Summary
Nasal embryonic LHRH factor (NELF) has been hypothesized to participate in the migration of GnRH and olfactory neurons into the forebrain, a prerequisite for normal hypothalamic-pituitary-gonadal function in puberty and reproduction. However, the biological functions of NELF, which has no homology to any human protein, remain largely elusive. Although mRNA expression did not differ, NELF protein expression was greater in migratory than postmigratory GnRH neurons. Pituitary Nelf mRNA expression was also observed and increased three-fold after exogenous GnRH administration. Contrary to a previous report, NELF displayed predominant nuclear localization in GnRH neurons, confirmed by mutagenesis of a putative nuclear localization signal resulting in impaired nuclear expression. NELF knockdown impaired GnRH neuronal migration of NLT cells in vitro. These findings and the identification of two putative zinc fingers suggest that NELF could be a transcription factor. Collectively, our findings implicate NELF as a nuclear protein involved in the developmental function of the reproductive axis.
Keywords: NELF, Nasal embryonic LHRH factor, GnRH neuron migration, Kallmann syndrome, hypogonadotropic hypogonadism
Introduction
Gonadotropin releasing hormone (GnRH) constitutes the pivotal reproductive peptide regulating the hypothalamic-pituitary-gonadal (HPG) axis in puberty and reproduction in mammals [Kramer and Wray 2000]. During embryologic development, GnRH neurons migrate along with olfactory neurons from the olfactory placode region outside the brain across the cribriform plate to enter the hypothalamus. GnRH is synthesized by a small number of GnRH neurons (~1,000-2,000 in humans; ~800-1,000 in mice) that are dispersed throughout the hypothalamus [Schwarting et al. 2007,Wierman et al. 2004]. The elucidation of factors that direct migration of GnRH neurons to their normal anatomic location is fundamental to understanding the initiation of mammalian puberty and normal reproduction.
Disruption of the GnRH neuron migratory pathway results in Kallmann syndrome (KS), which is characterized by impaired olfaction and GnRH deficiency [Kim et al. 2008a]. GnRH deficiency, also known as idiopathic hypogonadotropic hypogonadism (IHH), manifests as absent or incomplete pubertal development, low serum sex steroids, low serum gonadotropins, and subsequent infertility [Crowley et al. 1985]. Associated congenital anomalies include neurological abnormalities, midfacial defects, unilateral renal agenesis, and dental agenesis in some patients [Kim et al. 2008a]. There is sufficient evidence to implicate mutations of the KAL1 [Franco et al. 1991,Legouis et al. 1991], FGFR1 [Dode et al. 2003,Pitteloud et al. 2006,Tsai et al. 2005], and CHD7 [Kim et al. 2008b] genes with impaired GnRH and olfactory neuron migration in humans. Mutations in the FGF8 [Falardeau et al. 2008] and PROK2/PROKR2 [Dode et al. 2006,Pitteloud et al. 2007b] genes have also been identified in patients with IHH/KS.
Nasal embryonic LHRH factor (NELF) is an attractive candidate gene for a role in GnRH neuron migration, mammalian puberty, and the pathophysiology of KS. Mouse Nelf was originally cloned from a differential cDNA library screen of migratory versus nonmigratory GnRH neurons [Kramer and Wray 2000]. Expression was aligned along the plasma membrane of olfactory and GnRH neurons before they entered the hypothalamus, and levels were down-regulated when GnRH neurons reached the forebrain [Kramer and Wray 2001,Kramer and Wray 2000]. Antisense techniques reducing NELF protein expression resulted in a decreased number of GnRH neurons and of GnRH nerve fiber complexity and length [Kramer and Wray 2000]. Two previous reports [Miura et al. 2004,Pitteloud et al. 2007a] implicated NELF in KS, but bona fide evidence demonstrating human NELF mutations in monogenic KS was lacking until our recent description of human NELF mutations causing monogenic IHH/KS (Xu et al, revision submitted).
Since its characterization, there has been little advance in the biology of NELF, and the knockout mouse has not yet been published. The NELF protein lacks a classical N-terminal signal peptide and has no homology with any other known human protein. Further characterization of NELF’s subcellular localization and functional role in the GnRH neuronal migration pathway and the reproductive axis will impact our understanding of its biological function. Our findings demonstrate that NELF protein was more highly expressed in migratory vs. postmigratory GnRH neuronal cells. NELF knockdown dramatically impaired GnRH neuronal cell migration in vitro. A functional nuclear localization signal and two putative zinc fingers were identified, suggesting NELF is a nuclear protein, and could be a transcription factor. Somewhat surprisingly, pituitary Nelf expression was upregulated by hypothalamic GnRH administration, an observation which will need to be explored in future studies. Our findings indicate that nuclear NELF has a developmental role in the hypothalamus.
Materials and Methods
Human NELF Cloning
Putative human NELF sequence was obtained by blasting against the human genome database available at the Entrez site at NCBI. Probes for northern blot analysis were generated by RT-PCR from human lymphoblast RNA as described previously [Prasad et al. 1998]. The resulting 925bp and 1040bp fragments were cloned into the TA easy PCR cloning vector (Invitrogen) and correct inserts were identified and excised by restriction enzyme digestion. An ARPE (adult human retinal pigment epithelial cell line) cDNA library was screened using the 32P labeled NELF probes. Positive clones were initially confirmed by restriction enzyme digestion. A single clone was subsequently picked to confirm the identity by nucleotide sequencing. The insert was then excised out of the pSPORT 3.1 vector and inserted into pET 32a(+) vector (Novagen, EMD Biosciences, inc., San Diego, CA), such that the NELF fusion protein with a His Tag at the N-terminal end was synthesized from the construct. This fusion protein was then induced using IPTG in E. coli. A Ni-NTA agarose kit/column (Qiagen) was used to isolate the 6XHIS-tagged NELF protein from the bacterial lysate.
Northern Blot Analysis
6ug of RNA isolated from different tissues was separated on a 1% formaldehyde-agarose gel and transferred to a Hybond-N transfer membrane, as described previously [Prasad et al. 1998]. The human NELF cDNA was labeled with [α32P]dCTP. Blots were hybridized at 24°C in 6X SSPE, 50% formamide, 10X Denhardt’s solution, 2% SDS, and salmon sperm DNA (100ug/mL) and then washed at high stringency with a final wash of 0.5X SSPE, 0.5%SDS at 60°C for 30 minutes [Prasad et al. 1998].
Nelf mRNA Expression in Pituitary Cell Lines
The LβT2 cell line was kindly provided by Dr. PL Mellon [Thomas et al. 1996]. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal calf serum (FCS), and 1% (v/v) penicillin/streptomycin (GIBCO Invitrogen, Carlsbad, CA) at 37°C in humidified 5% CO2/95% air. Cells were treated for the indicated times with 10nM of the GnRH analog [des-Gly10, D-Ala6]-LH-RH ethylamide acetate hydrate (Sigma-Aldrich, St. Louis, MO). Total RNA from cultured cells was extracted using TRIzol Reagent (Invitrogen) and DNase treated using the Turbo DNA-free kit (Ambion, Austin, TX). cDNA was prepared by RT-PCR using Superscript II Reverse Transcriptase (Invitrogen) and amplified in triplicate by quantitative real-time PCR on an Applied Biosystems 7900HT (Foster City, CA) using Taqman Universal PCR Master Mix (Applied Biosystems) and the following gene-specific Taqman Gene Expression Assay primer/probe sets (Applied Biosystems): mouse Nelf mm00480341_m1; mouse Chd7 mm01219527_m1; mouse Fshb mm00433361_m1; eukaryotic 18S rRNA endogenous control (VIC/MGB Probe, Primer Limited). Measurement of target gene transcripts were calibrated to the expression levels of 18S transcript in each sample and expressed relative to Time Zero using the comparative CT method of calculation.
