Abstract
Saccharomyces cerevisiae has a global pattern of histone acetylation in which histone H3 and H4 acetylation levels are lower at protein-coding sequences than at promoter regions. The loss of Eaf3, a subunit of the NuA4 histone acetylase and Rpd3 histone deacetylase complexes, greatly alters the genomic profile of histone acetylation, with the effects on H4 appearing to be more pronounced than those on H3. Specifically, the loss of Eaf3 causes increases in H3 and H4 acetylation at coding sequences and decreases at promoters, such that histone acetylation levels become evenly distributed across the genome. Eaf3 does not affect the overall level of H4 acetylation, the recruitment of the NuA4 catalytic subunit Esa1 to target promoters, or the level of transcription of the genes analyzed for histone acetylation. Whole-genome transcriptional profiling indicates that Eaf3 plays a positive, but quantitatively modest, role in the transcription of a small subset of genes, whereas it has a negative effect on very few genes. We suggest that Eaf3 regulates the genomic profile of histone H3 and H4 acetylation in a manner that does not involve targeted recruitment and is independent of transcriptional activity.
Histone acetylases and deacetylases modify lysine residues on the N-terminal tails of histones, and these modifications correlate with the activation and repression of transcription, respectively (16, 49, 54). Acetylation affects chromatin structure by changing the electrostatic charge on histones and thus weakening histone-DNA (20) and nucleosome-nucleosome (39) interactions. As a consequence, hyperacetylated nucleosomes are more accessible to regulatory factors and are more active for transcription or recombination in vitro (36, 38, 44, 53, 57). Aside from their direct effect on nucleosome structure, acetylated histone tails interact directly with bromodomains (10, 22, 45), a structural motif found in many chromatin-modifying complexes. Such chromatin-modifying complexes preferentially associate with hyperacetylated nucleosomes (1, 9, 17, 18), and this preferential association can depend on which lysine within the histone tail is acetylated (1).
Histone acetylases and deacetylases can be targeted to specific promoters by interaction with DNA-bound transcriptional activators or repressors. Recruitment of these histone-modifying activities leads to promoter-localized acetylation or deacetylation and changes in gene expression. In the yeast Saccharomyces cerevisiae, activator-specific recruitment of the SAGA histone acetylase complex leads to increased acetylation at the HIS3 and HO promoters and correlates with transcriptional activation (8, 29, 32, 33). Conversely, Ume6 represses transcription by recruiting the Sin3-Rpd3 histone deacetylase complex to promoters, thereby generating a localized domain of deacetylated nucleosomes (9, 27, 50).
In addition to targeted recruitment, histone acetylases and deacetylases also act in an untargeted manner over the entire genome (32, 47, 59). Specifically, a loss of an acetylase leads to decreased acetylation throughout the genome, whereas a loss of a deacetylase leads to a global increase in acetylation. These globally acting enzymes generate a highly dynamic equilibrium of histone acetylation and deacetylation reactions, which can be locally perturbed by targeted acetylases and deacetylases (28). Following the dissociation of the recruiting activator or repressor from a promoter, targeted acetylation and deacetylation are reversed within minutes, and the steady-state level of acetylation is reset by global activities (28).
Esa1 is the only essential histone acetylase in yeast, and conditional esa1 alleles arrest at the G2/M checkpoint (7, 52). Esa1 is the catalytic subunit of the NuA4 complex that can be recruited to reconstituted chromatin templates by activators, creating a large domain of H4 hyperacetylation and resulting in activator-dependent transcription (2, 53, 56, 58). In vivo, Esa1 is recruited to ribosomal-protein (RP) gene promoters by Rap1 (47), and a loss of Esa1 function results in gene-specific decreases in transcription (13, 47). Esa1 is also recruited to double-strand breaks, and it plays an important role in double-strand break repair (5). In addition to being recruited to specific genomic locations, Esa1 is essential for the genome-wide acetylation of H4 (47, 59), but this nontargeted activity is not generally required for transcription (47).
To examine the biological roles of global and targeted acetylation, we looked for components of the NuA4 complex that are required for either global or targeted acetylation, but not both. Eaf3 (Esa1 associated factor 3) is an atypical component of NuA4 since it is not essential for cell cycle progression or catalytic activity (11). After the experimental work described here was largely completed, it was discovered that Eaf3 is also a component of the Rpd3 histone deacetylase complex (14, 19). Rpd3 is recruited to target promoters by the Ume6 repressor (26) and presumably by other DNA-binding proteins (35), and it also acts in a global manner throughout the genome (59). Here, we demonstrate that wild-type (WT) yeast cells have a global pattern of H3 and H4 acetylation in which acetylation is higher at promoters than at coding regions. Furthermore, we show that Eaf3 regulates the global pattern, but not the overall level, of histone acetylation in a manner that does not involve targeted recruitment of histone-modifying complexes and is independent of transcriptional activity.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The eaf3 deletion strain (7143) and the isogenic parent strain (4741) were obtained from Research Genetics. Esa1 was three-hemagglutinin (HA3) tagged in the eaf3 mutant and WT parent strain by using a two-step gene replacement construct (47) to generate JRY18 (MATa his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 esa1::HA3-ESA1) and JRY19 (MATa his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 eaf3::KAN esa1::HA3-ESA1).
