Abstract
The expression of human thymidine kinase 1 (hTK1) is highly dependent on the growth states and cell cycle stages in mammalian cells. The amount of hTK1 is significantly increased in the cells during progression to the S and M phases, and becomes barely detectable in the early G1 phase by a proteolytic control during mitotic exit. This tight regulation is important for providing the correct pool of dTTP for DNA synthesis at the right time in the cell cycle. Here, we investigated the mechanism responsible for mitotic degradation of hTK1. We show that hTK1 is degraded via a ubiquitin-proteasome pathway in mammalian cells and that anaphase-promoting complex/cyclosome (APC/C) activator Cdh1 is not only a necessary but also a rate-limiting factor for mitotic degradation of hTK1. Furthermore, a KEN box sequence located in the C-terminal region of hTK1 is required for its mitotic degradation and interaction capability with Cdh1. By in vitro ubiquitinylation assays, we demonstrated that hTK1 is targeted for degradation by the APC/C-Cdh1 ubiquitin ligase dependent on this KEN box motif. Taken together, we concluded that activation of the APC/C-Cdh1 complex during mitotic exit controls timing of hTK1 destruction, thus effectively minimizing dTTP formation from the salvage pathway in the early G1 phase of the cell cycle in mammalian cells.
Thymidine kinase (TK) is a crucial enzyme in the salvage pathway of dTTP synthesis for DNA replication. Mammalian cells contain two different TK isozymes, cytosolic TK1 (2) and mitochondrial TK2 (23), which do not share significant homology in their amino acid sequences. The expression of cytosolic TK1 is cell cycle-dependent, whereas the levels of the mitochondrial isozyme, TK2, remain stable throughout the cell cycle. The expressed level of cytosolic TK1 is significantly increased in the G1/S transition through its transcriptional and translational controls (1, 9, 24, 45). During progression of the G2/M phase, human TK1 (hTK1) is accumulated and becomes phosphorylated by cdc2 kinase on serine 13 (5, 6). hTK1 is then rapidly degraded at mitosis, so that the level of hTK1 protein is kept low in the early G1 phase (25). This tight regulation ensures its functional role in supplying dTTP to coordinate with DNA replication during cell cycle progression.
In this study, we investigated the molecular mechanism responsible for mitotic hTK1 proteolytsis. It is well established that the ubiquitin-proteasome pathway represents a fundamental mechanism for regulating protein level in the cell cycle (18). Ubiquitinylation of protein substrates is carried out by a series of enzymatic reactions catalyzed by E1, E2, and E3 enzymes (42). The SCF complex (Skp1-Cullin1/CDC53-F box) and the APC/C (anaphase-promoting complex/cyclosome) are two ubiquitin ligases (E3) that play a determining role in targeting proteins for cell cycle-dependent proteolysis (11, 34, 35). The SCF complex recognizes phosphorylated substrates, such as p27, cyclin D, cyclin E, E2F, IκB, and β-catenin, to promote their degradation in mammalian cells via the so-called F box proteins (16, 22). The interaction between F box protein and Skp1, one subunit of the SCF complex, bridges phosphorylated substrate and F box protein to Cullin1, which organizes this E3 complex and helps recruit E2 ubiquitin-conjugating enzyme through interaction with the Roc1/UBC subunit (16). In contrast to the SCF complex, substrate phosphorylation is not a requisite recognized by the APC/C complex (17, 32, 35). APC/C-mediated ubiquitinylation and degradation require two different activators, Cdc20 and Cdh1 (21, 35, 49). Cdc20-APC/C is activated during mitosis and targets cyclin B (53, 55) and securin (54) for proteolysis through binding to the destruction box (D box; RXXLXXXXN) at their N termini. APC/C-Cdh1 recognizes either a D box or the KEN box (37, 38). To date, human Cdc6, Aurora-A kinase, Cdc20, Cdc25A, and mouse ribonucleotide reductase R2 protein have been found to be the substrates of APC/C-Cdh1 in late mitosis and G0/G1 transition (4, 12, 20, 28, 36).
The C-terminal region of mouse TK1 (mTK1) or hTK1 has been shown to play a crucial role in determining its protein stability during mitosis (25, 46). Other reports, however, have suggested that the capability of substrate binding (47) and the multimerization status of the mTK1 subunit might contribute to the stability of mTK1 protein in a proteasome-independent manner (40). On the other hand, with expression of hTK1 in the yeast systems, we found that serine 13 phosphorylation plays a role in hTK1 degradation, which requires the functional activity of SCF (26). Despite these experimental results, the molecular event that targets hTK1 for mitotic degradation in mammalian cells is still unknown. In this study, we provided the first evidence that in mammalian cells, the ubiquitin-proteasome pathway controls the expression level of hTK1 dependent on the APC/C-Cdh1 E3 ligase, which interacts with hTK1 by recognizing the KEN box located at its C-terminal region. Overall, the results obtained from this study concluded that hTK1 is another new target for APC/C-Cdh1-mediated proteolysis at mitosis in mammalian cells.
MATERIALS AND METHODS
Materials.
Anti-hTK1 polyclonal antibody was prepared as described previously (5). Anti-Cdh1 antibody was kindly provided by Kristian Helin (European Institute of Oncology, Milan, Italy) (36). Anti-β-tubulin (TUB2.1), anti-FLAG (M2), and anti-Cdc27 (AF3.1) antibodies were obtained from Sigma Chemicals (St. Louis, Mo.), and anti-cyclin B1 (GNS1) and anti-Myc (9E10) antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, Calif.). Anti-hemagglutinin (HA; 12CA5) and anti-green fluorescent protein (GFP) antibodies were purchased from Roche (Mannheim, Germany) and Clontech (Palo Alto, Calif.), respectively, and anti-Skp1 antibody was obtained from Transduction Laboratories (Los Angeles, Calif.). Nocodazole, phosphocreatine, creatine kinase, bovine ubiquitin, ubiquitin aldehyde, Polybrene, and lactacystin were obtained from Sigma Chemicals. Methyl-ubiquitin, G418, and N-acetyl-Leu-Leu-norleucinal (LLnL) were purchased from Calbiochem (San Diego, Calif.). Okadiac acid was purchased from Roche.
Construction of expression plasmids and site-directed mutagenesis.
pCDNA3.1-hTK1 was generated by insertion of the EcoRI-XhoI fragment of the pBSK-hTK1 (6) into plasmid pCDNA3.1 (Invitrogen). cDNA encoding hTK1, devoid of the initiation ATG codon, was amplified by PCR with pBSK-hTK1 as the template and subcloned into the EcoRI site of plasmid pCVM2FLAG (Sigma), producing pFLAG-hTK1. The first methionine of hTK1 was deleted to eliminate alternative initiation of FLAG-hTK1 translation from the start codon of hTK1 (8), thus ensuring only the FLAG-hTK1 polypeptide derived from pFLAG-hTK1 is expressed in the cells. The glutathione S-transferase (GST)-fused hTK1 constructs cloned into pGEX-3X expression vectors were created as described previously (6). Mutations of hTK1 were generated by using the Quick-Change site-directed mutagenesis kit (Stratagene) with specific mutated primers. The following amino acid changes were made: the KEN box motif at positions 203 to 205 were mutated into AEN, KAA, and AAA, producing K mt, EN mt, KEN mt, respectively. For construction of retrovectors of hTK1, the SacI-XhoI restriction fragment of FLAG-fused wild-type or KEN box-mutated hTK1 in the corresponding pFLAG-hTK1 plasmid was individually inserted into the retroviral S2 vector (provided by Lih-Hwa Hwang, National Taiwan University, Taipei, Taiwan, Republic of China). pHA-Cdc20, pHA-Cdc6, and pHA-Cdh1 were kindly provided by Kristian Helin (European Institute of Oncology, Milan, Italy) (36). Mouse E1 and Xenopus UbcX expression plasmids were kindly provided by Tim Hunt (ICRF Clare Hall Laboratory, South Mimms, United Kingdom) (53). DNA fragments covering amino acids (aa) 1 to 120 of Cdc20, 1 to 125 of Cdh1, and 1 to 452 of Cullin1 were amplified by PCR with pHA-Cdc20, pHA-Cdh1, and pFLAG-Cullin1 as the templates and were cloned into pCMV2FLAG plasmid to generate pFLAG-Cdc20(1-120), pFLAG-Cdh1(1-125), and pFLAG-Cullin1(1-452). pFLAG-Cullin1(full length) was a gift from Zhen-Qiang Pan (The Mount Sinai School of Medicine, New York, N.Y.) (52). pCS2+ Myc-Cdc20 and pCS2+ Myc-Cdh1 were kindly provided by Marc W. Kirschner (Harvard Medical School, Boston, Mass.) (51). Human cyclin B1 was amplified by PCR from HeLa cDNA and inserted into the BamHI site of plasmid pCDNA3.1, yielding pCDNA3.1-hcyclinB1. pCDNA3.1-p27 was a gift provided by Joan Massague (Howard Hughes Medical Institute, Seattle, Wash.) (39). His-tagged Cdh1 of pET28a-Fzr/Cdh1 (provided by Jan-Michael Peters, Research Institute of Molecular Biology, Vienna, Austria) (27) was subcloned into the pCDNA3.1 vector, yielding pCDNA3.1-His-Cdh1. The DNA sequence of each plasmid used for this study was confirmed by dideoxynucleotide sequencing.