Quantitative RT-qPCR in Immortalized GnRH Neuronal Cell Lines
RNA was isolated from NLT, GN11 and GT1-7 cells using Tri Reagent (Molecular Research Center, INC, Cincinnati, Ohio). RT-qPCR was performed 3 times in triplicate for each cell line using the Cepheid Smart Cycler (Cepheid, Sunnyvale, CA) with the LightCycler RNA Amplification kit SYBR Green I (Roche, Switzerland). A standard curve was constructed using known amounts of 150 bp of Nelf exon 3 cDNA (common to all predicted Nelf splice variants in the mouse). Mouse Nelf primer sequences were as follows—forward: 5′ ccagagtcaccctgagaacc 3′and reverse: 5′ ccctctatagccggcttcc 3′. Results were observed directly and also normalized to the Ppia gene, encoding cyclophillin A, using the CT method. Similarly, a standard curve for Ppia was also performed. Primer sequences for Ppia were forward: 5′cacaaacggttcccagtttt 3′and reverse: 5′ tccacaatgttcatgccttc 3′.
Cell culture, Construction of pEGFP-NELF and Transient transfection
Three GnRH neuronal (NLT, GN11 and GT1-7) and three non-neuronal (COS-7, CHO, and HEK293) cell lines were grown on plates with DMEM, 1% L-glutamine, 1% antibiotic-antimycotic solution, and 10% FCS. Cells were plated onto 6-well plates 24 hours before transfection. Full length wild type NELF cDNA was inserted into the expression vector pEGFP-N1 (Clonetech, Mountain View, CA) using HindIII and BamHI restriction sites. Lipofectamine Plus Reagent (Invitrogen, Carlsbad, California) was used for cell transfection. Cells were lysed 24 hours after transfection and subjected either to immunoblotting, seeded on poly-L-lysine coated glass coverslips for immunofluorescence microscopy, or resuspended in DMEM with 1% FCS for chemomigration assays.
PSORT II and Site-directed mutagenesis
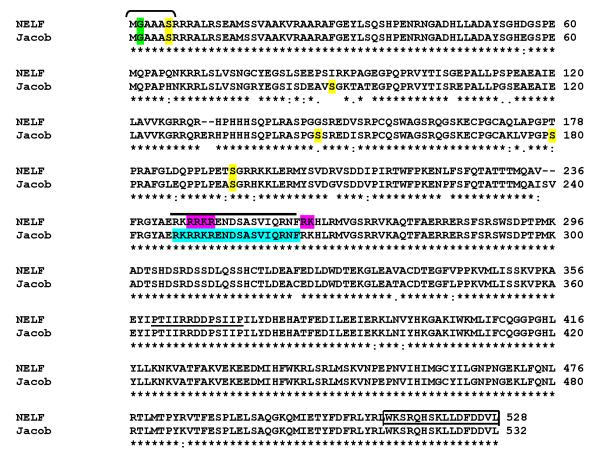
Two putative nuclear localization signals were identified by PSORT II (http://psort.ims.u-tokyo.ac.jp/form2.html). Mutations were made by the QuickChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA), and confirmed by DNA sequencing. The amino acids that were mutated are shown in Figure 5. Within the putative bipartite nuclear localization signal (NLS) from amino acids 243-260, the RRKR amino acids at positions were mutated to AAKA (3A mutant); and the RK at positions – were mutated to AA (2A mutant).
Figure 5.
Amino acid alignment of human NELF (NP_056352) and rat Jacob (AJ293697) using CLUSTAL 2.0.11 is shown. The locations of the myristoylation site (brackets above amino acids 1-7) and bipartite NLS of NELF (bold line above amino acids 243-260 of NELF) corresponding to the identical sequence in Jacob protein shaded in aqua). Included in the NLS is the sequence RRKR (mutated to AAKA—the 3A mutant) and adjacent to NLS is the RK sequence (mutated to AA—the 2A mutant), both shaded in magenta. In addition, the peptide sequence used to generate the anti-NELF antibody by Kramer et al [Kramer and Wray 2000] is shown (underline beginning at NELF amino acid 360) and the C-terminal location for our new anti-NELF antibody (boxed sequence at the very C-terminus), which was used for all figures in the current study. Shown, shaded in yellow, are potential sites of phosphorylation of the Jacob protein [Dieterich et al. 2008].
New anti-NELF antibody Generation
The anti-NELF antibody, generated against sequence PTIIRRDDPSIIP, was generously provided by Susan Wray. For this project, all data was obtained using a new NELF polyclonal antibody that was generated against the C-terminus of NELF using the sequence CWKSRQHSKLLDFDDVL (amino acids 513-528) with the addition of an N-terminal cysteine (Figure 5). Rabbits were injected with Freund’s adjuvant containing the peptide sequence. Following the second bleed, affinity purification was performed. The peptide sequence used to raise the antibody was conserved in 5 species (human, mouse, rat, zebrafish and gallus).
Nuclear/Cytoplasmic Separation and Immunoblotting
Cell lysates from GnRH neuronal cells (NLT, GN11 and GT1-7) and transfected cells were homogenized and fractionated with the Celytic Nuclear Extraction Kit (Sigma, St. Louis, MO) by differential velocity sedimentation to yield nuclear and cytosolic fractions, which were then subjected to western blot analysis. Nuclear purification using sucrose density gradients was used for confirmation [Blobel and Potter 1966]. Protein was quantitated using the BCA protein assay. Ten micrograms of protein/well were separated on 10%SDS acrylamide gels at 80V for 1.5 hours, and transferred to nitrocellulose membranes. Membranes were blocked using 5% milk and incubated with primary antibody anti-NELF 2μg/mL (kindly provided by Susan Wray [Kramer and Wray 2000] or the newly generated polyclonal anti-NELF antibody) , anti-actin (Sigma, St. Louis, MO), anti-GFP (Invitrogen 1:1000) or anti-Histone H1 (Santa Cruz Biotechnology 1:1000). Blotted membranes were washed in TBST, incubated with secondary peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA) at 0.8μg/mL, and detected by ECL Detection Reagents (Amersham Biosciences, United Kingdom) [Patel et al. 2001]. The size and amount of protein by densitometry was determined by software Kaleida Graph and NIH Image J.
Immunofluorescence microscopy and confocal microscopy
Cells were seeded on poly-L-lysine-coated glass coverslips, and fixed in 4% paraformaldehyde in PBS (pH=7.4) for 20 min at RT. Cells were washed with PBS, and permeabilized using 0.1% Triton X-100 in PBS for 8 min at RT. After blocking in PBS containing 10% BSA for 30 min at RT, cells were incubated with antibody (2-5 μg/mL in 2% BSA, 4°C for 12 hours), followed by three PBS washes and incubation with secondary antibody Alexa Fluor 594 (Invitrogen) (1 μg/mL in 2% BSA for 60 minutes). DAPI was used to stain the nuclei (2ug/mL in PBS for 15 minutes). Coverslips were mounted using VectaShield (Vector, Burlingame, CA) and viewed using laser-scan-confocal microscopy (Zeiss Axiovert LSM 510, Carl Zeiss, Jena, Germany). In addition, GnRH cells were fixed in paraformaldehyde and incubated in blocking buffer with 1 ug/ml of antibody following preincubation (60min at RT) with 1 ug/ml of NELF peptide CWKSRQHSKLLDFDDVL (amino acids 513-528) and cells were examined by laser-scan-confocal microscopy. No staining was observed.
The following experiment was performed to determine the presence of a functional nuclear localization signal (NLS). To detect a ≥3-fold difference with 80% power in the nuclear/cytoplasmic ratio of the wild type NLS vs. the 3A mutant, 50 cells/group were needed. For both the 3A mutant and the wild type, 50 cells were randomly selected by a blinded observer for analysis using confocal microscopy. First, the midplane of the cell was imaged. Then the nucleus and cytoplasm were individually outlined; and the GFP signal intensity was measured and converted to a numerical value. Standard pixel counts of GFP staining intensity were obtained using Zeiss LMS 510 Imaging software; and the nuclear/cytoplasmic ratio was calculated for 50 cells in each group.