Chromatin immunoprecipitation.
Formaldehyde-cross-linked chromatin was prepared as described previously (34, 47) and was immunoprecipitated with antibodies against the HA epitope (F7; Santa Cruz Biotechnology, Santa Cruz, Calif.), tetra-acetylated (lysines 5, 8, 12, and 16) H4 (Upstate Biotechnology), and diacetylated (lysines 9 and 14) H3 (Upstate Biotechnology). Quantitative PCR analyses were performed in real time by using an Applied Biosystems 7700 sequence detector and SYBR green as a detection agent, except for the experiment in Fig. 1B, in which radiolabeled PCR products were separated on polyacrylamide gels and quantified on a PhosphorImager. In each case, the apparent immunoprecipitation efficiency was calculated by dividing the amount of PCR product for the immunoprecipitated sample by the amount of PCR product for the corresponding input sample.
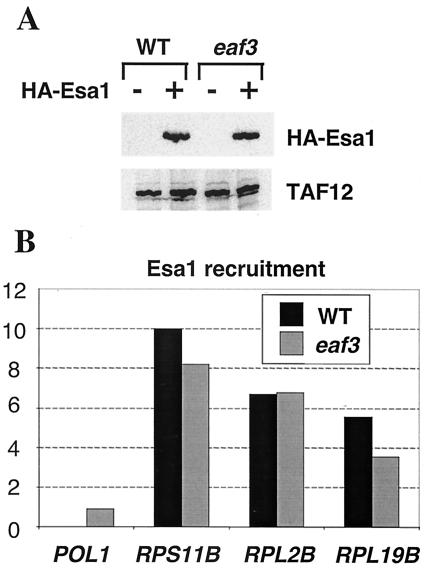
FIG. 1.
Esa1 recruitment to RP promoters is unaffected by the deletion of EAF3. (A) Protein from WT and eaf3 strains containing HA3-Esa1 and the untagged parent strains were probed by Western blot analysis with antibodies against the HA epitope and TAF12 (as a loading control). (B) Cross-linked chromatin from WT and eaf3 strains containing HA3-WT Esa1 was immunoprecipitated with antibody against the HA epitope. Esa1 recruitment (defined in arbitrary units as described in Materials and Methods) was analyzed by quantitative PCR with primers to the RPS11B, RPL2B, and RPL19B promoters and the POL1 coding sequence control.
For measuring Esa1 recruitment, the background cross-linking (defined as the apparent immunoprecipitation efficiency for the POL1 coding sequence in a WT strain) was subtracted from each value. In order to compare Esa1 recruitment across multiple experiments, recruitment at the RPS11B promoter in the WT strain was set to 10, and recruitment values for individual experiments were scaled relative to that for RPS11B. Two independent chromatin preparations were subjected to immunoprecipitation, and duplicate PCRs were performed across a titration of chromatin concentrations. The data were averaged following scaling to RPS11B. The experimental error for individual determinations was ±20%.
For measurements of H3 and H4 acetylation, immunoprecipitation efficiencies were scaled relative to that of RPS11B (arbitrarily set to 10) prior to averaging; background was not subtracted, as it is unclear what represents background acetylation. The eaf3/WT acetylation ratios were calculated by dividing the immunoprecipitation efficiency in the eaf3 strain by the immunoprecipitation efficiency in the WT strain for each PCR product. Four independent chromatin preparations were subjected to immunoprecipitation, and each PCR was run in triplicate. The experimental error for individual determinations is ±15%. However, the conclusion that promoter regions have lower levels of acetylation in eaf3 deletion strains is based on multiple promoter regions, and the difference is clearly significant as determined by a two-tailed, unpaired t test (P = 0.001). Furthermore, as the overall levels of histone acetylation are comparable in WT and eaf3 deletion strains, the more dramatic increases in protein-coding regions must be balanced out by corresponding decreases in noncoding regions.
Western blotting.
Cells were lysed with glass beads in radioimmunoprecipitation assay buffer containing protease inhibitors. For Western analysis, 30 μg of protein, an amount determined by the Bradford assay (6a), was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose. The filters were probed with antibodies against the HA epitope (12CA5 ascites fluid), TAF12 (a gift from Michael Green), and tetra-acetylated H4 and diacetylated H3 by standard techniques.
Transcriptional analysis.