Cell culture, synchronization, FACS analysis, and transfection.
LM-TK deficient (LM-TK−) and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) supplemented with 10% fetal bovine serum plus 100 μg of streptomycin per ml and 100 U of penicillin per ml (Life Technologies) at 37°C under 5% CO2. For G2/M arrest, nocodazole (Sigma) was added to subconfluent HeLa and LM-TK− cells at a final concentration of 0.5 μg/ml for 20 h. To obtain early G1-phase cells, mitotic phase-arrested cells were shaken off, washed with phosphate-buffered saline (PBS), and incubated in fresh medium for an additional 3 to 5 h. The synchronized cells were fixed in 70% (vol/vol) ethanol, and the cell cycle profile was examined by fluorescence-activated cell sorter (FACS) analysis with a Becton Dickinson FACScan flow cytometer and CellQuest software. For ectopic expression experiments, cells plated on a 60-mm-diameter dish were transiently transfected with a mixture containing 3 μg of DNA with 18 μg of Lipofectamine (Invitrogene, Life Technologies) according to the manufacturer's instructions.
Protein extraction, Western blotting, and immunoprecipitation.
Cell extracts were prepared and Western blotting was performed as described previously (6). Immunoprecipitations were performed as previous described (19). Equal amounts of protein in each cell lysate were incubated with anti-hTK1 antibodies at 4°C for 2 h, and the immunocomplexes were adsorbed onto protein A-Sepharose (Amersham Pharmacia), followed by being washed five times with lysis buffer and eluted with sample loading buffer at 95°C for 5 min.
RNA interference.
RNA interference was performed essentially as described previously (12, 13). Duplex small interfering RNA (siRNA) against Cdh1, corresponding to nucleotide sequence 5′-AATGAGAAGTCTCCCAGTCAG-3′, was kindly provided by Chi-Ying F. Huang (National Health Research Institute, Taipei, Taiwan, Republic of China). Transfection of siRNA was carried out with Oligofectamine (Invitrogene; Life Technologies) according to the manufacturer's instructions.
Expression and purification of recombinant proteins.
His-tagged mouse E1and Xenopus UbcX were expressed in Escherichia coli BL21 (DE3) and were purified under native condition with Ni-nitrilotriacetic acid (NTA) beads (Qiagen). His-tagged Cdh1 protein was purified from a stable His-Cdh1-expressing LM-TK− cell line, which was established by transfection with pCDNA3.1-His-Cdh1, followed by G418 selection. The wild type and three KEN box mutants of GST-hTK1 were expressed in E. coli JM109 and purified by glutathione 4B beads (Amersham Pharmacia) as described previously (5).
Pulse-chase experiment.
For metabolic labeling, cells were washed with PBS twice and incubated in 5 ml of methionine-free DMEM for 1 h of deprivation. Cells were then incubated in 2 ml of fresh methionine-free DMEM containing dialyzed 10% fetal bovine serum (FBS) and [35S]methionine (500 μCi; Amersham Pharmacia) for 30 min. To terminate labeling, complete DMEM was added for chasing prior to harvesting at the indicated time points. Immunoprecipitation of hTK1 was performed as described above, followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Retrovirus-mediated expression of Flag-tagged hTK1 in HeLa cells.
S2 retroviral vectors expressing Flag-tagged wild type or KEN box mutants of hTK1 were individually transfected to the packaging cell line PT67. Stable clones were selected in the medium supplemented with G418 antibiotic (400 μg/ml). Following propagation of each clone, viral supernatant from the individual culture was collected and stored at −80°C. To infect HeLa cells on a 6-cm-diameter dish, 3 ml of viral supernatant mixed with 5 μg of Polybrene per ml (Sigma) was incubated with cells for 48 h. Afterwards, HeLa cells were kept in the medium containing G418 for further selection. Each stable clone was propagated and confirmed by detection of FLAG-tagged hTK1 in the cell lysates by Western blotting with anti-hTK1 or anti-FLAG antibodies.
In vitro transcription and translation, in vitro binding assay, and GST pull-down assay.
An in vitro transcription and translation coupling system (TNT; Promega) using plasmids was employed to prepare [35S]methionine-labeled in vitro-translated proteins. For in vitro binding assay, the cold in vitro-translated proteins were prepared with unlabeled methionine. The in vitro binding assay was performed as described in previous studies (37). Cold in vitro-translated Myc-tagged proteins were bound to anti-Myc antibody-adsorbed beads. After the adsorption, 35S-labeled substrates were added to the binding buffer (50 mM HEPES [pH 7.7], 50 mM NaCl, 1 mM MgCl2, 1 mM EDTA, 1-mg/ml bovine serum albumin, 0.2% Tween 20). Following 2 h of incubation on the rotator at 4°C, samples were washed four times with the binding buffer and eluted with an SDS-PAGE sample loading buffer at 95°C for 5 min. GST pull-down assays were performed as described previously (19).
Preparation of cell extract for in vitro ubiquitinylation.
Early G1 cell extract was prepared as previously described (3). After washing twice with PBS and once with a hypotonic buffer (20 mM HEPES [pH 7.5], 5 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol [DTT]), cells were resuspended in a hypotonic buffer supplemented with a protease inhibitor cocktail (Sigma) and lysed by Dounce homogenization. After removal of cell debris by centrifugation at 2,000 × g, the supernatants were then recentrifuged at 14,000 × g at 4°C for 30 min to collect the clear cytoplasmic extracts, and aliquots of the extract were snap-frozen in liquid N2 and stored in 10% glycerol at −80°C.
Immunodepletion and purification of APC/C.
Immunodepletion of the SCF complex and APC/C from cell extracts by using anti-Skp1 and anti-Cdc27 antibodies, respectively, was performed as described previously (38, 48). Ten micrograms of antibody was preadsorbed to 15 μl of protein A-Sepharose at 4°C for 2 h and then mixed with 40 μl of cell extracts for 1 h. Beads were removed by high-speed centrifugation, and supernatants were collected for in vitro assays.
For purification of HeLa cells mitotic APC/C, mitotic cell extracts were prepared and immunoprecipitated with Cdc27 antibody covalently linked to protein A-Sepharose by dimethylpimelimidate (Sigma) as described previously (15, 50). The Cdc27 antibody-linked beads were incubated with 5 volumes of extracts for 2 h at 4°C and then removed by centrifugation. The beads were further washed three times with XB buffer (10 mM HEPES [pH 7.7], 1 mM MgCl2, 100 mM KCl, 1 mM DTT, 0.1 mM CaCl2, 0.2 μM okadaic acid) supplemented with 400 mM KCl and 0.5% NP-40 and four times with XB buffer. After removal of the residual washing buffer, the APC/C beads were used for in vitro ubiquitinylation experiments.
In vitro ubiquitinylation.
In vitro ubiquitinylation was carried out by incubating cell extracts with 1 μl of 35S-labeled hTK1 in a reaction mixture (20 μl) containing an energy regeneration system (2 mM ATP, 40 mM phosphocreatine, and 80-μg/ml creatine kinase), 1 μM ubiquitin aldehyde, and LLnL as described previously (30) at 30°C. The ubiquitinylated forms of hTK1 were analyzed by SDS-PAGE prior to autoradiography.