Micro-RNAi for NELF knockdown In GnRH Neuronal Cell Lines
Based on the NELF sequence, 21-23 bp top and bottom strand oligonucleotides were designed using Invitrogen’s RNAi Designer at www.invitrogen.com/rnai, and four regions were chosen. After annealing DNA oligonucleotides to generate a double stranded oligonucleotide, it was cloned into the pcDNA™ 6.2-GW/EmGFP-miR expression vector using T4 DNA ligase in the BLOCK-iT Pol II miR RNAi Expression Vector Kit (Invitrogen) to yield pcDNA 6.2-GW/EmGFP-miR-NELF. The NELF/GFP construct was also co-transfected. Following transformation, the desired colonies were selected with spectinomycin. Only one of four miRNA-NELF oligonucleotides reduced NELF/GFP protein expression by western blot analysis (~70% reduction) and immunofluorescence after cotransfection of the NELF/pEGFP and miRNA constructs into HEK293 cells vs. the random control (not shown). The sequence for the NELF miRNA construct was as follows: 5′- TGCTGTGTCCTCGAAGGTTGCATGCTGTTTTGGCCACTGACTGACAGCATGCA CTTCGAGGACA-3′. The selected NELF miRNA construct and the kit control (a random sequence) were transiently transfected into NLT cells to determine the effect of NELF knockdown upon GnRH neuron migration.
Chemomigration assay
Chemomigration using the Boyden chamber method has been reported by others [Allen et al. 2002,Giacobini et al. 2002] to study GnRH neuron migration using immortalized neuronal cells. A 48-well Boyden chamber with 3.2mm wells and 8uM pores (Neuroprobe, Cabin John, MD) precoated with gelatin was used for chemomigration as reported [Allen et al. 2002,Giacobini et al. 2002]. NLT cells were grown in 10%FCS to 90% confluency and trypsinized. In the first experiment all cells were utilized; and the in second experiment cells were sorted with fluorescence-activated cell sorting (FACS) to enrich for transfected cells.
The cells from both experiments were counted in a hemocytometer, and resuspended in 1%FCS. Then ~105 cells were added to the upper surface of the membrane for each well of the Boyden chamber. Following three hours of incubation at 37°C, the membrane of the Boyden chamber was removed from the apparatus. The top of the membrane (where the cells were initially loaded) was washed with phosphate buffered saline and scraped to remove unmigrated cells. The bottom of the membrane (containing the migrated cells) was fixed and stained using Diff-Quick (Biomap, Milano, Italy), then mounted onto a glass slide. Each well was individually photographed and all cells were counted by an observer blinded as to the identity of the well. The number of migrated cells from the wild type vs. knockdown were then compared for each experiment.
Statistics
Data were square-root transformed (RT-qPCR) or log transformed (for western blot analysis of endogenous NELF expression). NELF mRNA expression studies and endogenous NELF subcellular localization by western blot analysis were analyzed by ANOVA for a randomized complete block design. The Mann-Whitney test was used to analyze differences in migrate GnRH cells.
For quantification of GFP nuclear & cytoplasmic signal intensity, a mixed model ANOVA was used in which the factorial fixed effects of phenotype (wild type vs. 3A), and subcellular localization (nucleus vs. cytoplasm) were included as well as the random effects of transfection, picture nested within phenotype, and cell nested within picture and phenotype. Differences were considered significant at p<0.05.
Results
Human and Mouse NELF mRNA Expression
Nelf mRNA Expression Patterns
By northern blot analysis, human NELF expression was greatest in the brain, but was also demonstrated in the heart, liver, kidney, and spleen (Figure 1A). Low levels of expression were observed in the small intestine, skeletal muscle, and peripheral white blood cells. The cloned transcript was identical to NM_015537 in the NCBI nucleotide database except for a two amino acid deletion constituting exon 5 of AY_255128. Site-directed mutagenesis was used to add the missing sequence so that the full length protein totaled 530 amino acids, as was published [Miura et al. 2004]. However, the revised NCBI sequence (NM_015537) is consistent with our original clone containing 528 amino acids. Therefore, it appears that the full length NELF cDNA is 528 amino acids.
Figure 1.
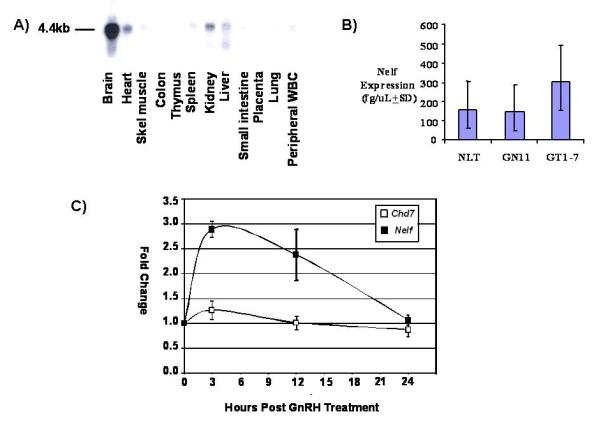
A) Northern blot analysis with a 32P labeled human NELF cDNA probe. Although not well see in the figure, low expression levels were observed in the small intestine, skeletal muscle, and peripheral WBCs. B) QRT-PCR of mouse Nelf mRNA expression in each of the three neuronal GnRH cell lines. Triplicate experiments (n=9 for each cell line) showed that all three GnRH cell lines express Nelf, but levels were not significantly different among cell lines. C) QRT-PCR of mouse Nelf mRNA expression following exogenous GnRH agonist treatment in LβT2 pituitary cells (n=3/group). Nelf expression increased three-fold at three hours, while Chd7 expression did not change after GnRH agonist treatment. Nelf expression was greater at both 3 and 12 hr vs. control (P<0.001). Although there was a statistically significant rise in Chd7 expression at 3 hr, this effect is only ~ 40% increase over baseline, questioning any biological significance.
Nelf mRNA Expression in Mouse Hypothalamic GnRH Neuronal & Pituitary Cells
It is very difficult to study GnRH neurons in vivo. Because of the small number of dispersed hypothalamic GnRH neurons, immortalized GnRH neuronal cell lines [Mellon et al. 1990,Radovick et al. 1991] were used to study mRNA and protein expression. Both had been generated using constructs containing the GnRH promoter cloned upstream to the SV40 T-antigen to produce transgenic mice. These mice developed tumors in the hypothalamus (GT1-7 cells) [Mellon et al. 1990] or along the olfactory neuron pathway (NLT and GN11 cells) [Radovick et al. 1991]. GT1-7 cells constitute mature, postmigratory neurons that secrete pulsatile GnRH [Mellon et al. 1990], while the less mature NLT and GN11 cells are migratory and secrete less GnRH [Radovick et al. 1991]. By RT-qPCR, Nelf expression did not differ (p= 0.0955) in migratory vs. postmigratory GnRH cells (Figure1B). Minimal Nelf mRNA expression was seen in CHO, COS-7 and HEK293 cells (not shown).
Since hypothalamic peptides may also be expressed and have reproductive functions in the pituitary gland, basal and GnRH-stimulated pituitary Nelf expression was examined. Nelf mRNA expression was detected in the primary pituitary gland and in two pituitary gonadotrope cell lines— immature α3 and mature LβT2 cells (not shown). LβT2 cells express gonadotrope-specific genes, including chorionic gonadotropin-alpha (Cga), follicle-stimulating hormone-beta (Fshb), luteinizing hormone-beta (Lhb), and GnRH receptor (Gnrhr), and as such are routinely utilized as a model system for mature gonadotropes [Horton and Halvorson 2004]. Following GnRH agonist treatment of pituitary LβT2 cells, a peak 3-fold increase in Nelf mRNA expression was observed three hours after treatment that was maintained ~2.5-fold at 12 hours (Figure 1C). This effect was not seen for Chd7 mRNA expression (Figure 1C), which encodes a protein also implicated in GnRH and olfactory neuron migration in mouse and human [Kim et al. 2008b].
