipitated the Myc-CHIP protein in the precipitated complex containing endogenous Smad1. When we coexpressed the constitutively active BMP type 1 receptor [BMPRIB(QD)], the anti-Smad1 antibody could pull down the Myc-CHIP protein (Fig. 2C, upper panel, compare two lanes on right). This result clearly showed that overexpressed Myc-tagged CHIP could form a complex with endogenous Smad1 with or without the signaling (Fig. 2C).
We further examined whether endogenous CHIP and Smad were associated without any overexpression. As shown in Fig. 2D, we used HepG2 cells that did not express any exogenous CHIP or Smad protein. We found that endogenous Smad1 and CHIP complex could be coimmunoprecipitated using anti-CHIP antibodies, while the preimmune serum did not. The above results reveal that Smad1 indeed interacted with CHIP.
CHIP interacts with the MH2 domain of Smad1 in vitro.
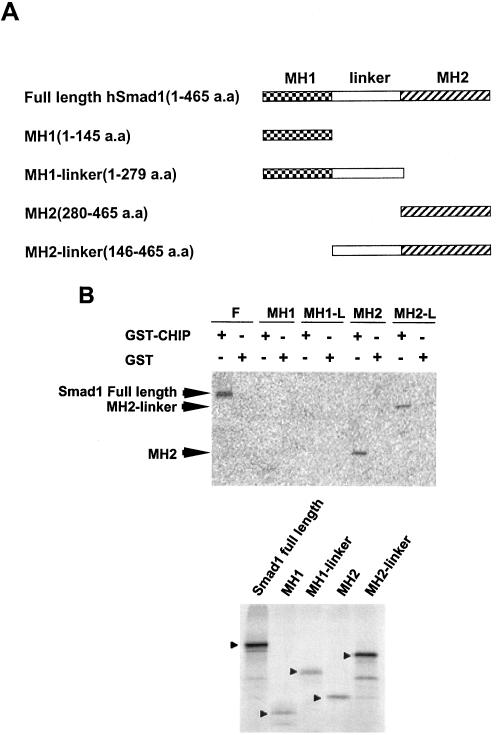
To characterize the necessary domain of Smad1 required for the interaction between CHIP and Smad1, we created a series of truncated mutants of Smad1 with different domains in a pcDNA6 vector (Fig. 3A). GST-CHIP or GST alone was incubated with 35S-labeled in vitro-translated deletion Smad1 proteins (Fig. 3B, bottom panel) to perform the binding assay. The data showed that full-length Smad1, MH2 domain, and MH2 + Linker of Smad1 could interact with GST-CHIP, while the MH1 domain and MH1 plus Linker of Smad1 did not, suggesting that CHIP was able to bind the carboxyl terminus of Smad1 through the MH2 domain (Fig. 3B, upper panel).
FIG. 3.
Identification of domains of Smad1 required for their interaction. (A) Diagram depicting the three domains: MH1, Linker region, MH2 within Smad1, and each expressing construct. (B) CHIP interacts with the MH2 domain of Smad1. Binding assay was performed with in vitro-translated 35S-labeled full-length or truncated Smad1 and GST-CHIP or GST protein alone. The lower panel shows the input of different truncated proteins used in the experiment.
CHIP causes degradation of Smad proteins in vivo.