Experiments involving reconstitution of in vitro ubiquitinylation were performed as described previously (14). The APC/C beads were activated by incubation with in vitro-translated Cdc20 or Cdh1 for 1 h at room temperature. The activated APC/C beads were then washed four times with XB buffer. Each ubiquitinylation assay was performed in a total volume of 5 μl. The reaction mixture contained an energy regeneration system, 1 μM ubiquitin aldehyde, 1.25-mg/ml ubiquitin, 200-μg/ml mouse E1, 100-μg/ml Xenopus UbcX, 100 ng of substrate, and 3 μl of APC/C beads. All reaction mixtures were incubated at 37°C for 1 h, after which, the mixtures were analyzed by 5 to 15% polyacrylamide gradient SDS-PAGE and Western blotting.
RESULTS
Human TK1 is unstable at mitosis and degraded via a ubiquitin-proteasome pathway.
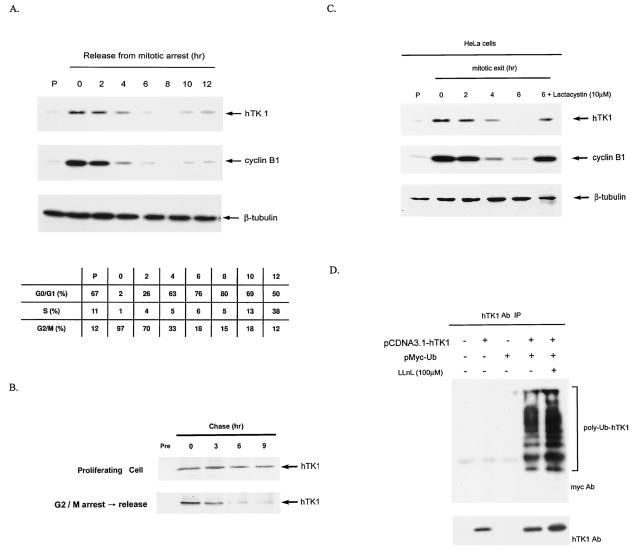
The fluctuation of endogenous hTK1 abundance during mitosis was examined in HeLa cells, which were synchronized by nocodazole to induce G2/M arrest and released into the next cell cycle. Compared with that in the asynchronous cells, the hTK1 levels were increased in G2/M-arrested cells, followed by a rapid degradation during release from the mitotic block (Fig. 1A). The FACS analysis showed that 76% of cells entered the G1 phase after being released from the mitotic block for 6 h, in which the expression levels of cyclin B1 were decreased in a way similar to that of hTK1. The levels of hTK1 and cyclin B1 were increased again as cells were released from G2/M block for 10 h and approached the next S phase. To ensure that the destabilization of hTK1 in mitotic exit phase is due to shorten of its half-life, we compared the half-lives of hTK1 in cells upon mitotic exit and in the proliferating state by using the pulse-chase experiment. As shown in Fig. 1B, the half-life of hTK1 was approximately 3 h in those cells released from G2/M arrest and was longer than 9 h in proliferating cells, confirming that a specific proteolytic mechanism in the mitotic exit phase confers the instability on hTK1 protein.
FIG. 1.
Mitotic degradation of hTK1 is via the ubiquitin-proteasomal pathway. (A) HeLa cells were arrested by nocodazole treatment for 20 h, followed by replenishment of fresh medium to release cells from the G2/M arrest. Extracts were prepared from HeLa cells released from G2/M arrest at the time points as indicated. The extract from asynchronized cells is indicated as the proliferating state (P). Expression of hTK1, cyclin B1, and β-tubulin was analyzed by Western blotting with anti-hTK1, anti-cyclin B1, and anti-β-tubulin antibodies, respectively. The cell cycle profile of corresponding cells was analyzed by flow cytometry as described in Materials and Methods and is expressed in the inset tables as percentages of cells in different phases. (B) HeLa cells of proliferating state or releasing from G2/M arrest were metabolically labeled with [35S]methionine (250 μCi/ml) for 30 min, followed by addition of fresh medium at the indicated time points prior to cell lysis. Cell extracts were immunoprecipitated by anti-hTK1 antibody, and the immunocomplexes were resolved by SDS-PAGE (12% polyacrylamide), followed by autoradiography. Normal rabbit immunoglobulin G (Pre) was used as a negative control. (C) Extracts were prepared from HeLa cells released from G2/M arrest. Cells were treated with lactacystin (10 μM) for 6 h during the mitotic exit phase. (D) LM-TK− cells were transfected with (+) or without (−) pCDNA3.1-hTK1 and pMyc-Ub as indicated. hTK1 was immunoprecipitated (IP) with anti-hTK1 antibody (Ab) and immunocomplexes were analyzed by SDS-PAGE and Western blotting with anti-hTK1 and anti-Myc antibodies.
To examine whether the degradation of hTK1 is dependent on proteasome activity, we treated HeLa cells with a specific proteasomal inhibitor, lactacystin, in the mitotic exit phase. Apparently, treatment of cells with lactacystin significantly inhibited hTK1 degradation (Fig. 1C, lane 6), with concomitant stabilization of cyclin B1. A similar observation could also be found in IMR-90 normal human diploid fibroblasts (data not shown). To further know whether hTK1 could be polyubiquitinylated in vivo, we then transfected cells with expression vectors of Myc-ubiquitin and hTK1 in TK-deficient mouse LM (LM-TK−) cells. Cell extracts were prepared for immunoprecipitation by hTK1 antibody, followed by Western blotting analysis with Myc antibody (Fig. 1D). The result clearly shows the existence of Myc-tagged polyubiquitinylated forms of hTK1 in the cells coexpressing hTK1 and Myc-ubiquitin. Treatment of these transfected cells with LLnL, another proteasome inhibitor, enhanced the extent of hTK1 ubiquitinylation. We also found that in HeLa cells, the extent of ubiquitinylation of endogenous hTK1 was increased following LLnL treatment (data not shown). These results provided clear evidence that the ubiquitin-protesome pathway controls the destruction of hTK1.
APC/C-Cdh1-dependent degradation of human TK1.
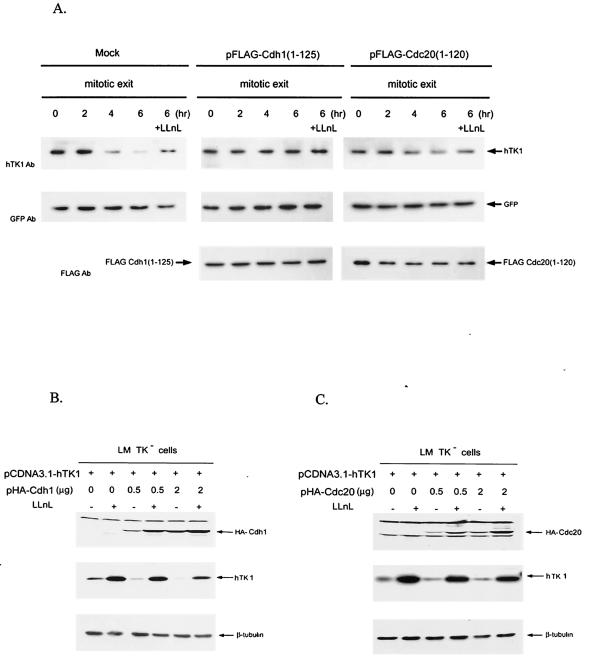
Because SCF and APC/C complexes are two major E3 ligases targeting substrates for cell cycle-dependent proteolysis, we then determined which of these two E3 complexes is necessary for mitotic degradation of hTK1. It has been shown that mutant proteins containing an amino-terminal 125 aa of Cdh1 and 120 aa of Cdc20 are potential dominant-negative mutants for APC/C-mediated protein degradation (37), and these mutants indeed inhibit the proteolysis of their corresponding APC/C substrates in vivo (51). On this basis, we transfected HeLa cells with the expression vector of a dominant-negative mutant of Cdc20 or Cdh1. Following transfection, cells were synchronized by nocodazole treatment with the subsequent release from mitotic block. We found that overexpression of the dominant-negative Cdh1 mutant clearly blocked the degradation of endogenous hTK1 during the mitotic exit phase, whereas the Cdc20 mutant had little effect on hTK1 stabilization (Fig. 2A). All cells were cotransfected with pEGFP, whose expression levels under this experimental condition were used as an internal control to indicate similar transfection efficiency in different transfected cells. Since LLnL-sensitive degradation of hTK1 at mitosis was abrogated in those cells expressing the dominant-negative form of Cdh1, it is evident that the functional activity of APC/C-Cdh1 complex is necessary for proteasome-dependent hTK1 at mitosis.