Total RNA from WT and eaf3 deletion strains grown in synthetic complete or yeast extract-peptone-dextrose medium to an A600 of 0.8 to 1.0 was prepared by the hot acid phenol method (23) and then purified on an RNeasy column (QIAGEN) in the presence of DNase I. First-strand cDNA was synthesized from 1.0 μg of total RNA of each sample by using an oligo(dT) primer and Superscript II reverse transcriptase (Invitrogen). To analyze transcription from individual genes, the resulting cDNA was subjected to real-time quantitative PCR analysis as described above.
For whole-genome expression analysis, first-strand cDNA from 1.0 μg of total RNA was incubated with E. coli DNA polymerase I and RNase H in the presence of deoxynucleoside triphosphates to produce second-strand cDNA. The resulting double-stranded cDNA was amplified by two rounds of primer extension followed by PCR as described previously (24) in the presence of amino-allyl-deoxyuridine (Sigma). This material was coupled with Cy3 or Cy5 dye (Amersham) and applied to microarrays spotted with yeast open reading frame PCR products. Microarray hybridization and washing were performed as described previously (24). Arrays were scanned on an Axon scanner, and the data were analyzed with the Axon GenePix 4.0 software. Four data sets, each representing one copy of the yeast genome, were analyzed as follows. Data were first filtered to remove those spots whose results were either too faint to be detected or for some other reason flagged as unreliable by the GenePix software. For the remaining 5,414 genes, background-subtracted Cy5 and Cy3 signals for all genes were normalized to the same median value. For each spot, a ratio of Cy5 fluorescence to Cy3 fluorescence was then obtained. For each gene, the median of these ratios from each of the four data sets was determined, and genes were ranked according to this median ratio. An unchanged gene in this analysis would have a ratio of 1. The observed ratios varied from 0.05 (EAF3) to 3.4 (PTR2), with a median of 1 and a standard deviation of 0.31. The ranked gene list was further analyzed with the Gene Ontology tools of the Saccharomyces Genome Database.
RESULTS
Eaf3 is not required for Esa1 recruitment to RP promoters.
Eaf3 is a stable component of the NuA4 acetylase complex that interacts directly with Esa1 but, unlike other components, is not required for cell growth (11). Deletion of Eaf3 does not affect NuA4 integrity or activity but does lead to a subtle decrease in the transcription of Esa1-regulated genes. Based on these results, we tested the hypothesis that Eaf3 plays a role in the targeted recruitment of Esa1 and the NuA4 complex. In previous work, RP promoters were identified as specific targets of HA3-tagged Esa1 (47). HA3-Esa1 is expressed at comparable levels in isogenic WT and eaf3 deletion strains (Fig. 1A), and as assayed by chromatin immunoprecipitation, Esa1 recruitment to RP promoters is not significantly affected by the deletion of Eaf3 (Fig. 1B). Thus, Eaf3 is not required for Esa1 recruitment to RP promoters.
Deletion of Eaf3 results in a dramatic increase in H4 acetylation at coding sequences and a slight decrease in acetylation at promoters.
To assess the role of Eaf3 in global and targeted acetylation, we compared acetylation levels in the WT and eaf3 strains at RP promoters and a variety of other promoters and coding sequences. H4 acetylation at the DYN1, MEC1, GLT1, MOT1, POL1, YLR454W, HSP104, and SSA4 coding sequences is dramatically higher (up to eightfold) in the eaf3 strain than in the WT strain (Fig. 2). H3 acetylation at these protein-coding sequences is also increased, although to a lesser extent (twofold). A loss of Eaf3 does not result in increased H3 or H4 acetylation at other coding sequences (DED1, FBA1, RPL2B, or RPS5) (see below). At all promoter sequences examined, both H4 and H3 acetylation are lower in the deletion strain (1.5- to 2-fold). For example, acetylation is decreased at the POL1 and HSP104 gene promoters but increased at the corresponding coding sequences.
FIG. 2.
Deletion of EAF3 results in increased acetylation at coding sequences and decreased acetylation at promoters. Cross-linked chromatin from WT and eaf3 deletion strains was immunoprecipitated with antibodies against tetra-acetylated H4 and diacetylated H3, and the acetylation status of the indicated promoters and coding sequences was determined as described in Materials and Methods. The data are presented as ratios of acetylation levels in the eaf3 deletion strain to those in the WT strain. CDS, coding sequences.
Although it appears that increased histone H4 acetylation due to a loss of Eaf3 occurs at some coding sequences and not at others, all genomic regions showing increased acetylation in the eaf3 deletion strain are located more than 1 kb from promoter sequences. To test the hypothesis that distance from promoters is important, we measured the level of histone acetylation at various locations across the large GLT1 and HSP104 genes. In accord with our hypothesis, H4 acetylation in the eaf3 mutant strain is lower at the promoter and proximal coding region, dramatically higher in the middle of the coding sequence (4.5-fold at GLT1 and 4- to 7-fold at two locations within HSP104), and relatively unaffected at the 3′ end of the gene (Fig. 3A). Similar results are observed for H3 acetylation, although the magnitude of the effect is lower than that for H4 acetylation. The marginal increase in acetylation at the 3′ end of the GLT1 coding sequence may be due to the influence of the promoter of the adjacent gene. Thus, the deletion of EAF3 leads to increased acetylation at coding regions of the genome that are greater than 1 kb from promoters. Acetylation at RP genes is affected in a manner similar to that of other genes, which is consistent with Eaf3 playing a role in global acetylation and not in targeted recruitment of the Esa1 complex.