NELF Protein Expression
Endogenous NELF Localizes to the Nucleus in Immortalized GnRH Neurons
NELF lacks a classical signal peptide and has no homology to any other human protein in the proteome. PSORT II predicts that NELF has ~70% probability of having a nuclear localization signal (NLS). To test this hypothesis, cell lysates from GnRH cells were separated into nuclear and cytoplasmic fractions and subjected to immunoblotting. Using the previously reported anti-NELF antibody [Kramer and Wray 2000], our findings were inconsistent (not shown). Therefore, a new polyclonal anti-NELF antibody was generated against a unique C-terminal amino acid sequence, which was identical in both human and mouse. This antibody was used for all of the data in the current study. No specific binding was observed when the antibody was incubated with the peptide prior to immunoblotting, indicating antibody specificity to the peptide sequence (not shown).
Protein from total cell lysates consistently revealed bands of the anticipated molecular weight (~63kDa) from all three cell lines, but greater expression was seen in migratory GN11 and NLT cells (Figure 2A). Subcellular fractionation indicated that endogenous NELF was enriched in the nuclear fraction (Figure 2B, lanes labeled N) vs. the cytoplasm (Figure 2B, lanes labeled C). Little NELF was detected in COS-7 cells (Figure 2B). For further confirmation and to exclude cytoplasmic contamination, nuclear isolation with sucrose density gradient fractionation was performed [Blobel and Potter 1966]. Nuclear marker histone H1 antibody staining was only observed in the nuclear fraction as shown in Figure 2C (lanes H, P1, and P2). The final nuclear pellet (P2), achieved by sedimentation through a 1.9 M sucrose cushion, demonstrated the most intense NELF and H1 staining [Blobel and Potter 1966]. Immunofluorescence microscopy also confirmed NELF’s predominantly nuclear localization in all three GnRH neuronal cell lines, as is shown in Figure 3A. These findings, consistent with the immunoblotting, demonstrate that endogenous NELF protein is more highly expressed in immature, migratory GnRH neurons, and that its localization is predominantly in the nucleus.
Figure 2.
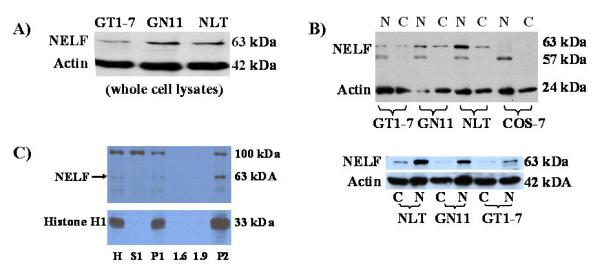
A) Protein expression by western blot analysis with our new anti-NELF antibody is greater in migratory NLT and GN11 cells than postmigratory GT1-7 cells. B) Following cellular fractionation, NELF expression is greater in the nucleus than the cytoplasm in all three GnRH cell lines as represented by a ~63 kDa band. A smaller ~57kDa nonspecific band was seen in the nuclear fraction, which could represent a NELF splice variant. Note absence of the 63 kDa band in COS-7 cells. The lower figure clearly shows greater nuclear expression in all three GnRH cell lines. Beta-actin was used as a loading control. C) Following sucrose density gradient separation, NELF expression was seen in the nucleus. The most intense expression of NELF was seen in the final nuclear pellet P2. Abbreviations are as follows: H = histone; S1 = supernatant 1; P = pellet 1; 1.6 M and 1.9 M sucrose layers; P2 = pellet 2, which comprises the nuclear pellet. A 100 kDa nonspecific band is seen in some fractions.
Figure 3.
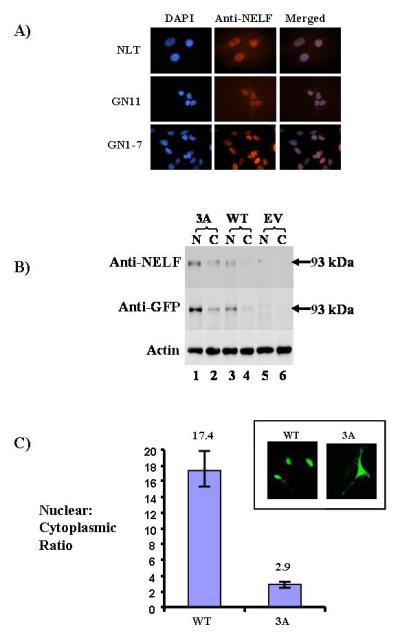
A) Immunofluorescence with confocal microscopy (63X) demonstrates predominant NELF expression in the nucleus in all three cell lines, as shown by DAPI and anti-NELF antibody staining. As a negative control, peptide used to generate the antibody was preincubated with the antibody, and no NELF staining was seen (not shown). B) Western blot analysis of the exogenous NELF/GFP fusion product in NLT cells is shown. The expected 93kDa band is shown using both anti-NELF and anti-GFP antibodies. Beta-actin is the loading control. C) Nuclear/cytoplasmic (N/C) ratios for wild type and 3A NELF mutant in NLT cells are shown. In the inset, immunofluorescence demonstrates nuclear GFP staining in the wild type, but diffuse staining in the 3A mutant, consistent with its reduced nuclear/cytoplasmic expression.
NELF Has a Functional Nuclear Localization Signal
To confirm NELF’s subcellular localization and identify important functional domains, full length human NELF cDNA was cloned into a pEGFP-N1 expression vector and transfected into COS-7 cells, which normally demonstrate minimal NELF expression. Western blot analysis demonstrated the expected 93kDa (GFP~30kDa; NELF~63kDa) wild type NELF/GFP fusion protein in both nuclear and cytoplasmic fractions with either anti-NELF or anti-GFP antibodies (Figure 3B, lanes 3-4). NELF/GFP fusion protein was absent in an empty vector (EV) pEGFP-N1 (Figure 3B, lanes 5-6).
PSORT II predicts two putative NELF nuclear localization signals (NLS)—RRKR at amino acid 245-248, and RK at residues 261-262 (Figure 5). To determine if these sequences mediated nuclear localization, RRKR was mutagenized to AAKA (termed the 3A mutant), and RK was changed to AA (the 2A mutant). Western blot analysis, using both anti-NELF and anti-GFP antibodies, demonstrated the expected ~93kDa band predominantly in the nucleus in the wild type. In contrast, the 3A mutant showed more localization in the cytoplasm vs. wildtype (Figure 3B, lanes 1-2 vs. 3-4). No bands were present in cells transfected with empty vector (Figure 3B, lanes 5-6). Cellular expression of the 2A mutant was similar to the WT (not shown), suggesting it has little or no role in nuclear localization.
Fluorescence microscopy was then performed on live NLT neuronal cells transfected with either empty vector, wild type, or the 3A mutant (Figure 3C). A diffuse GFP cellular signal was observed in the empty vector. Nuclear staining only was seen with the wild type; and both nuclear and cytoplasmic staining was seen for the 3A mutant (Figure 3C inset). The 2A mutant did not alter NELF subcellular localization (not shown). Colocalization of GFP and DAPI in fixed NLT cells confirmed the impaired nuclear localization of the 3A mutant vs. wild type (not shown).
Quantification of the nuclear and cytoplasmic expression was then analyzed by confocal microscopy in NLT cells transfected with either the wild type NELF or 3A mutant. The nuclear/cytoplasmic ratio was 17.4 for the wild type compared with. 2.9 for the 3A mutant (Figure 3C), which was highly significant. This represented a 6-fold (95% CI: 5.15, 6.98; p < 0.0001) reduction in nuclear expression for the 3A mutant. These findings indicate that the RRKR amino acid residues of NELF constitute a functional NLS, which is completely conserved in all species identified. In summary, both endogenous and ectopic NELF showed predominant localization in the nucleus, and mutagenesis of a NLS demonstrated a significant reduction in the nuclear/cytoplasmic ratio. Collectively, these data indicate that NELF is principally a nuclear protein.