FIG. 2.
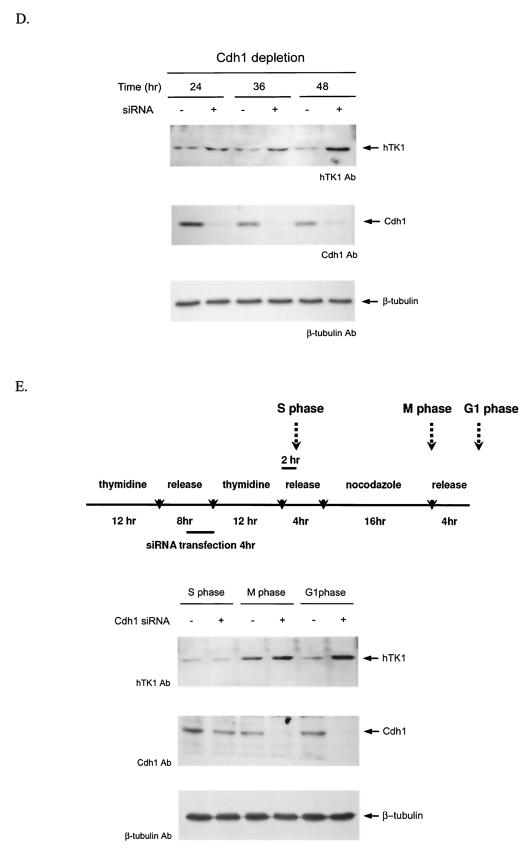
Mitotic degradation of hTK1 requires functional Cdh1. (A) HeLa cells were transfected with pFLAG-Cdc20(1-120), pFLAG-Cdh1(1-125) or empty vector (Mock) together with pEGFP as an internal control. Following transfection for 24 h, cells were synchronized as described in the legend to Fig. 1. One set of cells during release from the mitotic arrest was treated with LLnL (100 μM) for 6 h. Expression of hTK1, Cdc20(1-120), Cdh1(1-125), and GFP was analyzed by Western blotting with specific antibodies (Ab). (B) LM TK− cells were transfected with 1 μg of pCDNA3.1-hTK1 and different amounts of pHA-Cdh1 as indicated. Empty vector was added to have a final 3 μg of total DNA for transfection. Cells were treated without (−) or with (+) LLnL (100 μM) for 6 h before harvesting. HA-Cdh1, hTK1, and β-tubulin were detected by Western blot analysis. (C) LM-TK− cells transfected with pCDNA3.1-hTK1 and different amounts of pHA-Cdc20 as indicated were analyzed as described above. (D) HeLa cells were transfected without (−) or with (+) 0.8 μg of duplex siRNA against Cdh1 as described in Materials and Methods. After transfection at different time points, cells were extracted for Western blot analysis. (E) Scheme of the synchronization procedures for Cdh1 siRNA transfection experiment in HeLa cells (upper panel). Cell extracts were prepared after transfection at different phases as indicated and analyzed as described above.
To substantiate the role of Cdh1 in regulating the expressed level of hTK1 in mammalian cells, we next transfected expression vectors of hTK1 with the wild type of Cdh1 or Cdc20 into LM-TK− cells. Increasing amounts of Cdh1 expression vector in transfection significantly diminished the ectopic expression of hTK1, while treatment of transfected cells with LLnL restored the expressed level of hTK1 (Fig. 2B). To the contrary, the expressed level of hTK1 remained rather constant regardless of increasing amounts of Cdc20 expression (Fig. 2C). These results suggested that Cdh1, but not Cdc20, acts as a rate-limiting factor for regulating proteasome-dependent degradation of hTK1.
We then further tested the effect of depletion of endogenous Cdh1 on degradation of hTK1 in asynchronous and G1-phase cells by duplex siRNA. As shown in Fig. 2D, transfection with duplex siRNA against Cdh1 resulted in reduction of endogenous Cdh1 protein expression, which was accompanied with increasing accumulation of hTK1 after 24 to 48 h of transfection. To examine the effect of Cdh1 depletion on degradation of hTK1 in the G1 phase, HeLa cells were arrested in the G1/S phase by thymidine block and released for cell cycle progression. Four hours before the second round of thymidine block, cells were transfected with duplex siRNA against Cdh1. After double-thymidine blocks, cells were synchronized at M phase by nocodazole treatment and released to G1 progression as shown in Fig. 2E (upper panel). As expected, endogenous Cdh1 was significantly depleted in the cells with siRNA transfection at the M and G1 phases (Fig. 2E, lower panel). The expressed levels of hTK1 were increased at M phase regardless of siRNA transfection. In contrast, following G1 progression, depletion of endogenous Cdh1 clearly caused accumulation of hTK1 in the G1-phase cells (Fig. 2E). Thus, knockdown of Cdh1 prohibited hTK1 degradation in the G1 phase, resulting in accumulation of hTK1.
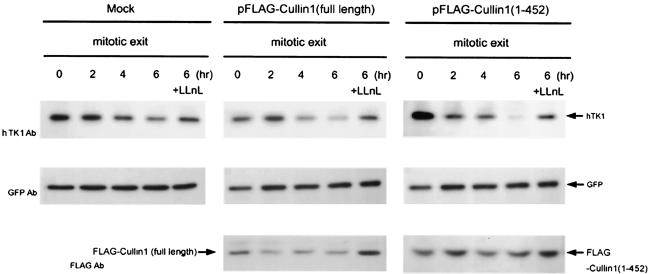
In the interest of determining whether the SCF complex is involved in hTK1 degradation in mammalian cells, we expressed Cul1(1-452), which lacks the Roc/UBC binding region while retaining its ability to bind to F box protein, as a dominant-negative mutant in cells to interfere with SCF-mediated degradation (12, 52). As shown in Fig. 3, endogenous hTK1 degradation in mitotic HeLa cells was unaffected by expressing dominant-negative mutant Cul1(1-452), while p27 is accumulated in response to this functional disruption (data not shown). Overexpression of full-length Cullin1 did not affect the expressed level of endogenous hTK1 either. Therefore, it is unlikely that the SCF complex plays a determining role in mitotic degradation of hTK1 in mammalian cells. Rather, the APC/C-Cdh1-mediated pathway is indeed necessary for controlling hTK1 degradation.
FIG. 3.
The function of SCF is not essential for mitotic degradation of hTK1. HeLa cells were transfected with pFLAG-Cullin1(full length), pFLAG-Cullin1(1-452), or empty vector (Mock) together with pEGFP as an internal control. Cell synchronization, LLnL treatment, and analysis of protein abundance were performed as described in the legend to Fig. 2.
The intact KEN box sequence is required for APC/C-Cdh1-mediated hTK1 degradation.