FIG. 3.
Effect of Eaf3 on acetylation across the GLT1 and HSP104 genes. (A) Ratios of H4 and H3 acetylation levels in the eaf3 deletion strain to those in the WT strain at the indicated regions of the 6.5-kb GLT1 and 2.7-kb HSP104 genes (the position of the upstream-most residues of the PCR products are indicated on the x axes, with +1 being the ATG initiation codon) were determined as described in the legend to Fig. 2. (B) Relative levels of H4 acetylation (arbitrary units; see Materials and Methods) for the indicated GLT1 and HSP104 regions. (C) Relative levels of H3 acetylation.
WT cells have higher levels of H3 and H4 acetylation at promoters than at coding regions.
To obtain additional insight into the effect of Eaf3 on the global acetylation profile, we reanalyzed the data to show the relative acetylation levels (quantitatively determined by immunoprecipitation efficiency and presented in arbitrary units) of various genomic regions in either the WT or eaf3 strain (Fig. 4). In WT cells, levels of H3 and H4 acetylation are not evenly distributed across the genome but rather are higher at promoters and lower at coding sequences. The seven promoters tested have an average H4 acetylation level of 20.6 (range, 4 to 30), whereas the eight coding sequences located at least 1 kb from a promoter have an average H4 acetylation level of 4.0 (range, 2 to 8). Similarly, H3 acetylation at the seven promoter regions has an average value of 15.4, whereas the average value at the eight coding regions is 6.2. This pattern is consistent with the two examples that have been reported (32, 59), although it was unclear from those examples whether the pattern would be commonly observed. There is variation in the acetylation level at the promoter sequences examined, and we suspect that this is due to regions of low nucleosome density (or less likely, transcription factors blocking acetylation sites at specific promoters). In any event, this pattern of high acetylation at promoters and low acetylation at coding regions is observed for a significant number of genomic regions that were essentially chosen at random, suggesting that this acetylation pattern is a general feature of the yeast genome.
FIG. 4.
The global pattern of H4 acetylation depends on Eaf3. Relative levels of H3 and H4 acetylation (arbitrary units) for the indicated promoter and coding regions in the WT and eaf3 deletion strains are shown. CDS, coding sequences.
Interestingly, the WT pattern of high acetylation near promoters and lower acetylation at coding sequences is absent in the eaf3 strain such that the acetylation level is more evenly distributed across the genome (Fig. 4). Although the change in H4 acetylation at coding sequences is much larger than at promoters (3- to 8-fold compared to 1.5- to 2-fold) (Fig. 2), the actual changes in acetylation are similar. For example, at the GLT1 gene (Fig. 3B), the 2-fold decrease in H4 acetylation at the promoter (primer −197) represents a decrease of 15 acetylation units (from 30 to 15), whereas the 4.5-fold increase in acetylation at the coding sequence (primer 3241) indicates an increase of 10 acetylation units (from 3 to 13). Similar results are observed for the HSP104 gene. Although the magnitude of the acetylation effect is smaller for H3 acetylation, the same trend is observed at both GLT1 and HSP104 (Fig. 3C). Thus, Eaf3 is responsible for maintaining the global histone acetylation profile in which promoters are relatively more acetylated than coding regions.
The overall level of acetylation is unchanged in an Eaf3 deletion strain.
Conditional esa1 alleles have dramatically reduced levels of H4 acetylation, as assayed either by Western blotting of bulk histones (7) or by chromatin immunoprecipitation at multiple loci (47, 59). Although we observed large changes in the histone acetylation profile for the eaf3 strain (Fig. 2 and 4), Western blots for acetylated histones H4 and H3 show that the overall level of acetylation in the eaf3 deletion strain is comparable to that of the WT strain (Fig. 5). This level of acetylation suggests that increased acetylation at coding sequences is counterbalanced by decreased acetylation at promoters. Thus, despite the change in the positioning of H4 acetylation, deletion of Eaf3 does not alter the overall level of H4 acetylation in the cell.
FIG. 5.
Eaf3 does not affect overall levels of H3 or H4 acetylation. Results of Western blot analysis of H4 and H3 acetylation in WT and eaf3 cell extracts are shown. Filters were probed with antibodies against acetylated H4 (acet-H4), acetylated H3, and TAF12 (as a loading control).
The altered pattern of global histone acetylation is independent of transcription.