NELF Knockdown Impairs GnRH Neuronal Cell Migration
Since Nelf was isolated from migrating GnRH neurons [Kramer and Wray 2000], NELF knockdown would be expected to impair GnRH neuron migration in vitro, but this has not been studied in immortalized, migratory GnRH neurons. An effective NELF miRNA, which localized to the cDNA at 1371-1391 (21bp in exon 11), was generated to analyze GnRH neuron migration by the Boyden chamber technique [Giacobini et al. 2002]. NELF knockdown resulted in a statistically significant (p<0.005) 3.2-fold reduction in NLT cell migration vs. control (Figure 4, left). In repeated experiments using fluorescence activated cell sorting (FACS) to enrich for GFP-transfected cells, a >12-fold reduction in GnRH neuron migration was observed (Figure 4, right). These data provide additional evidence that NELF functions as a stimulatory factor in GnRH neuron migration.
Figure 4.
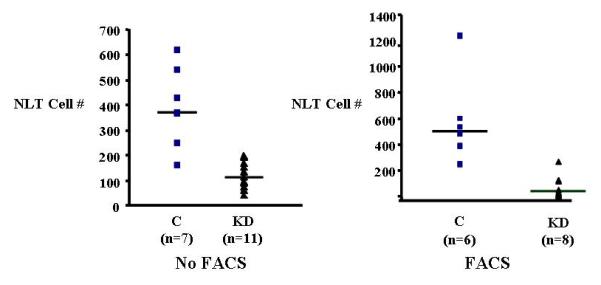
Boyden chamber NLT cell migration results are shown. On the left, NELF knockdown (KD) results in a three-fold reduction in migration compared with the control. On the right, when fluorescence-activated cell sorting (FACS) was done to enrich for transfected cells, GnRH neuron migration was reduced more than 12-fold.
Discussion
NELF Isolation and Expression Patterns
During embryonic development, GnRH neurons migrate along with olfactory neurons migrate from the olfactory placode region to cross the cribriform plate [Schwarting et al. 2007,Wierman et al. 2004]. Most GnRH neurons affecting reproduction arrive at their final destination in the hypothalamus, while olfactory neurons migrate to both the forebrain and the olfactory bulb [Schwarting et al. 2007,Wierman et al. 2004]. Disruption of this neural migration pathway results in the clinical disorder Kallmann syndrome. Understanding the function of genes involved in the GnRH/olfactory neuronal migration pathway, such as NELF, will provide further insight into the normal hypothalamic-pituitary-gonadal axis in mammalian puberty and reproduction.
However, the study of GnRH neuron development, migration, and function is challenging since there are only 1-2,000 neurons that are dispersed throughout the brain in primates and less in mice [Mellon et al. 1990,Radovick et al. 1991]. Therefore, the use of migratory (GN11 and NLT)[Radovick et al. 1991] and postmigratory (GT1-7)[Mellon et al. 1990] GnRH neuronal cell lines assumes primary importance in these studies. Although they are immortalized GnRH cells, they developed neurite projections, growth cones and multiple cell–cell contacts in culture. In addition, they possess many characteristics of their physiologic counterparts including mRNA expression and pulsatile secretion of GnRH; and they have regulatable fast sodium channels found in neurons, as well as the expression of neuronal (neuron-specific enolase, neurofilament protein), but not glial specific cell markers (glial fibrillary acidic protein, myelin basic protein, or myelin proteolipid protein) [Mellon et al. 1990]. These immortalized GnRH neuronal cells have been used extensively to study GnRH neuron function [Allen et al. 2002,Mellon et al. 1990,Nielsen-Preiss et al. 2007,Pierce et al. 2008,Radovick et al. 1991,Wierman et al. 2004].
Since Nelf was isolated from migratory GnRH neurons, it was anticipated that mRNA and protein expression would be greater in migratory NLT GnRH neuronal cells than postmigratory GT1-7 cells. Somewhat surprisingly, Nelf mRNA expression did not differ with regard to migratory status. Human NELF was also expressed in other tissues as described previously by others [Miura et al. 2004]. In the present study, northern analysis demonstrated robust mRNA expression in the brain, but some degree of expression was also seen in the heart, liver, kidney, and spleen; and low levels of expression were observed other tissues.
In contrast to transcription, NELF protein expression by western blot analysis and immunofluorescence microscopy was consistently greater in the migratory NLT and GN11 neurons than postmigratory GT1-7 cells. By western blot analysis, a ~63 kDa band was obtained, which is consistent with NELF’s predicted molecular weight of 59.8 kDa. A smaller 57 kDa band was also seen in the nuclear fraction (shown in Figure 2B), which could represent altered splicing. The lack of correlation between NELF mRNA expression and protein expression is not unprecedented [Chen et al. 2002,Esfandiari et al. 2003,Jinnah et al. 2004,Schwahn et al. 2001]. This could occur by “leaky” expression of splice variants [Jinnah et al. 2004] or by altered expression of proteins with multiple isoforms [Chen et al. 2002]. Alternatively, mRNA/protein discordance could occur by affecting the rate of protein translation [Esfandiari et al. 2003] or posttranslational modifications [Stamm et al. 2005]. NELF is predicted to have three isoforms in mouse A: (NM_001039386), B (NM_001039387), and C (NM_020276) and four isoforms in human: a (NM_001130969), b (NM_015537), c (NM_001130970), and d (NM_001130971). Therefore, the lack of correlation between mRNA and protein expression could potentially be explained by multiple isoforms. This will require more in-depth study in the future.
After completion of our studies, the characterization of the Jacob protein in the rat was reported [Dieterich et al. 2008], which indicated that human NELF and the Jacob protein have extremely high homology. These two protein sequences are aligned in Figure 5. From the 82% cDNA sequence and 94% protein sequence identity, it appears that Jacob and NELF are the corresponding orthologs of rat and human. The Jacob protein is expressed in the brain, particularly in the limbic system, including the thalamus, hypothalamus, and amygdala. Our finding of several different bands in the nucleus is consistent with the results of similar studies of the Jacob protein, where bands of variable size (62-80 kDa) were seen in nuclear fractions [Dieterich et al. 2008]. These investigators postulated that these different molecular weights were due to post-translational modifications [Dieterich et al. 2008], but they could also be due to the detection of multiple splice variants. The role of Jacob in GnRH neuron migration was not reported in this study. However, it is tempting to speculate, based upon our findings in mouse and human and the high sequence homology in rat, that Jacob will play a role in GnRH neuron migration.
Subcellular Localization of NELF
Our subcellular distribution of NELF protein contrasts rather dramatically with Kramer et al [Kramer and Wray 2001,Kramer and Wray 2000] who demonstrated NELF protein localization on the cell surface of GnRH and olfactory neurons. Using the complementary techniques of western blot analysis and immunofluorescence microscopy with a newly generated NELF-specific antibody in our study, NELF was found to be principally localized in the nucleus, with minimal staining was also seen in the cytoplasm. Further support for nuclear localization comes from mutagenesis of the completely conserved RRKR NLS sequence, which reduced nuclear localization 6-fold. This RRKR sequence is contained within a bipartite NLS of sequence RKRRKRENDSASVIQRNF (amino acids 243-260) predicted in PSORTII that is also completely conserved in the Jacob protein [Dieterich et al. 2008], shown in Figure 5. When these same four amino acids (RRKR) were deleted, these investigators also found that nuclear localization was impaired when the construct was transfected into COS-7 cells [Dieterich et al. 2008]. Therefore, our findings and those of Dieterich demonstrate agreement in protein localization of these orthologs.
NELF Has Two Putative Zinc Finger Domains
Additional features of NELF provide further evidence for a potential role for NELF in the nucleus. Zinc finger domains, which are commonly seen in transcription factors [Mendiratta et al. 2006], were detected in the NELF sequence. The H2C2 type putative zinc finger within amino acids 131-170 is highly conserved in five different species (human, mouse, rat, chicken and zebrafish), and the putative CHHC zinc finger at residues 411-463 was conserved in four (Figure 6). These putative zinc fingers indicate that NELF could be a transcription factor that could mediate the expression of other genes involved in GnRH migration.