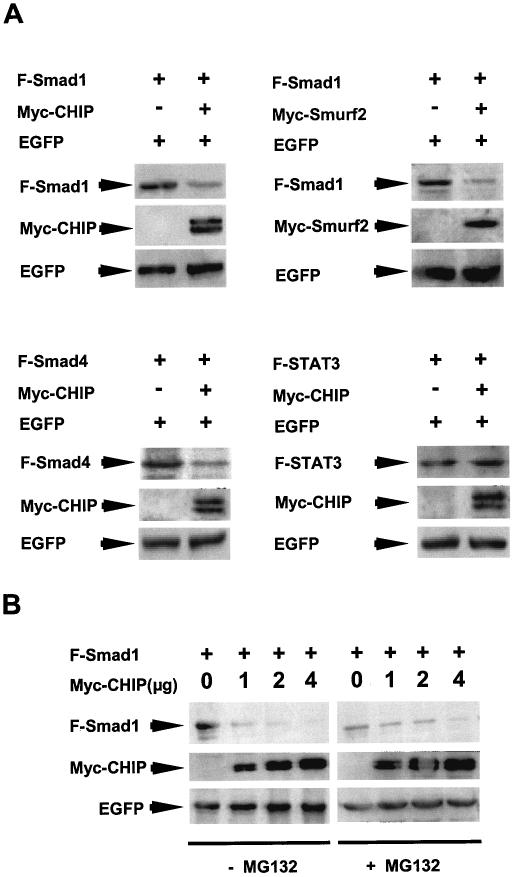
It has been reported previously that CHIP is not only a cochaperon protein but also is an E3 ubiquitin ligase involved in targeted protein degradation (28). Thus, it is important to know whether the expression of CHIP would have any effect on Smad protein turnover. As shown in Fig. 4, when the Myc-tagged CHIP protein was overexpressed, the level of the Smad1 protein was dramatically reduced, whereas the level of the coexpressed green fluorescent protein (EGFP) was not changed (Fig. 4A, upper left panel), indicating that this protein degradation was specific for Smad1. Similarly, the level of Smad4 protein also diminished when the CHIP protein was overexpressed (Fig. 4A, bottom left panel). As a control, overexpression of the Smurf2 protein also resulted in similar degradation of the Smad1 protein (Fig. 4A, upper right panel). In contrast, overexpression of CHIP had no effect on the unrelated protein (STAT3) level (Fig. 4A, bottom right panel), suggesting that the function of the interaction of CHIP with Smads might be similar to that of Smurf2, which mediates Smad protein degradation through the proteasome.
FIG. 4.
Effects of CHIP on Smad levels. (A) CHIP decreases the steady-state level of the Smad1 (upper left panel) and Smad4 (bottom left panel) proteins. The steady-state levels of Smads were detected by immunoblotting of the total cell lysate. Smurf2 (upper right panel) and unrelated protein STAT3 (bottom right panel) were used as controls. EGFP was always coexpressed in all experiments. (B) The decrease of Smad1 protein depends on the expression level of CHIP and can be inhibited by MG132. HEK293T cells transfected with F-Smad1 and Myc-CHIP were treated without (left) or with (right) 50 μM MG132 for 4 h before lysis of the cells. Immunoblot was performed with anti-Flag M2 monoclonal mouse antibody.
We next examined the possibility of whether observed CHIP-mediated Smad protein degradation can be inhibited by proteasome inhibitors. Since CHIP was an E3 ubiquitin ligase and was identified as a partner protein in initiating the ubiquitin pathway for protein degradation, we chose the proteasome inhibitor MG132, which also has been shown to be an inhibitor for protein degradation in the ubiquitination pathway. We coexpressed CHIP with Smad1 or Smad4 with or without MG132. We observed that if no MG132 was added, Smad1 protein completely diminished when the CHIP protein was expressed at increased levels (Fig. 4B, left panels). In contrast, when MG132 was added, the Smad1 protein was less degraded, with increased CHIP expression (Fig. 4B, right panels). As a control, the protein level of EGFP was not affected under these conditions (Fig. 4B, lower panels). Thus, it is possible that CHIP-mediated Smad1 degradation is dependent on proteasome that is sensitive to MG132.
CHIP mediates Smad1 ubiquitination.