Hypoacetylation of H3 in coding regions inhibits transcription in vivo (30), suggesting the possibility that the altered pattern of histone acetylation in the eaf3 strain might have transcriptional consequences. To address the relationship between the global pattern of acetylation and transcription, we first analyzed RNA levels of all genes tested here in WT and eaf3 deletion strains (Fig. 6A). Eaf3 has no significant effect on transcription, except possibly on that of DYN1 (a 1.6-fold effect), indicating that the altered pattern of histone acetylation in the eaf3 strain is unrelated to transcription.
FIG. 6.
Eaf3 positively affects the transcription of a small subset of yeast genes. (A) Ratios of RNA of the indicated genes in the eaf3 strain to RNA of the same genes in the WT strain as determined by quantitative reverse transcriptase PCR analysis; (B) numbers of genes showing more than the indicated decrease (n-fold) (light gray) or increase (n-fold) (dark gray) in transcription in the eaf3 strain with respect to the WT strain, as determined by whole-genome microarray analysis. Interpretable hybridization signals were obtained for 5,414 genes.
To examine this issue more generally, we compared the transcriptional profiles of the WT and eaf3 strains on microarrays representing the yeast genome (Fig. 6B). Approximately 0.9% (49 out of 5,414 genes with a measurable signal) of yeast genes show a threefold or greater decrease in RNA levels in the eaf3 strain, whereas only one gene (PTR2) shows a threefold increase. With a twofold cutoff, 286 genes (5%) are positively affected by Eaf3, whereas 14 (0.3%) are negatively affected. Very limited numbers of genes show fourfold (n = 14)- and fivefold (n = 7)-decreased RNA levels in the eaf3 strain. Genes involved in mating and pheromone response (e.g., MFA1, AGA1, and GPA1) are preferentially up-regulated by Eaf3, whereas genes encoding transporters (e.g., PTR2, FET3, and OPT2) and small nucleolar RNAs are preferentially down-regulated by Eaf3. Thus, Eaf3 plays a positive, but quantitatively modest, role in the transcription of a small subset of genes, whereas it has a negative effect on very few genes. Thus, the role of Eaf3 in maintaining the global pattern of histone acetylation does not correlate with its modest and very selective effect on transcription.
DISCUSSION
The level of histone acetylation in yeast cells is higher at promoters and lower at coding regions in a manner that is independent of transcription.
In principle, a nontargeted histone acetylase should generate equal levels of histone acetylation throughout the genome except at locations where acetylases or deacetylases are specifically targeted. However, we show here that WT yeast cells have a distinct pattern of global H3 and H4 acetylation in which promoter regions are more acetylated than coding regions (Fig. 4). This conclusion is based on the clear distinction between seven promoter regions and eight coding sequences located at least 1 kb from a promoter. These genomic regions represent a diverse set of genes, and they were chosen primarily for technical convenience. Thus, it is highly likely that the patterns of H3 and H4 acetylation, with high levels at promoters and low levels at coding regions, occur throughout most of the yeast genome, and this pattern is consistent with the results of previous analysis of H3 acetylation at the HIS3 and PHO5 loci (32, 59).
Although H3 and H4 acetylation levels are low at all eight coding sequences located more than 1 kb from a promoter, low-level acetylation is not typically observed at coding sequences located closer to a promoter. In part, this observation reflects the resolution of chromatin immunoprecipitation experiments, which is determined by the size of cross-linked DNA fragments. For this reason, high-level acetylation at the promoter region will appear to spread towards the proximal coding region; i.e., the apparent domain of high acetylation is larger than the actual domain (27). However, analysis of comparable samples for occupancy by specific DNA-binding proteins (TATA-binding protein and Rap1) indicate that sequences 500 bp away from the site of association contribute minimally to the overall signal (47). Thus, we suspect that the actual domain of high acetylation is larger than the promoter region itself and in fact spreads into the nearby coding region.
Interestingly, the global pattern of H3 and H4 acetylation that distinguishes the promoter from coding regions is independent of transcriptional activity. The loss of Eaf3 essentially abolishes the global histone acetylation pattern, whereas it has very modest and highly specific transcriptional effects. Although many aspects of chromatin structure are mechanistically linked to transcription, yeast promoter regions are more accessible to nuclear proteins than coding regions in a manner that is independent of transcriptional activity (40).
Eaf3 regulates the global pattern of H3 and H4 acetylation.
Although initially identified as a component of the NuA4 complex (11), Eaf3 is also a component of the Rpd3 histone deacetylase complex (14, 19). Our results demonstrate that Eaf3 is required for the normal pattern of global H3 and H4 acetylation. In an eaf3 deletion strain, histone acetylation is decreased at promoters and increased at coding sequences (Fig. 2) such that histone acetylation levels are evenly distributed across the genome (Fig. 4). Eaf3 does not affect NuA4 activity in vitro (11), nor does it significantly affect the recruitment of Esa1 to RP promoters (Fig. 1) or the overall level of H4 acetylation (Fig. 5) in vivo. In addition, Eaf3 does not affect the transcription of genes repressed by targeted recruitment and localized histone deacetylation mediated by Rpd3. Thus, our results strongly suggest that Eaf3 affects the global distribution of the NuA4 and/or the Rpd3 complex, not the catalytic activities of these complexes.
