Abstract
Phosphoinositide 3-kinases (PI3Ks) can be divided into three distinct classes (I, II, and III) on the basis of their domain structures and the lipid signals that they generate. Functions have been assigned to the class I and class III enzymes but have not been established for the class II PI3Ks. We have obtained the first evidence for a biological function for a class II PI3K by expressing this enzyme during Drosophila melanogaster development and by using deficiencies that remove the endogenous gene. Wild-type and catalytically inactive PI3K_68D transgenes have opposite effects on the number of sensory bristles and on wing venation phenotypes induced by modified epidermal growth factor (EGF) receptor signaling. These results indicate that the endogenous PI3K_68D may act antagonistically to the EGF receptor-stimulated Ras-mitogen-activated protein kinase pathway and downstream of, or parallel to, the Notch receptor. A class II polyproline motif in PI3K_68D can bind the Drk adaptor protein in vitro, primarily via the N-terminal SH3 domain of Drk. Drk may thus be important for the localization of PI3K_68D, allowing it to modify signaling pathways downstream of cell surface receptors. The phenotypes obtained are markedly distinct from those generated by expression of the Drosophila class I PI3K, which affects growth but not pattern formation.
Phosphoinositide 3-kinases (PI3Ks) signal to activate downstream molecules through the generation, within cellular membranes, of specific lipids that are phosphorylated at the D-3 position of the inositol ring. These lipids function as second messengers and appear to be involved in processes such as mitogenesis, cell survival, growth, differentiation, adhesion, motility, and vesicle trafficking (reviewed in reference 67). Hence, PI3Ks have been the focus of intensive research. In multicellular organisms, PI3Ks occur as a family of enzymes (40, 67). These can be assigned to three separate classes (class I, class II, and class III) based on their domain structures, differences in catalytic activity towards defined substrates, and modes of regulation. For many of the downstream events mediated by PI3K activation, the exact nature of the PI3K involved has not been defined.
The class I PI3Ks are heterodimers that associate with cell surface receptors via an adaptor subunit (reviewed in reference 67). Two class I PI3K subclasses have been identified. The prototypical class IA PI3Ks are composed of a catalytic p110 subunit and a regulatory subunit. The regulatory subunit contains two SH2 domains that bind phosphotyrosine residues in specific motifs on activated tyrosine kinase receptors or receptor substrates; this interaction results in translocation of the associated catalytic activity to its lipid substrates at the membrane. A number of distinct adaptor subunits (50 to 85 kDa) and p110 subunits (p110α, p110β, and p110δ) have been identified. These show some variation in tissue distribution. The use of isoform-specific inhibitory antibodies has identified distinct roles for different p110 isoforms in regulating cell motility and cytoskeletal changes or in mitogenesis, but these are dependent on both receptor and cell type (reviewed in reference 67). PI3Kγ represents a separate subclass of PI3Ks (class IB) that is activated by Gβγ subunits via a distinct p101 regulatory subunit (63).
The third class of PI3Ks are homologues of Vps34. The VPS34 gene was originally identified through studies of mutants defective in sorting proteins to the yeast vacuole, indicating an involvement of this PI3K in protein trafficking (28). Vps34 also appears to act in an osmotic stress-activated pathway (22). TheVps34 catalytic subunit associates with a protein kinase, Vps15, which appears to constitutively localize Vps34 to the Golgi membrane and may play a role in activation of the lipid kinase. Like the class I heterodimers, the Vps34-Vps15 holoenzyme forms an evolutionarily conserved complex of regulatory and catalytic subunits responsible for its distinct subcellular localization and substrate specificity. Analogous sorting mechanisms exist in higher eukaryotes, and homologues of Vps34 have been identified in a range of multicellular organisms, including humans (69) and flies (39).
Class II PI3Ks were originally identified by sequence homology with other PI3Ks (40). In Drosophila melanogaster (PI3K_68D) (40, 44) and Caenorhabditis elegans (CE05832, encoded by the F39B1.1 gene), a single species is present, while in mammals three forms have been identified, i.e., PI3K-C2α (also termed mcpk and p170) (20, 44, 68), PI3K-C2β (2, 11), and PI3K-C2γ (43, 47). PI3K-C2α and PI3K-C2β show an essentially ubiquitous distribution, while the expression of PI3K-C2γ is restricted, principally to the liver. These proteins have a modular core comprising four PI3K homology regions: HR4, a Ras binding domain analogous to those in Raf and RalGDS that is a member of the ubiquitin-like structural superfamily; HR3, a C2 domain; HR2, a helical domain that is a member of the ARM repeat structural superfamily; and HR1, a catalytic domain (40; Flybase report for Pi3K68D/CG11621, http://fly.ebi.ac.uk:7081/.bin/fbidq.html?FBgn0015278). In contrast to class I PI3Ks, a Ras-binding function has not been demonstrated for a class II PI3K (2). Class II PI3Ks are larger than other PI3Ks (PI3K_68D is 210 kDa) due to extensions at both the amino and carboxy termini. The C terminus contains a PX (NADPH oxidase homology) domain (48) and a further, characteristic, C2 domain. Within the N-terminal region some, but not all, class II members also contain one or more polyproline motifs which may mediate interactions with SH3 or WW domains. The functions of the class II-specific sequences are unclear. In some molecules, C2 domains mediate interactions that are regulated by calcium ions (58). Both C2 domains in class II PI3Ks, however, lack the critical aspartate residues that form the calcium binding pocket (40). Thus, analysis of the primary sequence has provided few clues as to the function and regulation of this class of enzymes. In particular, a regulatory subunit has not been identified, and little is known about how these PI3Ks are activated or the biological processes that they control.