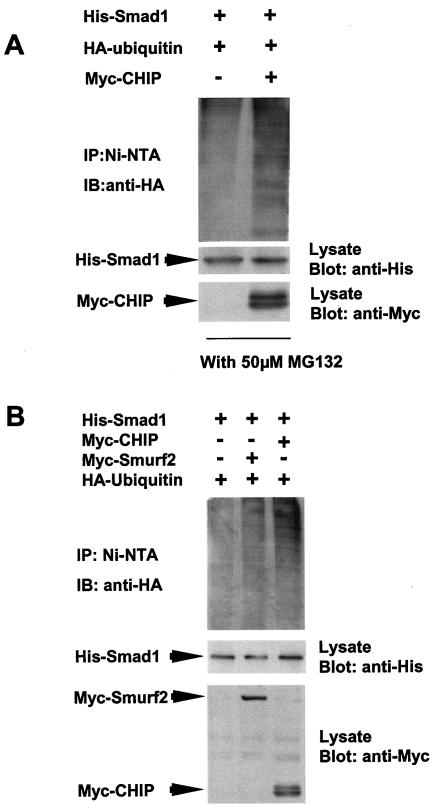
To show direct evidence for the ubiquitination of Smad1 mediated by CHIP, we overexpressed His-Smad1, Myc-CHIP, and HA-ubiquitin in 239T cells in the presence of MG132. We precipitated the complex with Ni-NTA beads for His-Smad1 and detected the complex with anti-HA antibody (for ubiquitin). The results showed that a stronger smear of ubiquitinated Smad1 could be seen in the presence of CHIP (Fig. 5A, upper panel, right lane), while the smear was fainter in the absence of CHIP (upper panel, left lane). Since Smurf2 mediated Smad degradation through ubiquitination, we used Smurf2 as a control. Smurf2 and CHIP were overexpressed in 293T cells in the presence of MG132 (Fig. 5B, upper panel, right lane). The data showed that in the presence of Smurf2 or CHIP, ubiquitinated Smad1 was detected simultaneously. However, without CHIP or Smurf2, the ubiquitinated smear of Smad1 was hardly detected (Fig. 5A and B, upper panels, left lanes). Altogether, the results suggest that CHIP could mediate the degradation of Smad1 through ubiquitination.
FIG. 5.
CHIP mediates the ubiquitination of Smad1. (A) Overexpressed ubiquitin precipitated with Smad1 in the presence of CHIP. The HEK293T cells were transfected with His-Smad1, HA-ubiquitin, and Myc-CHIP as indicated with the treatment of 50 μM MG132 for 4 h before harvest. The cell lysates were precipitated with Ni-NTA. The complex was eluted and blotted with anti-HA antibody. The smear indicates the ubiquitination of Smad1 (top panel, right lane). (B) Both CHIP and Smurf2 mediate the ubiquitination of Smad1. The experiment was carried out as for panel A. The smear can be seen in lanes 2 and 3 in the top panel. The expression of CHIP and that of Smurf2 were observed as indicated in the lower panel.
CHIP inhibits transcription of TGF-β/BMP response gene.
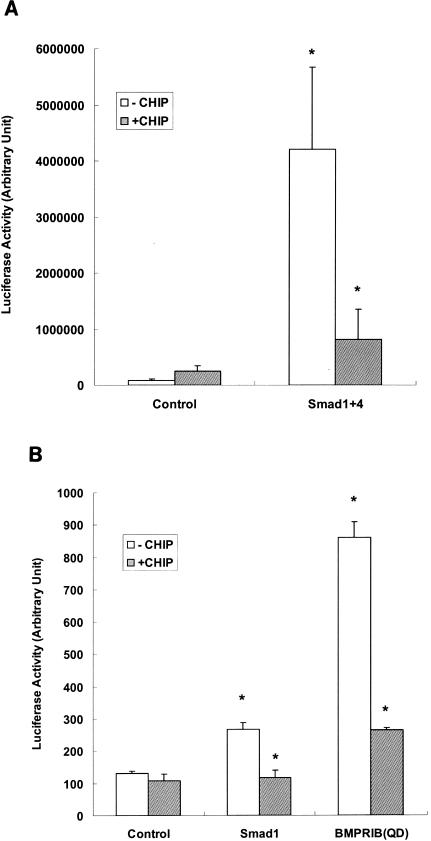
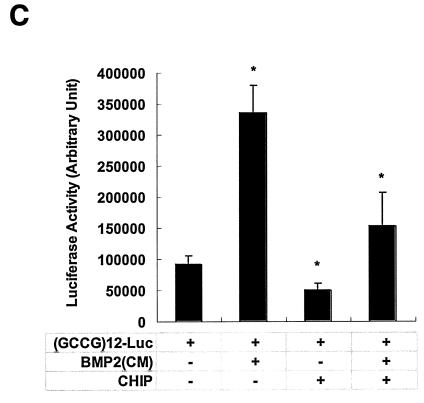
If CHIP indeed mediates Smad protein degradation, then Smad-dependent transcription activities should also be reduced or inhibited. We examined this possibility using the (GCCG)12-Luc construct (1), which contained the Smad binding element linked with the luciferase reporter gene. As controls, expression with the reporter or with CHIP alone yielded little transcriptional activity. When Smad1 and Smad4 were transiently transfected into 293T cells, the reporter construct generated high levels of luciferase activities (Fig. 6A). However, when CHIP