Our results do not directly address the relative contributions of the NuA4 and Rpd3 complexes in the Eaf3-dependent pattern of global histone acetylation. Eaf3-dependent effects on global acetylation patterns appear more pronounced for H4 than for H3, suggesting that the Eaf3-dependent distribution of NuA4 throughout the genome plays a role. Eaf3 in the Rpd3 complex cannot account for a preferential effect on H4 because the global and recruited Rpd3 complex affects H3 and H4 equally (9, 27, 50, 55, 59), and the four other Rpd3-like histone deacetylases do not specifically affect global H4 acetylation (48). On the other hand, Eaf3 affects global acetylation of H3 in a manner qualitatively similar to that of H4. As the NuA4 complex does not acetylate histone H3 in vitro (2), this observation suggests that the Eaf3-dependent distribution of the Rpd3 complex also plays a role in the global pattern of histone acetylation.
The above arguments suggest that Eaf3 might affect the global histone acetylation pattern via both the NuA4 and Rpd3 complexes. However, we cannot exclude the possibility that the apparent preferential effect of Eaf3 on H4 acetylation might be related to the antibodies used to analyze H3 and H4, not bona fide levels of histone acetylation. We also cannot exclude the possibility that the H3 acetylation profile arises indirectly from the H4 acetylation profile because the SAGA complex, which globally acetylates H3 (32, 59), preferentially associates with acetylated nucleosomes in vitro (1, 18) and in vivo (9). Thus, Eaf3 might affect global histone acetylation primarily, and possibly exclusively, through either the NuA4 or the Rpd3 complex.
Potential mechanisms for Eaf3-dependent control of the global histone acetylation profile.
As NuA4 and the Rpd3 complex have opposing enzymatic activities, Eaf3-dependent effects on their genomic distribution must be reciprocal in order to account for the observed pattern of histone acetylation. Specifically, our results require that Eaf3 confer a preferential association of NuA4 with promoter regions and/or a preferential association of Rpd3 with coding regions. We will consider potential models for NuA4 and Rpd3 separately, although the models are not mutually exclusive.
In WT yeast cells, promoter regions are generally more accessible to nuclear proteins than coding regions, and this preferential accessibility is determined by a general property of the promoter DNA sequence and not by defined sequence elements in the promoter (40). In the context of NuA4, Eaf3 might recognize the structural difference between promoters and coding regions by facilitating the interaction of NuA4 with DNA, which is preferentially accessible in promoter regions. Alternatively, Eaf3 might recognize some feature of chromatin (e.g., nucleosome conformation or nonhistone protein) that is distinct between promoters and coding regions. We disfavor the idea that the Eaf3-dependent pattern of H4 acetylation reflects Eaf3-dependent interactions of NuA4 with transcriptional regulatory proteins associated with promoters. Esa1 association with promoters in vivo does not correlate directly with transcriptional activity (47), and the level of H4 acetylation at promoters analyzed here is unrelated to transcriptional activity.
Eaf3 contains a chromodomain, a structural domain that can bind methylated lysines in histones (3, 25, 37). Coding regions are preferentially methylated on histone H3 at lysines 4 (specifically the trimethylated form) and 36 (31, 43, 51). These observations suggest the possibility that Eaf3 might bias the distribution of the Rpd3 complex towards coding regions. In this regard, it is interesting that the levels of methylation of H3-lysine 36 are low at promoter-proximal regions and higher at more-downstream positions (31). However, as the levels of H3-lysine 36 methylation and H3-lysine 4 trimethylation in coding regions are directly related to transcriptional activity (31, 43, 51), the pattern of H3 methylation does not easily explain the Eaf3-dependent effect on histone acetylation. In addition, it is unclear if the Eaf3 chromodomain binds methylated histones (or shows specificity towards particular methylated lysines) or contributes to the ability of Eaf3 to discriminate between promoter and coding sequences.
Biological role of Eaf3 and the global acetylation profile.
Depletion of the H4-specific acetylase Esa1 results in an extensive loss of acetylation and a G2/M arrest (7, 21, 47). Global H4 acetylation is the essential function of Esa1, because a NuA4 subcomplex containing Esa1, Epl1, and Yng2 that mediates global H4 acetylation and is sufficient for cell viability exists but is unlikely to be recruited to promoters (6). Although global H4 acetylation appears to be required for viability, our results indicate that alteration of the global H3 and H4 acetylation profile in cells from which Eaf3 has been removed has no significant growth effect and only a very modest effect on transcription. The subtle alterations in transcription in the eaf3 deletion strain likely reflect a change in the global acetylation profile rather than a change in targeted recruitment. As some Eaf3-dependent transcriptional effects are also observed in Esa1-depleted strains (13, 47), some Esa1-dependent effects on gene expression might also reflect the global activity, rather than promoter-specific recruitment, of the NuA4 complex.