Figure 6.
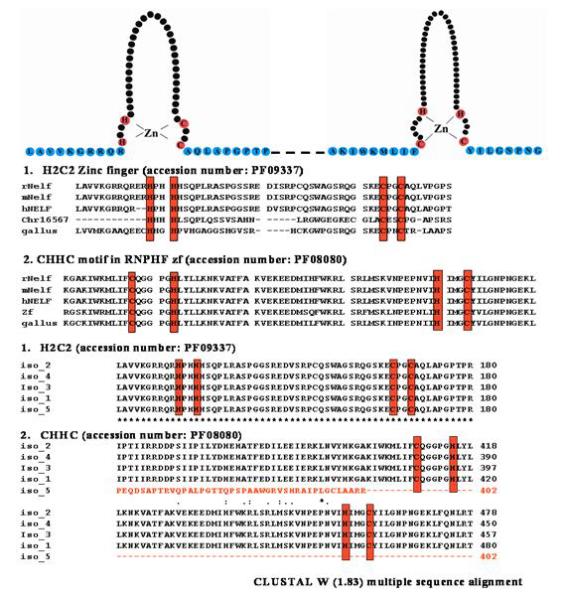
Schematic presentation of putative zinc finger domains of NELF are shown. Two zinc finger domains with HH-CC and CH-HC consensus are shown consecutively. These zinc finger domains were represented in the Pfam program of PF08080 (http://www.sanger.ac.uk/cgi-bin/Pfam/getacc?PF08080) and PF09337 (http://www.sanger.ac.uk/cgi-bin/Pfam/getacc?PF09337). Red colored circles represent cysteine or histidine residues binding directly with zinc ions. The alignment of NELF proteins containing two putative zinc finger domains from five different species of human, mouse, rat, chicken and zebrafish. The fully conserved putative zinc finger domains of HH-CC and CH-HC are boxed in red.
NELF was also found to have a myristoylation consensus sequence by PSORTII, which was identical to that identified in the Jacob protein (Figure 5). As reported by Dieterich et al [Dieterich et al. 2008], mutagenesis of the glycine at position 2 to alanine blocked the incorporation of 3H-myristic acid compared with control, thereby confirming a functional myristoylation site. Myristoylation is an important feature of the NELF protein, which could clearly affect its function. Myristoylation, typically an irreversible protein modification, is known to be involved a variety of cellular processes and can occur during translation or following translation [Farazi et al. 2001]. N-myristoylated proteins play a role in a wide variety of signal transduction cascades. Although NELF was nearly completely nuclear in our mouse GnRH neuronal cell lines, myristoylation could potentially account for extranuclear NELF localization identified by Kramer et al [Kramer and Wray 2000].
The Function of NELF in GnRH Neuron Migration
NELF’s putative role in GnRH neuron migration is supported by several lines of evidence. Nelf was first identified in a differential screen of migratory vs. nonmigratory GnRH neurons [Kramer and Wray 2000]. High levels of mRNA and protein expression were found in the forebrain, olfactory epithelium, and olfactory pit of mouse embryos. Maximal expression occurred between E12.5-E14.5 in the olfactory epithelium, a time when GnRH and olfactory neurons are migrating. NELF staining of GnRH neurons was also detected between E11.5-17.5, but not postnatally or in adult mice [Kramer and Wray 2001,Kramer and Wray 2000].
In the present study, NELF expression was identified in both migratory, as well as postmigratory GnRH neuronal cells, suggesting it could have some function once migration is completed. Following NELF knockdown in NLT cells, there was a three-fold reduction in migration of GnRH neuronal cells, which was even more marked (>12-fold) with FACS enrichment of transfected cells. Therefore, findings from the present study extend those of previous studies indicating a role for NELF in the GnRH neuron migration process. Similar results have been reported in zebrafish, where nelf knockdown impaired GnRH neuron migration [Palevitch et al. 2009].
Other Potential Roles of NELF
NELF could have additional roles in reproduction in addition to GnRH neuron migration. Activation of the hypothalamic-pituitary-gonadal axis is orchestrated by hypothalamic GnRH, which stimulates the pituitary to result to synthesize and secrete FSH and LH. These pituitary gonadotropins then stimulate the gonads to produce sex steroids and gametes. Our findings demonstrate Nelf mRNA expression in the pituitary gland and in two pituitary gonadotrope cell lines (αT3 and LβT2). When LβT2 cells were treated with a GnRH agonist, pituitary Nelf mRNA expression increased in a time-dependent fashion. Maximal expression (3-fold) occurred at 3 hours and was maintained more than 2-fold at 12 hours. It is tempting to speculate that NELF could have some role in pituitary gonadotrope function, but this will have to be confirmed in future studies. There is precedence for a nuclear protein to have both hypothalamic and pituitary functions. DAX1, a nuclear steroid receptor, is expressed in the hypothalamus, pituitary, and adrenal [Zanaria et al. 1994]. Mutations in the NR0B1 gene encoding DAX1 cause both hypothalamic and pituitary dysfunction in patients with normosmic IHH [Habiby et al. 1996]. In addition, steroidogenic factor-1 also plays an important physiologic role in both the hypothalamus and pituitary, as well as the adrenal [Achermann et al. 2001].
Clinical Correlations
To confirm a relevant, clinical translational role for NELF in human puberty, mutations should be present in patients with pubertal disorders. There have only been two human NELF mutations reported. The first was a heterozygous missense Thr480Ala variant, but no in vitro analysis was performed, so its functional significance is unknown [Miura et al. 2004]. In another report, a heterozygous NELF splice mutation (c.1159-14_- 22del ) was identified in a KS patient; but, a coexistent FGFR1 mutation was also present [Pitteloud et al. 2007a]. In this patient, KS was only present in the patient who had heterozygous FGFR1 and NELF mutations, suggesting the possibility of digenic disease. Therefore, no NELF mutations causing monogenic IHH or KS have been reported. We have recently reported the first verified human NELF mutations in monogenic IHH confirmed vitro (Xu et al; revision submitted), as well as two new digenic patterns in patients with KS.
Conclusions
Results from this study further solidify an important role for NELF in GnRH neuronal migration, a prerequisite for normal function of the hypothalamic-pituitary-gonadal axis. NELF is predominantly a nuclear protein, probably a transcription factor, more highly expressed in migratory GnRH neurons. Although this has not yet been studied, it is tempting to speculate that NELF could regulate the expression of downstream targets modulating neuron migration such as netrin/DCC [Schwarting et al. 2001,Schwarting et al. 2004], HGF/Met [Giacobini et al. 2002], anosmin-1 [Cariboni et al. 2004], FGFR1 [Gill et al. 2004], Gas6/Ark [Allen et al. 2002], neuropilins [Cariboni et al. 2007], and/or others. The nuclear protein HSPB3 has been shown to interact with NELF [Stelzl et al. 2005].
NELF is expressed in immature and mature GnRH neurons, as well as immature and mature pituitary cells, indicating differential developmental functions in these important reproductive tissues. Therefore, the genes NELF regulates could differ in more mature GnRH neurons. The next challenge will be to identify downstream targets of NELF for both the migratory and postmigratory functions. It will also be important to identify and characterize all of the isoforms in relevant tissues. Finally, clinical translational support for NELF’s function in human puberty has been demonstrated in IHH/KS patients possessing NELF mutations. In conclusion, its role in hypothalamic GnRH neuron migration, as well as its involvement in IHH/KS indicate that NELF has functional importance in mammalian puberty and reproduction.
Table 1.