was coexpressed with these Smad proteins, luciferase activity was reduced to the same basal level, suggesting that CHIP blocked the transcriptional activities of Smad1 and Smad4. The same results were obtained when the (CAGA)12-MLP-Luc reporter construct (23) was used (data not shown). With Mv1Lu cells, we carried out another luciferase assay using the (GCCG)12-Luc reporter system, and we observed the same result. Furthermore, overexpression of Smad1 or the constitutively active form of BMPRIB(QD) could stimulate the transcriptional activity of Smad1 (Fig. 6B, open columns). However, the presence of the CHIP protein could inhibit such activity significantly (P ≤ 0.01) (Fig. 6B). These experiments were repeated four times in triplicate. Similar results were obtained when COS-7 cells were used (data not shown). When we used the medium containing BMP2 (BMP2CM), we could stimulate the reporter 3.5-fold (Fig. 6C) over the control that had no BMP2 (P ≤ 0.01). This BMP2-induced transcription activity was almost abolished when CHIP was expressed (Fig. 6C), indicating that CHIP could block Smad1/Smad4-mediated gene transcription in response to BMP2.
FIG. 6.
CHIP expression inhibits the transcription activity of Smad1/Smad4. (A) The effect of CHIP on Smad1- and Smad4-mediated transcription. HEK293T cells were transfected with plasmids encoding F-Smad1 and F-Smad4 as indicated together with (GCCG)12-MLP-Luc, which could still respond to Smad1/Smad4 or Smad2/Smad4 complex. Twenty-four hours after transfection, cells were extracted and measured for luciferase activity according to the manufacture's protocol (Promega). (B) CHIP blocks BMPRIB(QD)-mediated gene transcription. A constitutive active receptor BMPRIB(QD) and various expression vectors were cotransfected into Mv1Lu cells. Cells were harvested after transfection of 24 h and luciferase assays were performed. All assays were carried out in triplicate, and the averages are shown with the standard deviation. (C) CHIP blocked BMP2-mediated gene transcription. BMP2 conditioned medium [BMP2(CM)] was generated by transfecting BMP2 expression vector into CHO cells for 48 h. BMP2(CM) was added to the cells transfected with reporter construct as before. CHO cell medium was used as control. This experiment was carried out in 239T cells. Arbitrary luciferase activity is shown. Values presented are means ± standard deviations (n = 3); asterisk, P ≤ 0.01.
pBS/U6/CHIPi stimulates Smad1/Smad4-mediated gene transcription.