Although a loss of Eaf3 and the global histone acetylation profile cause only subtle phenotypes in S. cerevisiae, Eaf3 homologues appear to be more important in other organisms. Mutation of the Schizosaccharomyces pombe homologue, Alp13, causes temperature-sensitive growth, sterility, and a defect in cell polarity as well as defects in chromosome stability (42, 46). Multicellular organisms appear to have multiple Eaf3-like proteins with nonredundant functions (41). Overexpression of human MRG15 results in abnormal nuclear morphologies and cell death, and a related protein (MORF4) lacking the chromodomain induces senescence in a subset of immortal cell lines (4). Drosophila MSL3 is essential for male viability by virtue of its role in dosage compensation (15), and Caenorhabditis elegans MRG1 is required maternally for primordial germ cells to initiate mitotic proliferation (12). It remains to be determined whether these more significant biological functions are due to the alteration of global acetylation profiles or some other function of the Eaf3 homologues.
Acknowledgments
We thank Michael Green for the TAF12 antibody, Joseph Geisberg for advice on real-time PCR, Frank Gibbons for help with statistical analysis of the microarray data, and Simon Boulton for critical reading of the manuscript.
This work was supported by a postdoctoral fellowship from the Human Frontiers Science Program to J.L.R. and by research grants (GM 30186 and GM 53720) from the National Institutes of Health to K.S.
REFERENCES
- 1.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. [DOI] [PubMed] [Google Scholar]
- 2.Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant, C. J. Brandl, L. Pillus, J. L. Workman, and J. Cote. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATF-related cofactor Tra1p. EMBO J. 18:5108-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 4.Bertram, M. J., N. G. Bérubé, X. Hang-Swanson, Q. Ran, J. K. Leung, S. Bryce, K. Spurgers, R. J. Bick, A. Baldini, Y. Ning, L. J. Clark, E. K. Parkinson, J. C. Barrett, J. R. Smith, and O. M. Pereira-Smith. 1999. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol. Cell. Biol. 19:1479-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qui, K. E. Harmon, P. C. Megee, P. A. Grant, M. M. Smith, and M. F. Christman. 2002. Acetylation of histone H4 by Esa1 is required for double-strand break repair. Nature 419:411-415. [DOI] [PubMed] [Google Scholar]
- 6.Boudreault, A. A., D. Cronier, W. Selleck, N. Lacoste, R. T. Utley, S. Allard, J. Savard, W. S. Lane, S. Tan, and J. Cote. 2003. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 17:1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Clarke, A. S., J. E. Lowell, S. J. Jacobson, and L. Pillus. 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19:2515-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 9.Deckert, J., and K. Struhl. 2002. Targeted recruitment of Rpd3 histone deacetylase represses transcription by inhibiting recruitment of Swi/Snf, SAGA, and TATA binding protein. Mol. Cell. Biol. 22:6458-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 11.Eisen, A., R. T. Utley, A. Nourani, S. Allard, P. Schmidt, W. S. Lane, J. C. Lucchesi, and J. Cote. 2001. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J. Biol. Chem. 276:3484-3491. [DOI] [PubMed] [Google Scholar]
- 12.Fujita, M., T. Takasaki, N. Nakajima, T. Kawano, Y. Shimura, and H. Sakamoto. 2002. MRG-1, a mortality factor-related chromodomain protein, is required maternally for primordial germ cells to initiate mitotic proliferation in C. elegans. Mech. Dev. 114:61-69. [DOI] [PubMed] [Google Scholar]
- 13.Galarneau, L., A. Nourani, A. A. Boudreault, Y. Zhang, L. Heliot, S. Allard, J. Savard, W. S. Lane, D. J. Stillman, and J. Cote. 2000. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell 5:927-937. [DOI] [PubMed] [Google Scholar]
- 14.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 15.Gorman, M., M. I. Kuroda, and B. S. Baker. 1993. Regulation of the sex-specific binding of the maleless dosage compensation protein to the male X-chromosome in Drosophila. Cell 72:39-49. [DOI] [PubMed] [Google Scholar]
- 16.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 17.Hassan, A. H., K. E. Neely, and J. L. Workman. 2001. Histone acetyltransferase complexes stabilize Swi/Snf binding to promoter nucleosomes. Cell 104:817-827. [DOI] [PubMed] [Google Scholar]
- 18.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 19.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 20.Hong, L., G. P. Schroth, H. R. Matthews, P. Yau, and E. M. Bradbury. 1993. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J. Biol. Chem. 268:305-314. [PubMed] [Google Scholar]
- 21.Howe, L., D. Auston, P. Grant, S. John, R. G. Cook, J. L. Workman, and L. Pillus. 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 15:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hudson, B. P., M. A. Martinez-Yamout, H. J. Dyson, and P. E. Wright. 2000. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J. Mol. Biol. 304:355-370. [DOI] [PubMed] [Google Scholar]