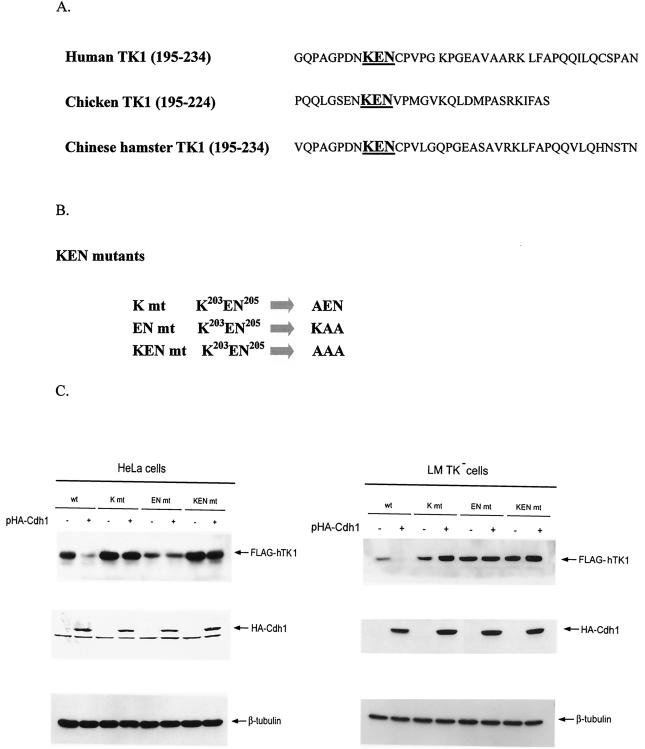
By sequence analysis, we found a putative KEN box is located at the C-terminal region of hTK1, and this KEN box motif is highly conserved in the TK1 sequence of human, chicken, and Chinese hamster cells (Fig. 4A). To test whether this KEN box motif could be crucial for targeting hTK1 for proteolysis, mutants carrying mutations with 1, 2, or 3 aa to alanine in this KEN box motif were generated as shown in Fig. 4B. We then analyzed whether expression levels of these mutants are responsive to Cdh1 overexpression. It appeared that the expressed levels of these three KEN box mutants are independent of Cdh1 overexpression in HeLa and LM-TK− cells (Fig. 4C). In contrast, the wild-type hTK1 is markedly decreased in cells overexpressing full-length Cdh1, which is consistent with the results shown in Fig. 2B. Thus, the KEN box at the C-terminal region of hTK1 serves as a degradation signal recognized by the APC/C activator, Cdh1.
FIG. 4.
Cdh1-dependent degradation of hTK1 requires the KEN box motif located at the C-terminal region. (A) Sequence alignment of the C-terminal region covering aa 195 to 234 of hTK1 with the corresponding regions of other species. The KEN box is indicated in boldface. (B) Schematic representation of mutants carrying a mutation in the KEN box sequence. (C) Extracts were prepared from HeLa cells (left panel) and LM-TK− cells (right panel) transfected with pFLAG-hTK1 (wild type [wt]) or KEN box mutants (K mt, EN mt, and KEN mt) with (+) or without (−) pHA-Cdh1, followed by SDS-PAGE separation and Western blot analysis as described in the legend to Fig. 2B.
KEN box mutants of hTK1 are resistant to degradation in the mitotic exit phase.
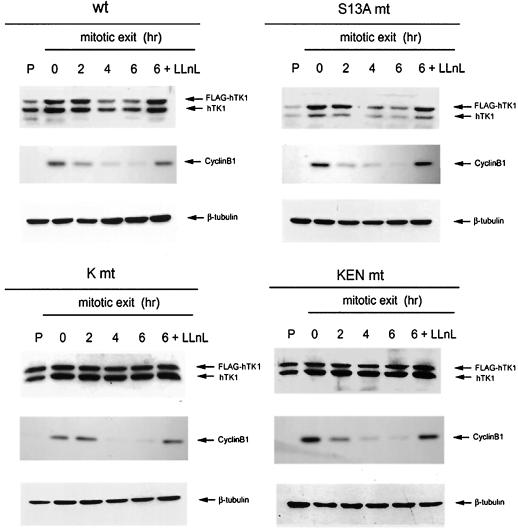
To ascertain whether the KEN box is involved in degradation of hTK1 at the time of mitotic exit, we established HeLa cell lines stably expressing various FLAG-tagged hTK1 mutants by retrovirus infection followed by clonal selection. Using this expression system in HeLa cells, we were able to examine mitotic degradation of FLAG-hTK1 in each stable cell line at an expressed level comparable with that of endogenous hTK1. As shown in Fig. 5, wild-type FLAG-hTK1 expressed in the stable cell line displayed a similar degradation pattern to endogenous hTK1 during the mitotic exit phase in a proteasome-dependent manner. A mutation converting serine 13 to alanine did not cause stabilization of hTK1 (Fig. 5, right upper panel), suggesting phosphorylation of serine 13 is not a requisite signal for hTK1 mitotic degradation in mammalian cells. In contrast, the levels of expression of FLAG-tagged K mt and KEN mt were rather stable in these stable cell lines under the same experimental conditions. Because of the increase in stability, an elevated expression level was also observed in these two cell lines, compared to that in the cells expressing wild-type FLAG-hTK1. Particularly, in terms of proliferation status, the levels of K mt and KEN mt of FLAG-hTK1 were higher than that of the wild type. However, it should be noted that endogenous hTK1 also became stabilized in the cells expressing hTK1 KEN box mutants. Because cyclin B1 got rapidly degraded in these cells, it is unlikely that lack of FLAG-hTK1 mutants and endogenous hTK1 degradation is due to the impairment of APC/C function in these cells. Since it is well established that TK1 forms dimer or tetramer (31, 40, 43), we speculated that multimerization between FLAG-KEN box mutant hTK1 and endogenous hTK1 might affect the efficiency of degradation for the endogenous counterparts. If this is true, it is doubtful whether the S13A mutation could really effectively abrogate the phosphorylation signal in the dimer or tetramer form of FLAG-hTK1 (S13A) containing endogenous hTK1 subunits. Nevertheless, these results still support the idea that KEN box is a necessary signal for mitotic degradation of hTK1.
FIG. 5.
The KEN box is a necessary signal for mitotic degradation of hTK1. Stable cell lines expressing the wild type (wt), Ser13A-mutated (S13A mt), and KEN box-mutated (K mt and KEN mt) FLAG-hTK1 were prepared as described in Materials and Methods. Mitotic arrest of cells and analysis of protein abundance were performed as described in the legend to Fig. 1A.
hTK1 interacts with Cdh1 in vivo and in vitro.
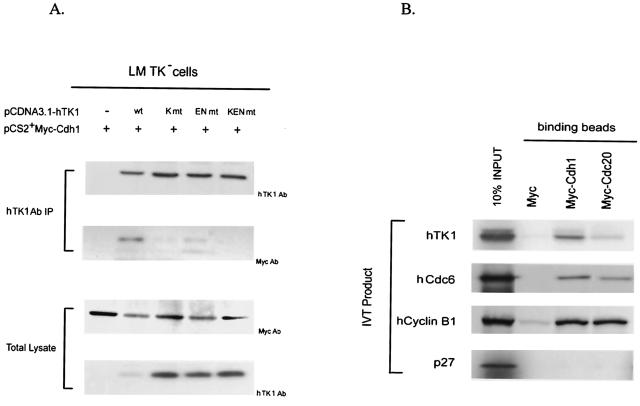
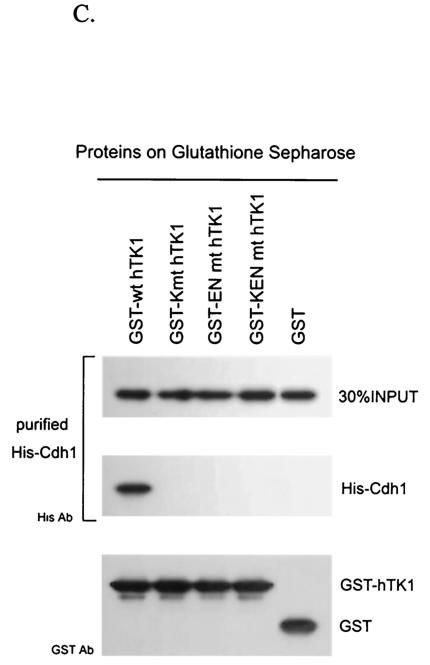
To know whether Cdh1 interacts with hTK1 in vivo, we transfected pCS2+Myc-Cdh1 with an expression vector of either wild-type or KEN box mutant hTK1 into LM TK− cells. After preincubation of transfected cells with LLnL for 6 h, the interactions between Cdh1 and various forms of hTK1 were examined by coimmunoprecipitation with hTK1 antibody. As shown in Fig. 6A, Myc-Cdh1 was clearly detected in the hTK1 (wild type) immunocomplex, but not in three mutated hTK1 precipitants (Fig. 6A, upper panel). Furthermore, endogenous hTK1 in HeLa cells also exhibited interaction with ectopically expressed Myc-Cdh1 (data not shown). To confirm the specificity of interaction between hTK1 and Cdh1, we incubated in vitro-translated Myc-Cdc20 or Myc-Cdh1 bound to Myc antibody-adsorbed beads with 35S-labeled in vitro-translated hTK1 (wild type). As shown in Fig. 6B, hTK1, human cyclin B1, and human Cdc6 (hCdc6) all could be pulled down by Myc-Cdh1 beads, but not by the control beads. Myc-Cdc20 could also pull down significant amounts of human cyclin B1 and hCdc6, but only a small amount of hTK1 protein, indicating a weak interaction between Cdc20 and hTK1. As a negative control, p27 protein did not exhibit interaction with either Cdh1 or Cdc20. To determine the effect of KEN box mutation on the interaction between hTK1 and Cdh1 in vitro, we compared the binding capacities of GST fusion proteins of hTK1 (wild type) and three KEN box mutants to His-Cdh1 purified from a cell line stably expressing His-Cdh1 by GST pull-down assay. The results clearly revealed that His-Cdh1 beads only pulled down hTK1 (wild type) protein, but not the KEN box mutants (Fig. 6C). Altogether, data from these in vivo and in vitro experiments conclusively indicated that hTK1 directly interacts with Cdh1 via its KEN box signaling motif.