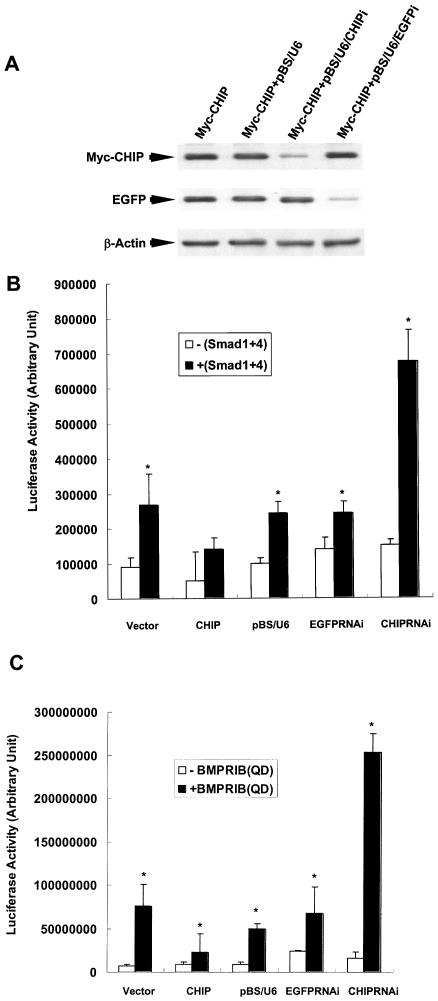
We next reasoned that if overexpression of CHIP could inhibit the transcriptional activities mediated by the Smad1 and Smad4 complex, we should expect stronger Smad1/Smad4-mediated gene transcription when endogenous CHIP was knocked down by double-stranded RNA, which was reported as a powerful tool to abolish gene expression in cultured cells (24). To investigate the functional role of endogenous CHIP, we designed and generated a pBS/U6/CHIPi construct, which could produce hairpin RNA in cells. As showed in Fig. 7A, in cells transfected with the pBS/U6/CHIPi vector, the expression of the CHIP protein was significantly reduced. However, pBS/U6/CHIPi had no effect on the protein level of EGFP, demonstrating that pBS/U6/CHIPi had a specific effect by inhibiting CHIP gene expression. As additional negative controls, we further showed that the backbone vectors pBS/U6 and pBS/U6/EGFPi had no effect on expression of CHIP, although pBS/U6/EGFPi strongly inhibited EGFP expression. Thus, these RNAi-mediated disruptions were specific. Consistent with our model, pBS/U6/CHIPi significantly enhanced the luciferase activities mediated by Smad1 and Smad4 (P ≤ 0.01) (Fig. 7). Furthermore, when constitutively active BMPRIB(QD) could be inhibited by expression of the CHIP protein (P ≤ 0.01), pBS/U6/CHIPi abolished the effect of CHIP (P ≤ 0.01) (Fig. 7C). In contrast, the control plasmids, pBS/U6 and pBS/U6/EGFPi, had no effect on luciferase activities, indicating that vector-based RNAi for CHIP specifically blocked the role of CHIP. All of the data demonstrated that the expression of CHIP and that of pBS/U6/CHIPi had opposite effects on Smad1/Smad4-mediated gene transcription, strongly suggesting that CHIP is a negative regulator of Smad1/Smad4-mediated signal transduction.
FIG. 7.
pBS/U6/CHIPi stimulates the transcription activity of Smad1/Smad4. (A) pBS/U6/CHIPi specifically blocked CHIP expression. HEK293T cells were transfected with plasmids as indicated, and the cell lysates were used for Western blots with indicated antibody. (B) pBS/U6/CHIPi stimulated the transcription activity of Smad1/Smad4. The plasmid pBS/U6/CHIPi and the other indicated vectors were transfected into HEK239T cells. (C) pBS/U6/CHIPi restored the transcription activity induced by expression of BMPRIB(QD). A constitutively active receptor, BMPRIB(QD), and various expression vectors were cotransfected into HEK293T cells. Cells were harvested after transfection of 24 h, and luciferase assays were performed. All the assays were carried out in triplicate. Values presented are means ± standard deviations (n = 3). Asterisk, P ≤ 0.01.
DISCUSSION
Smad proteins play essential roles in signal transduction and gene regulation induced by TGF-β superfamily cytokines, which are regulated at a variety of levels (17, 20). Activation of Smad proteins is controlled by specific phosphorylation by ligand-bound receptors, whereas inhibition or “turn-off” of Smad-mediated signaling can be regulated by protein turnover of Smads. It has been shown that Smurf proteins have a function in specifically targeting Smad proteins for ubiquitin-mediated degradation, which may represent a tight control of the signal process. However, it is unclear whether there is any other general regulatory mechanism that could control Smad protein levels in the absence of ligand.