- 23.Iyer, V., and K. Struhl. 1996. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5208-5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs, S. A., S. D. Taverna, Y. Zhang, S. D. Briggs, J. Li, J. C. Eissenberg, C. D. Allis, and S. Khorasanizadeh. 2001. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 20:5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadosh, D., and K. Struhl. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365-371. [DOI] [PubMed] [Google Scholar]
- 27.Kadosh, D., and K. Struhl. 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18:5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katan-Khaykovich, Y., and K. Struhl. 2002. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 16:743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krebs, J. E., M.-H. Kuo, C. D. Allis, and C. L. Peterson. 1999. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 13:1412-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kristjuhan, A., J. Walker, N. Suka, M. Grunstein, D. Roberts, B. R. Cairns, and J. Q. Svejstrup. 2002. Transcriptional inhibition of genes with severe histone H3 hypoacetylation in the coding region. Mol. Cell 10:925-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krogan, N. J., M. Kim, A. Tong, A. Golshani, G. Cagney, V. Canadien, D. P. Richards, B. K. Beattie, A. Emili, C. Boone, A. Shilatifard, S. Buratowski, and J. Greenblatt. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23:4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo, M.-H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 33.Kuo, M.-H., J. Zhou, P. Jambeck, M. E. A. Churchill, and C. D. Allis. 1998. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12:627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-612. [DOI] [PubMed] [Google Scholar]
- 35.Kurdistani, S. K., D. Robyr, S. Tavazoie, and M. Grunstein. 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31:248-254. [DOI] [PubMed] [Google Scholar]
- 36.Kwon, J., K. B. Morshead, J. R. Guyon, R. E. Kingston, and M. A. Oettinger. 2000. Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Mol. Cell 6:1037-1048. [DOI] [PubMed] [Google Scholar]
- 37.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 38.Lee, D. Y., J. J. Hayes, D. Pruss, and A. P. Wolffe. 1993. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72:73-84. [DOI] [PubMed] [Google Scholar]
- 39.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 40.Mai, X., S. Chou, and K. Struhl. 2000. Preferential accessibility of the yeast his3 promoter is determined by a general property of the DNA sequence, not by specific elements. Mol. Cell. Biol. 20:6668-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marin, I., and B. S. Baker. 2000. Origin and evolution of the regulatory gene male-specific lethal-3. Mol. Biol. Evol. 17:1240-1450. [DOI] [PubMed] [Google Scholar]
- 42.Nakayama, J., G. Xiao, K. Noma, A. Malikzay, P. Bjerling, K. Ekwall, R. Kobayashi, and S. I. Grewal. 2003. Alp13, an MRG family protein, is a component of fission yeast Clr6 histone deacetylase required for genomic integrity. EMBO J. 22:2776-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 44.Nightingale, K. P., R. E. Wellinger, J. M. Sogo, and P. B. Becker. 1998. Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26 gene in chromatin. EMBO J. 17:2865-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Owen, D. J., P. Ornaghi, J. C. Yang, N. Lowe, P. R. Evans, P. Ballario, D. Neuhaus, P. Filetici, and A. A. Travers. 2000. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5. EMBO J. 19:6141-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radcliffe, P., D. Hirata, D. Childs, L. Vardy, and T. Toda. 1998. Identification of novel temperature-sensitive lethal alleles in essential beta-tubulin and nonessential alpha 2-tubulin genes as fission yeast polarity mutants. Mol. Biol. Cell 9:1757-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed] [Google Scholar]
- 48.Robyr, D., Y. Suka, I. Xenarios, S. K. Kurdistani, A. Wang, N. Suka, and M. Grunstein. 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109:437-446. [DOI] [PubMed] [Google Scholar]
- 49.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 50.Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner, and M. Grunstein. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831-835. [DOI] [PubMed] [Google Scholar]
- 51.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 52.Smith, E. R., A. Eisen, W. Gu, M. Sattah, A. Pannuti, J. Zhou, R. G. Cook, J. C. Lucchesi, and C. D. Allis. 1998. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. USA 95:3561-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steger, D. J., A. Eberharter, S. John, P. A. Grant, and J. L. Workman. 1998. Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc. Natl. Acad. Sci. USA 95:12924-12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 55.Suka, N., Y. Suka, A. A. Carmen, J. Wu, and M. Grunstein. 2001. Highly specific antibodies determine histone acetylation in yeast heterochromatin and euchromatin. Mol. Cell 8:473-479. [DOI] [PubMed] [Google Scholar]
- 56.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 57.Vettese-Dadey, M., P. A. Grant, T. R. Hebbes, C. Crane-Robinson, C. D. Allis, and J. L. Workman. 1996. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 15:2508-2518. [PMC free article] [PubMed] [Google Scholar]
- 58.Vignali, M., D. J. Steger, K. E. Neely, and J. L. Workman. 2000. Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J. 19:2629-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]