FIG. 6.
hTK1 interacts with Cdh1 via the KEN box. (A) LM-TK− cells were transfected with pCDNA3.1-hTK1 of the wild type (wt) or KEN box mutants (K mt, EN mt, and KEN mt) together with pCS2+Myc-Cdh1. After 24 h, cells were treated with LLnL (100 μM) for 6 h before harvesting. hTK1 was immunoprecipitated as described in the legend to Fig. 1D and analyzed by Western blotting with specific antibodies. (B) In vitro-translated 35S-labeled hTK1, hCdc6, human cyclin B1, and p27 were individually incubated with anti-Myc antibody-coated beads containing the unlabeled in vitro-translated Myc-Cdh1, Myc-Cdc20, or Myc. The left lane indicates 10% of the 35S-labeled in vitro-translated product in each binding assay. (C) Purified His-Cdh1 proteins were subjected into GST pull-down assays using glutathione-Sepharose beads bound with GST, GST-hTK1 (wild type), and three KEN box mutants as described in Materials and Methods. The pull-down samples were analyzed by SDS-PAGE and Western blotting with specific antibodies.
In vitro polyubiquitinylation of hTK1 requires APC/C-Cdh1 complex via a KEN box.
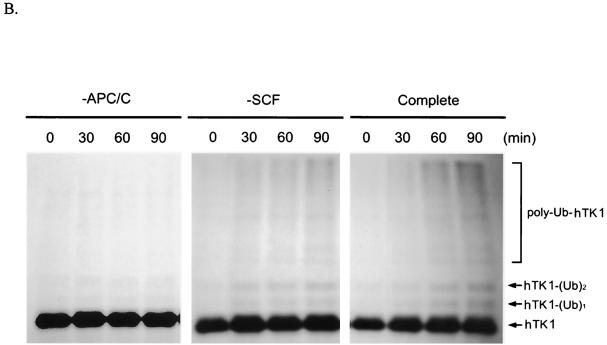
In vitro polyubiquitinylation assays were used to demonstrate whether the APC/C complex is a direct E3 for hTK1. For this purpose, we prepared early G1 extracts from LM-TK− cells and used specific antibodies against Cdc27 and Skp1 to individually deplete APC/C and SCF complex, respectively, from the extracts as described previously (40, 50). The success of depletion was confirmed by Western blot analysis of Cdc27 and Skp1 in these depleted extracts (Fig. 7A). With these extracts, we carried out an in vitro polyubiquitinylation reaction using 35S-labeled in vitro-translated hTK1 (wild type) as a substrate in the presence of LLnL and ubiquitin-aldehyde, a known inhibitor of deubiquitinylation. As shown in Fig. 7B, the complete and SCF-depleted cell extracts were able to catalyze the formation of mono-, di-, and polyubiquitinylated forms of hTK1 in a time-dependent manner. In contrast, no ubiquitinylated form of hTK1 was seen in the reaction using APC/C-depleted extract. Because addition of methyl-ubiquitin (Me-Ub), the specific ubiquitin competitor, to the complete extract decreased the formation of polyubiquitinylated hTK1 (data not shown), we knew that the accumulated high-molecular-weight forms of hTK1 are derived from polyubiquitinylation.
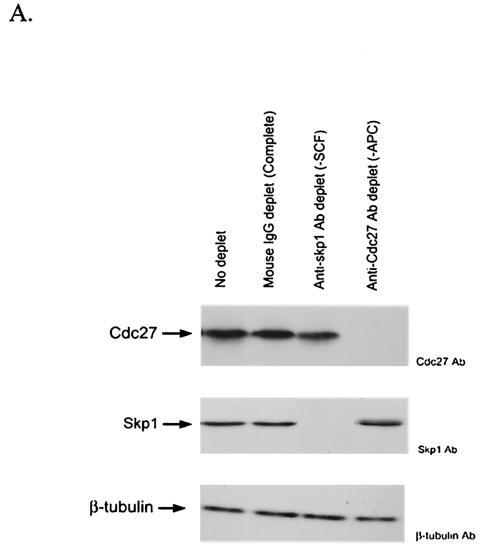
FIG. 7.
APC/C-Cdh1 is the direct ubiquitin E3 ligase confers polyubiquitinylation of hTK1. (A) The early G1-phase extracts after depletion by control mouse immunoglobulin G (IgG), Cdc27, and Skp1 antibodies as described in Materials and Methods (representing complete, APC/C-negative, and SCF-negative extract, respectively) were subjected to Western blotting analysis with anti-Cdc27, anti-Skp1, and anti-β-tubulin antibodies. (B) An in vitro ubiquitinylation reaction was carried out with in vitro-translated 35S-labeled hTK1 using the complete, SCF-depleted (−SCF), or APC-depleted (−APC) extracts for the time indicated. The reaction products were analyzed by SDS-PAGE (9% polyacrylamide) prior to autoradiography. The hTK1-(Ub)1, hTK1-(Ub)2, and poly-Ub-hTK1 indicate the mono-, di-, and polyubiquitinylation of hTK1. (C) Reconstitution of in vitro ubiquitinylation of hTK1 was performed with His-hTK1 (wild type) or KEN mutant as a substrate in each reaction mixture containing E1, E2 (UbcX), APC/C, and Cdc20 or Cdh1 as indicated. The samples were analyzed by SDS-PAGE (5 to 15% polyacrylamide) and Western blotting with anti-hTK1 antibody.
To further demonstrate whether APC/C-Cdh1 is the direct E3 ligase responsible for polyubiquitinylation of hTK1, we used a reconstitution system containing purified APC/C, mouse E1, Xenopus UbcX, and in vitro-translated Cdc20 or Cdh1 to examine ubiquitinylation activity of His-hTK1. As shown in Fig. 7C, wild-type His-hTK1 can be polyubiquitinylated in a reaction mixture containing complete enzyme components and the activator Cdh1. No ubiquitinylated ladders occurred if APC/C or Cdh1 was omitted in the reaction (lane 1 and 2), and complex containing Cdc20 did not support polyubiquitinylation of His-hTK1. Mutation of the KEN box abolished in vitro polyubiquitinylation of His-hTK1 by the reconstituted complex (lane 5). A similar observation can also be seen when human E2C/UbcH10 was used as the E2 ligase for the reconstitution experiment (data not shown). Thus, these results clearly demonstrated that the APC/C-Cdh1 is the ubiquitin E3 ligase of hTK1 by recognizing the KEN box in mammalian cells.
DISCUSSION
In this study, we provided the first evidence that hTK1 is ubiquitinylated and degraded via the proteasome pathway in mammalian cells, and rapid degradation of hTK1 during mitotic exit is dependent on the ubiquitin E3 ligase activity of the APC/C through binding to the KEN box motif. In particular, we found that Cdh1 acts as a rate-limiting and necessary factor for regulating mitotic degradation of hTK1. Because hTK1 is capable of interacting with Cdh1 via the KEN box, we concluded that hTK1 is recognized by APC/C-Cdh1 complex for its mitotic degradation.