We now present a biochemical study in which protein levels of Smad1 and Smad4 can be regulated by the CHIP protein, which is a U-box-dependent E3 ubiquitin ligase involved in degradation of a number of different proteins (2, 9, 21). We have provided evidence that the CHIP protein could physically interact with the Smad1 and Smad4 proteins under a variety of in vitro or in vivo conditions. Intriguingly, we found that this interaction was only slightly affected by the constitutively active BMP receptor, suggesting that the CHIP-Smad interaction might have important functions in regulation of the Smad protein levels that are independent of activation. However, it is clear that the presence of CHIP can negatively affect Smad-mediated signaling and transcription. We demonstrated that expression of CHIP resulted in specific degradation of both Smad1 and Smad4. Consistent with a hypothesis that CHIP is a possible negative regulator of TGF-β/BMP signal transduction, we have further demonstrated that expression of the CHIP protein blocked transcriptional activity generated by the Smad1/Smad4 complex. Most importantly, we have shown that pBS/U6/CHIPi, which specifically blocked endogenous CHIP expression, could enhance Smad1/Smad4-mediated gene transcription, which was opposite to the overexpression of CHIP. These data have suggested a novel mechanism by which the cellular effects of TGF-β/BMP signals can be potentially regulated through the CHIP-Smad interaction.
The CHIP protein was initially shown as a cochaperon interacting with HSP70 (2). It has been reported that CHIP participated in degradation of a number of important proteins, such as the glucocorticoid receptor, cystic fibrosis transmembrane conductance receptor, Parkin, and Erk (8, 9, 19). There is more evidence that CHIP has multiple functions as an E3 ligase mediating the degradation of several proteins (27). In our preliminary experiments using mammalian cells, we did not show if CHIP more likely interacts with different Smad1 forms (folded or unfold, phosphorylated or unphosphorylated). It is possible that protein folding of Smads may be a factor in the CHIP-induced degradation, and a chaperon (HSP70) might be also involved. The detailed mechanism of the interrelationship of HSP70, CHIP, and Smad in vivo is an interesting subject but is beyond the scope of the present studies and will need to be investigated further in the future. It should be pointed out that since CHIP is capable of mediating degradation of Smads, the interaction of CHIP and Smad1 was clearly observed only when the proteasome inhibitor MG132 was present. Furthermore, we suspected that CHIP-mediated Smad1 degradation might involve protein ubiquitination, which is consistent with the fact that CHIP is also an E3 ubiquitin ligase. Indeed, we observed that the ubiquitination of Smad1 was enhanced dramatically in the presence of the CHIP protein, indicating that the ubiquitination of CHIP-Smad1 complexes occurred (Fig. 5). Therefore, we concluded that CHIP was involved in the ubiquitination of Smad1.
Another unanswered question is the specificity of CHIP-mediated Smad protein degradation. We have shown that CHIP interacts with Smad1 and Smad4, which is the complex activated in response to BMP signals (18, 20). We have preliminary evidence suggesting that other R-Smads, such as Smad2 and Smad3, might also be the targets for CHIP-mediated interaction and degradation (unpublished results; in this report we focused on Smad1 and Smad4), implying that CHIP could also interfere with TGF-β signal transduction. However, CHIP does not mediate TGF-β receptor degradation (data not shown). These observations may suggest that CHIP mediates a varied spectrum of protein degradation compared with the Smurf family proteins, which prefer to target different kinds of Smad proteins and TGF-β receptors (6, 20). It was known that there were two classes of E3 ligating enzymes containing HECT domain and RING finger domain in the substance selection in the ubiquitination process (26). It was reported that Smurf1 and Smurf2 contained HECT domains. Patterson and colleagues reported that CHIP contained a U box (9), which is a modified RING finger domain. Here we demonstrate that CHIP mediates the ubiquitination of Smad1, suggesting that both forms of E3 (CHIP as U-box E3 and Smurfs as HECT E3) can be involved in the degradation of Smad proteins, which is similar to the ubiquitination of p53 (26), implying the necessity of a tight control of important regulatory proteins in general. Furthermore, we have observed that CHIP was mainly located in the cytoplasm (data not shown), indicating a possibility that the Smad-CHIP interaction may occur predominantly within the cytoplasm and then Smads may quickly be moved into the proteasome and degraded. Thus, we hypothesize that CHIP regulates constitutive levels of Smad proteins. These observations shed new light on the regulation of Smad-mediated TGF-β/BMP signal transduction, suggesting that Smad proteins can be controlled at a variety of levels using different mechanisms. However, our results in the present study have raised more questions. It is of interest to further investigate the mechanism of control during the degradation of Smads by CHIP. We speculate that the phosphorylation of Smads or CHIP may also be involved. Although our data provided evidence that the interaction of CHIP with Smads was ligand independent, we cannot exclude the possibility that these proteins may be prephosphorylated, or controlled by other signaling pathways. Further studies of the CHIP-Smad interaction should shed light on the network of control of Smad proteins.