Abbreviations
| 3A | 3A mutation made in the nuclear localization signal |
| CHD7 | chromodomain helicase DNAbinding protein 7 |
| EV | empty vector |
| FACS | fluorescence-activated cell sorting |
| FGF8 | fibroblast growth factor-8 |
| FGFR1 | fibroblast growth factor receptor-1 |
| Fshb | follicle stimulating hormone-beta |
| GFP | green fluorescent protein |
| GN11 | GnRH neuronal cells that are migratory |
| GnRH | gonadotropin releasing hormone |
| GT1-7 | GnRH neurons that are postmigratory |
| IHH | idiopathic hypogonadotropic hypogonadism |
| KAL1 | Kallmann syndrome-1 |
| KS | Kallmann syndrome |
| LHRH | luteinizing hormone releasing hormone |
| NELF | nasal embryonic LHRH factor |
| NLS | nuclear localization signal |
| NLT | GnRH neuronal cells that are migratory |
| PROK2 | prokineticin-2 |
| PROKR2 | prokineticin receptor-2 |
Acknowledgments
We are grateful to Susan Wray for providing us with NELF antibody. L.C.L. was funded by NIH grants HD33004 and HD040287, as well as the Medical College of Georgia Research Institute. We also acknowledge support to L.C.L. from Dean D.D. Miller, Institute of Molecular Medicine and Genetics Director R. Yu, and Ob/Gyn Chair A.A. Murphy at MCG for the completion of these studies.
L.C.L. was supported from NIH grants HD33004 and HD040287 for this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have nothing to disclose.
References
- Achermann JC, Meeks JJ, Jameson JL. Phenotypic spectrum of mutations in DAX-1 and SF-1. Mol Cell Endocrinol. 2001;185:17–25. doi: 10.1016/s0303-7207(01)00619-0. [DOI] [PubMed] [Google Scholar]
- Allen MP, Xu M, Linseman DA, Pawlowski JE, Bokoch GM, Heidenreich KA, Wierman ME. Adhesion-related kinase repression of gonadotropin-releasing hormone gene expression requires Rac activation of the extracellular signal-regulated kinase pathway. J Biol Chem. 2002;277:38133–40. doi: 10.1074/jbc.M200826200. [DOI] [PubMed] [Google Scholar]
- Blobel G, Potter VR. Nuclei from rat liver: isolation method that combines purity with high yield. Science. 1966;154:1662–5. doi: 10.1126/science.154.3757.1662. [DOI] [PubMed] [Google Scholar]
- Cariboni A, Hickok J, Rakic S, Andrews W, Maggi R, Tischkau S, Parnavelas JG. Neuropilins and their ligands are important in the migration of gonadotropin-releasing hormone neurons. J Neurosci. 2007;27:2387–95. doi: 10.1523/JNEUROSCI.5075-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariboni A, Pimpinelli F, Colamarino S, Zaninetti R, Piccolella M, Rumio C, Piva F, Rugarli EI, Maggi R. The product of X-linked Kallmann’s syndrome gene (KAL1) affects the migratory activity of gonadotropin-releasing hormone (GnRH)-producing neurons. Hum Mol Genet. 2004;13:2781–91. doi: 10.1093/hmg/ddh309. [DOI] [PubMed] [Google Scholar]
- Chen G, Gharib TG, Huang CC, Taylor JM, Misek DE, Kardia SL, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM, Beer DG. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1:304–13. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- Crowley WF, Jr., Filicori M, Spratt DI, Santoro NF. The physiology of gonadotropin-releasing hormone (GnRH) secretion in men and women. Recent Prog Horm Res. 1985;41:473–531. doi: 10.1016/b978-0-12-571141-8.50015-9. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Karpova A, Mikhaylova M, Zdobnova I, Konig I, Landwehr M, Kreutz M, Smalla KH, Richter K, Landgraf P, Reissner C, Boeckers TM, Zuschratter W, Spilker C, Seidenbecher CI, Garner CC, Gundelfinger ED, Kreutz MR. Caldendrin-Jacob: a protein liaison that couples NMDA receptor signalling to the nucleus. PLoS Biol. 2008;6:e34. doi: 10.1371/journal.pbio.0060034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dode C, Levilliers J, Dupont JM, De Paepe A, Le Du N, Soussi-Yanicostas N, Coimbra RS, Delmaghani S, Compain-Nouaille S, Baverel F, Pecheux C, Le Tessier D, Cruaud C, Delpech M, Speleman F, Vermeulen S, Amalfitano A, Bachelot Y, Bouchard P, Cabrol S, Carel JC, Delemarre-Van De Waal H, Goulet-Salmon B, Kottler ML, Richard O, Sanchez-Franco F, Saura R, Young J, Petit C, Hardelin JP. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet. 2003;33:463–5. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- Dode C, Teixeira L, Levilliers J, Fouveaut C, Bouchard P, Kottler ML, Lespinasse J, Lienhardt-Roussie A, Mathieu M, Moerman A, Morgan G, Murat A, Toublanc JE, Wolczynski S, Delpech M, Petit C, Young J, Hardelin JP. Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006;2:e175. doi: 10.1371/journal.pgen.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfandiari F, Green R, Cotterman RF, Pogribny IP, James SJ, Miller JW. Methyl deficiency causes reduction of the methyl-CpG-binding protein, MeCP2, in rat liver. Carcinogenesis. 2003;24:1935–40. doi: 10.1093/carcin/bgg163. [DOI] [PubMed] [Google Scholar]
- Falardeau J, Chung WC, Beenken A, Raivio T, Plummer L, Sidis Y, Jacobson-Dickman EE, Eliseenkova AV, Ma J, Dwyer A, Quinton R, Na S, Hall JE, Huot C, Alois N, Pearce SH, Cole LW, Hughes V, Mohammadi M, Tsai P, Pitteloud N. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest. 2008;118:2822–31. doi: 10.1172/JCI34538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J Biol Chem. 2001;276:39501–4. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- Franco B, Guioli S, Pragliola A, Incerti B, Bardoni B, Tonlorenzi R, Carrozzo R, Maestrini E, Pieretti M, Taillon-Miller P, Brown CJ, Willard HF, Lawrence C, Persico M, Camerino G, Ballabio A. A gene deleted in Kallmann’s syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature. 1991;353:529–36. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- Giacobini P, Giampietro C, Fioretto M, Maggi R, Cariboni A, Perroteau I, Fasolo A. Hepatocyte growth factor/scatter factor facilitates migration of GN-11 immortalized LHRH neurons. Endocrinology. 2002;143:3306–15. doi: 10.1210/en.2002-220146. [DOI] [PubMed] [Google Scholar]
- Gill JC, Moenter SM, Tsai PS. Developmental regulation of gonadotropin-releasing hormone neurons by fibroblast growth factor signaling. Endocrinology. 2004;145:3830–9. doi: 10.1210/en.2004-0214. [DOI] [PubMed] [Google Scholar]
- Habiby RL, Boepple P, Nachtigall L, Sluss PM, Crowley WF, Jr., Jameson JL. Adrenal hypoplasia congenita with hypogonadotropic hypogonadism: evidence that DAX-1 mutations lead to combined hypothalmic and pituitary defects in gonadotropin production. J Clin Invest. 1996;98:1055–1062. doi: 10.1172/JCI118866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton CD, Halvorson LM. The cAMP signaling system regulates LHbeta gene expression: roles of early growth response protein-1, SP1 and steroidogenic factor-1. J Mol Endocrinol. 2004;32:291–306. doi: 10.1677/jme.0.0320291. [DOI] [PubMed] [Google Scholar]
- Jinnah HA, Harris JC, Nyhan WL, O’Neill JP. The spectrum of mutations causing HPRT deficiency: an update. Nucleosides Nucleotides Nucleic Acids. 2004;23:1153–60. doi: 10.1081/NCN-200027400. [DOI] [PubMed] [Google Scholar]
- Kim HG, Bhagavath B, Layman LC. Clinical Manifestations of Impaired GnRH Neuron Development and Function. Neurosignals. 2008a;16:165–182. doi: 10.1159/000111561. [DOI] [PubMed] [Google Scholar]