It has been previously shown that deletion of the carboxyl-terminal 40 aa of hTK1 completely abolishes cell cycle regulation and stabilizes the protein throughout the cell cycle (25). The KEN box identified in this study is located in this C-terminal region, thus explaining how this region confers the degradation signal. By comparing the amino acid sequences of cytosolic TK1 among different eukaryotic cells, we found a KEN box or KEN-like box is present in C-terminal region of chicken, Chinese hamster, and mouse TK1. Notably, the vaccinia virus and related viral TK proteins are highly homologous to cytosolic hTK1, but lack this KEN box in their C-terminal regions. Therefore, it is logical to assume that absence of the KEN box signal in the C-terminal region of viral TK may provide a mechanism allowing its escape from the cell cycle-dependent degradation in the host.
Unlike mammalian cells, both budding and fission yeasts lack their cytosolic TK1. Our laboratory has previously used the inducible expression systems in yeast to study the degradation mechanism of hTK1 (26). Through this approach, we demonstrated that degradation of hTK1 is impaired in Saccharomyces cerevisiae carrying a temperature-sensitive mutation in proteasomal gene pre1-1 or the SCF complex gene cdc34 or cdc53, while mutation in cdc16, an essential component of APC/C in yeast, does not affect hTK1 proteolysis. In that study, we also found that mutation of serine 13 to alanine, which disrupts its phosphorylation, stabilizes the expression of hTK1 in budding and fission yeasts. These results suggested that the SCF-mediated pathway contributes to hTK1 degradation via the phosphorylation signal in the yeast system. However, in this study, we found that expression of a dominant-negative form of Cullin1 did not result in accumulation of hTK1 in the mitotic cells (Fig. 3), indicating that in mammalian cells, the SCF is not essential for targeting mitotic degradation of hTK1. This situation is similar to the case of Cdc6, in which degradation is via the SCF complex in yeast, yet requires the APC/C-Cdh1 complex in human cells (33, 36). In addition, we found that there is no difference in the stabilities of the wild-type and mutant form (S13A) of FLAG-tagged-TK1 at mitosis in the stable cell lines (Fig. 5), suggesting that serine 13 phosphorylation is probably not a necessary signal for its mitotic degradation. Previously, Littlepage and Ruderman (28) showed that mutation of serine 53 to aspartic acid, which mimics the phosphorylation state, indeed abrogates Aurora-A kinase degradation in the mitotic exit phase, indicating dephosphorylation could control timing of its destruction. In the case of hTK1, we found that conversion of serine 13 to either alanine or aspartic acid did not have any effect on the level of hTK1 expressed in response to Cdh1 overexpression (data not shown), suggesting that neither phosphorylation nor dephosphorylation of serine 13 is crucial in Cdh1-mediated degradation of hTK1. At this point, we have speculated that yeasts, lacking endogenous TK, apparently do not need TK to supply dTTP during DNA replication; therefore, it is possible that the ectopically expressed hTK1 is readily phosphorylated in yeast and is targeted for degradation by a not-yet-identified F box protein to avoid the unnecessary synthesis of dTTP prior to the onset of activation of APC/C-Cdh1 complex. After all, it remains to be seen whether this phosphorylation-dependent degradation pathway for hTK1 does exist in mammalian cells under other physiological conditions, such as inhibition of DNA synthesis in the S phase.
It has been shown that the dTTP pool in S-phase cells is 20 times larger than that in G0-phase cells, which coincides with the level of expression of TK1 (44). Presumably, this fluctuation is important for maintaining genome stability, since reports have shown that elevation of dTTP concentration increases in vitro and in vivo mutation rates (10, 29). Extending from these observations, it is noteworthy that dTTP has been identified as an allosteric effector for ribonucleotide reductase, directing its substrate specificity (7). An abnormal increase in the dTTP level has been shown to promote the catalytic efficiency of ribonucleotide reductase for converting rGDP to dGDP, which can result in increase of the dGTP pool (41). Given that a larger dGTP pool is mutagenic, it is proposed that a balance of deoxynucleoside triphosphate (dNTP) pools is important for the fidelity of DNA repair and replication. Apparently, restricting dTTP level in cells by TK1 degradation is a way of controlling the balance of dNTP pools in the G1 phase. While this article was in preparation, a report showed that mouse ribonucleotide reductase R2 (mR2) protein is targeted to the APC/C-Cdh1 complex through KEN box-mediated binding and gets degraded in late mitosis (4). Considering the coordination between the supply of dTTP and regulation of ribonucleotide reductase activity in providing the correct pool of dNTP, information gained from these two reports together suggests that during mitotic exit, a common proteolytic controller, APC/C-Cdh1, is utilized to degrade two dNTP pool regulators, mR2 and TK1, by which an unwanted supply of dNTP can be stopped concurrently at entry into the G1 phase.
Acknowledgments
We thank Kristian Helin for kindly providing HA-tagged Cdc20, Cdh1, and Cdc6 plasmids and anti-Cdh1 antibody and Marc W. Kirschner for providing Myc-tagged Cdc20 and Cdh1 constructs. We are grateful to Zhen-Qiang Pan, Joan Massague, Lih-Hwa Hwang, and Chi-Ying F. Huang for providing Cullin1, p27 expression plasmids, retroviral S2 vector, and siRNA against Cdh1, respectively. We acknowledge Tim Hunt, Joan V. Ruderman, and Jan-Michael Peters for providing mouse E1, Xenopus UbcX, wild-type and C114S mutant human E2C/UbcH10, and His-tagged Cdc20 and Cdh1 expression plasmids. We greatly appreciate Jane Kirk, Michael Brandeis, Michele Pagano, and Kathleen Gould for helpful suggestions in immunodepletion and in vitro ubiquitinylation experiments.
This study was supported by grants NSC90-2320-B-002-065 and NSC 92-3112-B-002-016 from the National Science Council, Taiwan, Republic of China.
REFERENCES
- 1.Bello, L. J. 1974. Regulation of thymidine kinase synthesis in human cells. Exp. Cell Res. 89:263-274. [DOI] [PubMed] [Google Scholar]
- 2.Bradshaw, H. D., Jr., and P. L. Deininger. 1984. Human thymidine kinase gene: molecular cloning and nucleotide sequence of a cDNA expressible in mammalian cells. Mol. Cell. Biol. 4:2316-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandeis, M., and T. Hunt. 1996. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 15:5280-5289. [PMC free article] [PubMed] [Google Scholar]
- 4.Chabes, A. L., C. M. Pfleger, M. W. Kirschner, and L. Thelander. 2003. Mouse ribonuclotide reductase R2 protein: a new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc. Natl. Acad. Sci. USA 100:3925-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, Z. F., D. Y. Huang, and N. C. Hsue. 1994. Differential phosphorylation of human thymidine kinase in proliferating and M phase-arrested human cells. J. Biol. Chem. 269:21249-21254. [PubMed] [Google Scholar]
- 6.Chang, Z. F., D. Y. Huang, and L. M. Chi. 1998. Serine 13 is the site of mitotic phosphorylation of human thymidine kinase. J. Biol. Chem. 273:12095-12100. [DOI] [PubMed] [Google Scholar]
- 7.Chimploy, K., and C. K. Mathews. 2001. Mouse ribonucleotide reductase control: influence of substrate binding upon interaction with allosteric effectos. J. Biol. Chem. 276:7093-7100. [DOI] [PubMed] [Google Scholar]
- 8.Chou, W. L., and Z. F. Chang. 2001. Cap-independent translation conferred by the 5′-untranslated region of human thymidine kinase mRNA. Biochim. Biophys. Acta 1519:209-215. [DOI] [PubMed] [Google Scholar]
- 9.Coppock, D. L., and A. B. Pardee. 1987. Control of thymidine kinase mRNA during the cell cycle. Mol. Cell. Biol. 7:2925-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dare, E., L. H. Zhang, D. Jenssen, and V. Bianchi. 1995. Molecular analysis of mutations in hprt gene of V79 hamster fibroblasts: effects of imbalances in the dCTP, dGTP, and dTTP pools. J. Mol. Biol. 252:514-521. [DOI] [PubMed] [Google Scholar]