Acknowledgments
This work was supported by the 985 Program of Tsinghua University and grants from the Chinese National Academic Foundation (no. 39970369, no. 30070703, and no. 30030050), 973 Project (2001CB510006 and 2002CB513000), and Chinese Young Investigator 863 Project to Z.J.C and X.Y.F. and a special fund from Tsinghua Yuan-Xing Pharmaceuticals, Shen-Zhen, China.
We thank Xiaofan Wang at Duke University for kindly providing the BMP, BMPRIB(QD), and Smad constructs. We thank Masahiro Kawabata for (GCCG)12-Luc reporter constructs and Ying Zhang for Myc-Smurf2 and HA-ubiquitin plasmids. We thank Xinhua Feng for kindly providing Smad plasmids and helpful suggestions and Yang Shi for kindly providing pBS/U6 and pBS/U6/EGFPi. Above all, this study would not be possible without support and encouragement from Nanming Zhao, Zhonghe Zhai, Haimeng Zhou, Xiufang Zhang, Xiangyang Yang, and other colleagues at Tsinghua University. Finally, we thank Michael Wolfgang for reading the manuscript.
REFERENCES
- 1.Akiyoshi, S., H. Inoue, J. Hanai, K. Kusanagi, N. Nemoto, K. Miyazono, and M. Kawabata. 1999. c-Ski acts as a transcriptional co-repressor in transforming growth factor-beta signaling through interaction with smads. J. Biol. Chem. 274:35269-35277. [DOI] [PubMed] [Google Scholar]
- 2.Ballinger, C. A., P. Connell, Y. Wu, Z. Hu, L. J. Thompson, L. Y. Yin, and C. Patterson. 1999. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19:4535-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonni, S., H. R. Wang, C. G. Causing, P. Kavsak, S. L. Stroschein, K. Luo, and J. L. Wrana. 2001. TGF-beta induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat. Cell Biol. 3:587-595. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. R., Y. Kang, P. M. Siegel, and J. Massague. 2002. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell 110:19-32. [DOI] [PubMed] [Google Scholar]
- 5.Derynck, R., Y. Zhang, and X. H. Feng. 1998. Smads: transcriptional activators of TGF-beta responses. Cell 95:737-740. [DOI] [PubMed] [Google Scholar]
- 6.Ebisawa, T., M. Fukuchi, G. Murakami, T. Chiba, K. Tanaka, T. Imamura, and K. Miyazono. 2001. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 276:12477-12480. [DOI] [PubMed] [Google Scholar]
- 7.Feng, X. H., Y. Y. Liang, M. Liang, W. Zhai, and X. Lin. 2002. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-beta-mediated induction of the CDK inhibitor p15(Ink4B). Mol. Cell 9:133-143. [DOI] [PubMed] [Google Scholar]
- 8.Imai, Y., M. Soda, S. Hatakeyama, T. Akagi, T. Hashikawa, K. I. Nakayama, and R. Takahashi. 2002. CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol. Cell 10:55-67. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, J., C. A. Ballinger, Y. Wu, Q. Dai, D. M. Cyr, J. Hohfeld, and C. Patterson. 2001. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 276:42938-42944. [DOI] [PubMed] [Google Scholar]
- 10.Kavsak, P., R. K. Rasmussen, C. G. Causing, S. Bonni, H. Zhu, G. H. Thomsen, and J. L. Wrana. 2000. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol. Cell 6:1365-1375. [DOI] [PubMed] [Google Scholar]
- 11.Kim, R. H., D. Wang, M. Tsang, J. Martin, C. Huff, M. P. de Caestecker, W. T. Parks, X. Meng, R. J. Lechleider, T. Wang, and A. B. Roberts. 2000. A novel smad nuclear interacting protein, SNIP1, suppresses p300-dependent TGF-beta signal transduction. Genes Dev. 14:1605-1616. [PMC free article] [PubMed] [Google Scholar]