- Kim HG, Kurth I, Lan F, Meliciani I, Wenzel W, Eom SH, Kang GB, Rosenberger G, Tekin M, Ozata M, Bick DP, Sherins RJ, Walker SL, Shi Y, Gusella JF, Layman LC. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet. 2008b;83:511–9. doi: 10.1016/j.ajhg.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer PR, Wray S. Nasal embryonic LHRH factor (NELF) expression within the CNS and PNS of the rodent. Brain Res Gene Expr Patterns. 2001;1:23–6. doi: 10.1016/s1567-133x(01)00004-7. [DOI] [PubMed] [Google Scholar]
- Kramer PR, Wray S. Novel gene expressed in nasal region influences outgrowth of olfactory axons and migration of luteinizing hormone-releasing hormone (LHRH) neurons. Genes Dev. 2000;14:1824–34. [PMC free article] [PubMed] [Google Scholar]
- Legouis R, Hardelin JP, Levilliers J, Claverie JM, Compain S, Wunderle V, Millasseau P, Le Paslier D, Cohen D, Caterina D, Bougueleret L, Delemarre-Van de Waal H, Lutfalla G, Weissenbach J, Petit C. The candidate gene for the X-linked Kallmann syndrome encodes a protein related to adhesion molecules. Cell. 1991;67:423–35. doi: 10.1016/0092-8674(91)90193-3. [DOI] [PubMed] [Google Scholar]
- Mellon PL, Windle JJ, Goldsmith PC, Padula CA, Roberts JL, Weiner RI. Immortalization of hypothalamic GnRH neurons by genetically targeted tumorigenesis. Neuron. 1990;5:1–10. doi: 10.1016/0896-6273(90)90028-e. [DOI] [PubMed] [Google Scholar]
- Mendiratta G, Eriksson PR, Shen CH, Clark DJ. The DNA-binding domain of the yeast Spt10p activator includes a zinc finger that is homologous to foamy virus integrase. J Biol Chem. 2006;281:7040–8. doi: 10.1074/jbc.M511416200. [DOI] [PubMed] [Google Scholar]
- Miura K, Acierno JS, Jr., Seminara SB. Characterization of the human nasal embryonic LHRH factor gene, NELF, and a mutation screening among 65 patients with idiopathic hypogonadotropic hypogonadism (IHH) J Hum Genet. 2004;49:265–8. doi: 10.1007/s10038-004-0137-4. [DOI] [PubMed] [Google Scholar]
- Nielsen-Preiss SM, Allen MP, Xu M, Linseman DA, Pawlowski JE, Bouchard RJ, Varnum BC, Heidenreich KA, Wierman ME. Adhesion-related kinase induction of migration requires phosphatidylinositol-3-kinase and ras stimulation of rac activity in immortalized gonadotropin-releasing hormone neuronal cells. Endocrinology. 2007;148:2806–14. doi: 10.1210/en.2007-0039. [DOI] [PubMed] [Google Scholar]
- Palevitch O, Abraham E, Borodovsky N, Levkowitz G, Zohar Y, Gothilf Y. Nasal embryonic LHRH factor plays a role in the developmental migration and projection of gonadotropin-releasing hormone 3 neurons in zebrafish. Dev Dyn. 2009;238:66–75. doi: 10.1002/dvdy.21823. [DOI] [PubMed] [Google Scholar]
- Patel KG, Liu C, Cameron PL, Cameron RS. Myr 8, a novel unconventional myosin expressed during brain development associates with the protein phosphatase catalytic subunits 1alpha and 1gamma1. J Neurosci. 2001;21:7954–68. doi: 10.1523/JNEUROSCI.21-20-07954.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A, Bliesner B, Xu M, Nielsen-Preiss S, Lemke G, Tobet S, Wierman ME. Axl and Tyro3 modulate female reproduction by influencing gonadotropin-releasing hormone neuron survival and migration. Mol Endocrinol. 2008;22:2481–95. doi: 10.1210/me.2008-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitteloud N, Acierno JS, Jr., Meysing A, Eliseenkova AV, Ma J, Ibrahimi OA, Metzger DL, Hayes FJ, Dwyer AA, Hughes VA, Yialamas M, Hall JE, Grant E, Mohammadi M, Crowley WF., Jr. Mutations in fibroblast growth factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci U S A. 2006;103:6281–6. doi: 10.1073/pnas.0600962103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, Dwyer A, Plummer L, Hughes V, Seminara S, Cheng YZ, Li WP, Maccoll G, Eliseenkova AV, Olsen SK, Ibrahimi OA, Hayes FJ, Boepple P, Hall JE, Bouloux P, Mohammadi M, Crowley W. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007a;117:457–63. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitteloud N, Zhang C, Pignatelli D, Li JD, Raivio T, Cole LW, Plummer L, Jacobson-Dickman EE, Mellon PL, Zhou QY, Crowley WF., Jr. Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci U S A. 2007b;104:17447–52. doi: 10.1073/pnas.0707173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad PD, Wang H, Kekuda R, Fujita T, Fei YJ, Devoe LD, Leibach FH, Ganapathy V. Cloning and functional expression of a cDNA encoding a mammalian sodium-dependent vitamin transporter mediating the uptake of pantothenate, biotin, and lipoate. J Biol Chem. 1998;273:7501–6. doi: 10.1074/jbc.273.13.7501. [DOI] [PubMed] [Google Scholar]
- Radovick S, Wray S, Lee E, Nicols DK, Nakayama Y, Weintraub BD, Westphal H, Cutler GB, Jr., Wondisford FE. Migratory arrest of gonadotropin-releasing hormone neurons in transgenic mice. Proc Natl Acad Sci U S A. 1991;88:3402–6. doi: 10.1073/pnas.88.8.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwahn U, Paland N, Techritz S, Lenzner S, Berger W. Mutations in the X-linked RP2 gene cause intracellular misrouting and loss of the protein. Hum Mol Genet. 2001;10:1177–83. doi: 10.1093/hmg/10.11.1177. [DOI] [PubMed] [Google Scholar]
- Schwarting GA, Kostek C, Bless EP, Ahmad N, Tobet SA. Deleted in colorectal cancer (DCC) regulates the migration of luteinizing hormone-releasing hormone neurons to the basal forebrain. J Neurosci. 2001;21:911–9. doi: 10.1523/JNEUROSCI.21-03-00911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarting GA, Raitcheva D, Bless EP, Ackerman SL, Tobet S. Netrin 1-mediated chemoattraction regulates the migratory pathway of LHRH neurons. Eur J Neurosci. 2004;19:11–20. doi: 10.1111/j.1460-9568.2004.03094.x. [DOI] [PubMed] [Google Scholar]
- Schwarting GA, Wierman ME, Tobet SA. Gonadotropin-releasing hormone neuronal migration. Semin Reprod Med. 2007;25:305–12. doi: 10.1055/s-2007-984736. [DOI] [PubMed] [Google Scholar]
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–68. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Thomas P, Mellon PL, Turgeon J, Waring DW. The L beta T2 clonal gonadotrope: a model for single cell studies of endocrine cell secretion. Endocrinology. 1996;137:2979–89. doi: 10.1210/endo.137.7.8770922. [DOI] [PubMed] [Google Scholar]
- Tsai PS, Moenter SM, Postigo HR, El Majdoubi M, Pak TR, Gill JC, Paruthiyil S, Werner S, Weiner RI. Targeted expression of a dominant-negative fibroblast growth factor (FGF) receptor in gonadotropin-releasing hormone (GnRH) neurons reduces FGF responsiveness and the size of GnRH neuronal population. Mol Endocrinol. 2005;19:225–36. doi: 10.1210/me.2004-0330. [DOI] [PubMed] [Google Scholar]
- Wierman ME, Pawlowski JE, Allen MP, Xu M, Linseman DA, Nielsen-Preiss S. Molecular mechanisms of gonadotropin-releasing hormone neuronal migration. Trends Endocrinol Metab. 2004;15:96–102. doi: 10.1016/j.tem.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Zanaria E, Muscatelli F, Bardoni B, Strom TM, Guioli S, Guo W, Lalli E, Moser C, Walker AP, McCabe ER, Meitinger T, Monace AP, Sassone-Corsi P, Camerino G. An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature. 1994;372:635–41. doi: 10.1038/372635a0. [DOI] [PubMed] [Google Scholar]