- 11.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 12.Donzelli, M., M. Squatrito, D. Ganoth, A. Hershko, M. Pagano, and G. F. Draetta. 2002. Dual mode of degradation of Cdc 25A phosphatase. EMBO J. 21:4875-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 14.Fang, G., H. Yu, and M. W. Kirschner. 1998. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 12:1871-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual, p. 521-523. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 16.Harper, J. W. 2002. A phosphorylation-driven ubiquitination switch for cell-cycle control. Trends Cell Biol. 12:104-107. [DOI] [PubMed] [Google Scholar]
- 17.Harper, J. W., J. L. Burton, and M. J. Solomon. 2002. The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 16:2179-2206. [DOI] [PubMed] [Google Scholar]
- 18.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 19.Huang, D. Y., and Z. F. Chang. 2001. Interaction of human thymidine kinase 1 with p21Waf1. Biochem. J. 356:829-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, J. N., I. Park, E. Ellingson, L. E. Littlepage, and D. Pellman. 2001. Activity of the APCCdh1 form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc20p. J. Cell Biol. 154:85-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irniger, S. 2002. Cyclin destruction in mitosis: a crucial task of Cdc20. FEBS Lett. 532:7-11. [DOI] [PubMed] [Google Scholar]
- 22.Jackson, P. K., A. G. Eldridge, E. Freed, L. Furstenthal, J. Y. Hsu, B. K. Kaiser, and J. D. Reimann 2000. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10:429-439. [DOI] [PubMed] [Google Scholar]
- 23.Johansson, M., and A. Karlsson. 1997. Cloning of the cDNA and chromosome localization of the gene of human thymidine kinase2. J. Biol. Chem. 272:8454-8458. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, L. F., L. G. Rao, and A. J. Muench. 1982. Regulation of thymidine kinase enzyme level in serum-stimulated mouse 3T6 fibroblasts. Exp. Cell Res. 138:79-85. [DOI] [PubMed] [Google Scholar]
- 25.Kauffman, M. G., and T. J. Kelly. 1991. Cell cycle regulation of thymidine kinase: residues near the carboxyl terminus are essential for the specific degradation of the enzyme at mitosis. Mol. Cell. Biol. 11:2538-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ke, P. Y., C. C. Yang, I. C. Tsai, and Z. F. Chang. 2003. Degradation of human thymidine kinase is dependent on serine-13 phosphorylation: involvement of the SCF-mediated pathway. Biochem. J. 370:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer, E. R., C. Geiffers, G. Holzl, M. Hengstschlager, and J. M. Peters. 1998. Activation of the human anaphase-promoting complex by proteins of the CDC20/Fizzy family. Curr. Biol. 8:1207-1210. [DOI] [PubMed] [Google Scholar]
- 28.Littlepage, L. E., and J. V. Ruderman. 2002. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 16:2274-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martomo, S., and C. K. Methaws. 2002. Effects of biological DNA precursor pool asymmetry upon accuracy of DNA replication in vitro. Mutat. Res. 499:197-211. [DOI] [PubMed] [Google Scholar]
- 30.Montagnoli, A., F. Fiore, E. Eytan, A. C. Carrano, G. F. Draetta, A. Hershko, and M. Pagano. 1999. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes Dev. 13:1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munch-Petersen, B., G. Tyrsted, and L. Cloos. 1993. Reversible ATP-dependent transition between two forms of human cytosolic thymidine kinase with different enzymatic properties. J. Biol. Chem. 268:15621-15625. [PubMed] [Google Scholar]
- 32.Page, A. M., and P. Hieter. 1999. The anaphase-promoting complex: new subunits and regulators. Annu. Rev. Biochem. 68:583-609. [DOI] [PubMed] [Google Scholar]
- 33.Perkins, G., L. S. Drury, and J. F. Diffley. 2001. Separate SCFCDC4 recognition elements target Cdc6 for proteolysis in S phase and mitosis. EMBO J. 20:4836-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters, J. M. 1998. SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr. Opin. Cell Biol. 10:759-768. [DOI] [PubMed] [Google Scholar]
- 35.Peters, J. M. 2002. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell 9:931-943. [DOI] [PubMed] [Google Scholar]
- 36.Petersen, B. O., C. Wagener, F. Marinoni, E. R. Kramer, M. Melixetian, E. L. Denchi, C. Gieffers, C. Matteucci, J. M. Peters, and K. Helin. 2000. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 14:2330-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfleger, C. M., and M. W. Kirschner. 2000. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 14:655-665. [PMC free article] [PubMed] [Google Scholar]
- 38.Pfleger, C. M., E. Lee, and M. W. Kirschner. 2001. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 15:2396-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polyak, K., M. H. Lee, H. E. Bromage, A. Koff, J. M. Roberts, P. Tempst, and J. Massague. 1994. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 78:59-66. [DOI] [PubMed] [Google Scholar]
- 40.Posch, M., C. Hauser, and C. Seiser. 2000. Substrate binding is a prerequisite for stabilisation of mouse thymidine kinase in proliferating fibroblasts. J. Mol. Biol. 300:493-502. [DOI] [PubMed] [Google Scholar]
- 41.Reichard, P., R. Eliasson, R. Ingemarson, and L. Thelander. 2000. Cross-talk between the allosteric effector-binding sites in mouse ribonucleotide reductase. J. Biol. Chem. 275:33021-33026. [DOI] [PubMed] [Google Scholar]
- 42.Scheffner, M., U. Nuber, and J. M. Huibregtse. 1995. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 373:81-83. [DOI] [PubMed] [Google Scholar]
- 43.Sherley, J. L., and T. J. Kelly. 1988. Human cytosolic thymidine kinase. Purification and physical characterization of the enzyme from HeLa cells. J. Biol. Chem. 263:375-382. [PubMed] [Google Scholar]
- 44.Spyrou, G., and P. Reichard. 1988. Dynamic of the thymidine triphosphate pool during the cell cycle of synchronized 3T3 mouse fibroblast. Mutat. Res. 200:37-43. [DOI] [PubMed] [Google Scholar]
- 45.Stuart, P., M. Ito, C. Stewart, and S. E. Conrad. 1985. Induction of cellular thymidine kinase occurs at the mRNA level. Mol. Cell. Biol. 5:1490-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sutterluety, H., S. Bartl, J. Karlseder, E. Wintersberger, and C. Seiser. 1996. Carboxy-terminal residues of mouse thymidine kinase are essential for rapid degradation in quiescent cells. J. Mol. Biol. 259:383-392. [DOI] [PubMed] [Google Scholar]
- 47.Sutterluety, H., and C. Seiser. 1997. Thymidine inhibits the growth-arrest-specific degradation of thymidine kinase protein in transfected L fibroblasts. J. Mol. Biol. 265:153-160. [DOI] [PubMed] [Google Scholar]
- 48.Tsvetkov, L. M., K. H. Yeh, S. J. Lee, H. Sun, and H. Zhang. 1999. p27Kip ubiquitination and degradation is regulated by the SCFSkp2 complex through phosphorylated Thr187 in p27. Curr. Biol. 9:661-664. [DOI] [PubMed] [Google Scholar]
- 49.Visintin, R., S. Prinz, and A. Amon. 1997. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278:460-463. [DOI] [PubMed] [Google Scholar]
- 50.Vorlaufer, E., and J. M. Peters. 1998. Regulation of the cyclin B degradation system by an inhibitor of mitotic proteolysis. Mol. Biol. Cell 9:1817-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wan, Y., X. Liu, and M. W. Kirschner. 2001. The anaphase-promoting complex mediates TGF-beta signaling by targeting SnoN for destruction. Mol. Cell 8:1027-1039. [DOI] [PubMed] [Google Scholar]
- 52.Wu, K., S. Y. Fuchs, A. Chen, P. Tan, C. Gomez, Z. Ronai, and Z. Q. Pan. 2000. The SCFHOS/β-TRCP-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol. Cell. Biol. 20:1382-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamano, H., C. Tsurumi, J. Gannon, and T. Hunt. 1998. The role of the destruction box and its neighbouring lysine residues in cyclin B for anaphase ubiquitin-dependent proteolysis in fission yeast: defining the D-box receptor. EMBO J. 17:5670-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zur, A., and M. Brandeis. 2000. Securin degradation is mediated by fzy and fzr, and is required for complete chromatid separation but for cytokinesis. EMBO J. 20:792-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zur, A., and M. Brandeis. 2002. Timing of APC/C substrate degradation is determined by fzy/fzr specificity of destruction boxes. EMBO J. 21:4500-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]