- 12.Kurokawa, M., K. Mitani, K. Irie, T. Matsuyama, T. Takahashi, S. Chiba, Y. Yazaki, K. Matsumoto, and H. Hirai. 1998. The oncoprotein Evi-1 represses TGF-beta signalling by inhibiting Smad3. Nature 394:92-96. [DOI] [PubMed] [Google Scholar]
- 13.Lin, X., M. Liang, and X. H. Feng. 2000. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J. Biol. Chem. 275:36818-36822. [DOI] [PubMed] [Google Scholar]
- 14.Liu, F., C. Pouponnot, and J. Massague. 1997. Dual role of the Smad4/DPC4 tumor suppressor in TGFbeta-inducible transcriptional complexes. Genes Dev. 11:3157-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo, R. S., and J. Massague. 1999. Ubiquitin-dependent degradation of TGF-beta-activated smad2. Nat. Cell Biol. 1:472-478. [DOI] [PubMed] [Google Scholar]
- 16.Luo, K., S. L. Stroschein, W. Wang, D. Chen, E. Martens, S. Zhou, and Q. Zhou. 1999. The Ski oncoprotein interacts with the Smad proteins to repress TGFbeta signaling. Genes Dev. 13:2196-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massague, J. 2000. How cells read TGF-beta signals. Nat. Rev. Mol. Cell Biol. 1:169-178. [DOI] [PubMed] [Google Scholar]
- 18.Massague, J., and D. Wotton. 2000. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 19:1745-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meacham, G. C., C. Patterson, W. Zhang, J. M. Younger, and D. M. Cyr. 2001. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3:100-105. [DOI] [PubMed] [Google Scholar]
- 20.Moustakas, A., S. Souchelnytskyi, and C. H. Heldin. 2001. Smad regulation in TGF-beta signal transduction. J. Cell Sci. 114:4359-4369. [DOI] [PubMed] [Google Scholar]
- 21.Murata, S., Y. Minami, M. Minami, T. Chiba, and K. Tanaka. 2001. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2:1133-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakao, A., M. Afrakhte, A. Moren, T. Nakayama, J. L. Christian, R. Heuchel, S. Itoh, M. Kawabata, N. E. Heldin, C. H. Heldin, and P. ten Dijke. 1997. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature 389:631-635. [DOI] [PubMed] [Google Scholar]
- 23.Shen, X., P. P. Hu, N. T. Liberati, M. B. Datto, J. P. Frederick, and X. F. Wang. 1998. TGF-beta-induced phosphorylation of Smad3 regulates its interaction with coactivator p300/CREB-binding protein. Mol. Biol. Cell 9:3309-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sui, G., C. Soohoo, B. Affar el, F. Gay, Y. Shi, and W. C. Forrester. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verschueren, K., J. E. Remacle, C. Collart, H. Kraft, B. S. Baker, P. Tylzanowski, L. Nelles, G. Wuytens, M. T. Su, R. Bodmer, J. C. Smith, and D. Huylebroeck. 1999. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. J. Biol. Chem. 274:20489-20498. [DOI] [PubMed] [Google Scholar]
- 26.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2:169-178. [DOI] [PubMed] [Google Scholar]
- 27.Wiederkehr, T., B. Bukau, and A. Buchberger. 2002. Protein turnover: a CHIP programmed for proteolysis. Curr. Biol. 12:R26-R28. [DOI] [PubMed] [Google Scholar]
- 28.Wotton, D., R. S. Lo, S. Lee, and J. Massague. 1999. A Smad transcriptional corepressor. Cell 97:29-39. [DOI] [PubMed] [Google Scholar]
- 29.Wrana, J. L. 2000. Regulation of Smad activity. Cell 100:189-192. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, Y., C. Chang, D. J. Gehling, A. Hemmati-Brivanlou, and R. Derynck. 2001. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 98:974-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu, H., P. Kavsak, S. Abdollah, J. L. Wrana, and G. H. Thomsen. 1999. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400:687-693. [DOI] [PubMed] [Google Scholar]