Studies on class II PI3Ks in mammalian cells have focused on signaling pathways which have been shown, using inhibitors, to involve a PI3K; some of these do not seem to be mediated by the known class I enzymes. Thus, PI3K-C2α has been reported to lie downstream of the monocyte chemotactic peptide-1 chemokine receptor (66), the insulin receptor (12), and the epidermal growth factor (EGF) receptor (EGFR) (3). PI3K-C2β can be activated following stimulation of the integrin αIIbβ3 with fibrinogen (74) and has been reported to be a target of the activated receptors for EGF and platelet-derived growth factor (3) and to associate with the stem cell factor receptor, c-kit (4). In each of these cases the biological process involved appears to require signaling by both class I and class II PI3Ks.
The three classes of PI3Ks differ in substrate specificity. In vitro, class I PI3Ks have a broad substrate specificity, phosphorylating the phosphoinositides (PIs) PI, PI4P, and PI(4,5)P2 at the D-3 position. In vivo, however, the major lipid signal generated by the class I enzymes is believed to be PI(3,4,5)P3, which can interact with specific pleckstrin homology domains in target proteins such as phosphoinositide-dependent kinase 1 and protein kinase B. These protein kinases appear to mediate many of the processes in which class IA PI3Ks have been implicated (reviewed in reference 67). In contrast, class III enzymes only phosphorylate PI to generate PI3P, consistent with a short activation loop which may be insufficient to accommodate the more highly phosphorylated PI substrates (70). PI3P can interact with FYVE fingers. This motif is commonly found in proteins which function at endosomes (reviewed in reference 67). The class II enzymes lack the critical lysine residue which, in class I enzymes, contacts the 5′ phosphate of PI(4,5)P2 (70); hence, they can generate PI3P and PI(3,4)P2 in vitro, but neither the signal generated in vivo nor its protein targets have been defined. Each of the three classes of PI3K can thus generate a different spectrum of 3-phosphorylated lipids, clearly providing the potential to signal to distinct targets.
Functional analyses of PI3Ks have been complicated by the presence of multiple members of each class in mammalian cells. In contrast, Drosophila has only one member of each of the three classes (40). These are named after their chromosomal location: class I is Dp110/PI3K_92E, class II is PI3K_68D, and class III is DVps34/PI3K_59F. Analysis of the function of each Drosophila PI3K may, therefore, indicate the prototypical function of each class of enzyme. Of particular interest are the class I and class II PI3Ks, since these are restricted to multicellular organisms and may thus participate in signaling between cells.
As an unbiased method of identifying cellular processes involving class II PI3K signaling, we have looked at the effects of expressing PI3K_68D within Drosophila imaginal disks. These are sheets of epithelial cells that are set aside within the embryo and which proliferate, grow, and pattern during larval life. Reorganization of these cells during metamorphosis generates the adult cuticular structures, such as the wings and eyes (16). Many of these structures are not essential for viability under laboratory conditions, and hence severe developmental defects can still be studied. Drosophila thus represents an ideal system in which to generate phenotypes by targeted expression. In this study we have used both wild-type and catalytically inactive versions of PI3K_68D in order to identify effects that are dependent on the lipid signal. We found that ectopic expression of wild-type and mutated versions of PI3K_68D in the larval imaginal disks affected the development of the wing veins, the wing margin, and the number of external sense organs. These results suggested a role for this PI3K in signaling pathways affecting patterning, and we describe genetic interactions with the EGF receptor and Notch pathways. In contrast, class I PI3Ks affect growth but not patterning (37). These results provide the first evidence for different roles for class I and class II PI3Ks in vivo and indicate that the lipid signals produced by these separate classes have distinct biological targets.
MATERIALS AND METHODS
Fly stocks.
Gal4 was expressed ubiquitously under the control of the heat shock promoter; in the wing pouch (especially the dorsal region) by using Gal4-MS1096 (13), an insertion in the Beadex gene (42); in a stripe along the A/P boundary and at lower levels in the A compartment by using Gal4-ptc (559.1) (29); and in epidermal cells by using Gal4-69B (9). UAS-Dp110 (37) and UAS-tor4021-EGFR (21) have been described previously. UAS-aos (54) was kindly provided by Mandy Simcox. An activated form of the EGFR (EGFRElpE1) hypomorphic (drk10626) (60) and antimorphic (drke0A) (59) alleles of drk, the loss-of-function mutations in Notch (N264-39 and notchoid), and the deficiencies Df(3L)vin2, Df(3L)vin3, Df(3L)vin4, Df(3L)vin6, and Df(3L)vin7 (1, 17) were obtained from Kathy Matthews at the Bloomington Stock Center. Df(3L)vin66 was provided by Christian Lehner. Dp110A, a deletion of the PI3K_92E gene, has been described previously (72).
Plasmid constructs.
Constructs were verified by their restriction sites and by sequence analysis with an Applied Biosystems 373A automated DNA sequencer. All PCR-amplified sequences were confirmed by sequencing. Sequences of the oligonucleotides used for PCR and site-directed mutagenesis are available upon request from the corresponding author.
PI3K_68D constructs.
All numbering of amino acids or nucleotides is from the initiating methionine (M1) or ATG (A1TG) in the PI3K_68D cDNA.
Plasmids carrying PI3K_68D (WT-PI3K_68D) and a catalytically inactive variant (KD-PI3K_68D) were generated in the pUAST vector, in which cDNAs are expressed under the control of five Gal4 binding sites (9). An N-terminal tag for the c-myc epitope (EQKLISEEDL) was incorporated. This is recognized by the monoclonal antibody 9E10 (23). The tag was introduced by using the BamH1 cassette in pSK (pSK-68D) (40) as a template. The myc-tagged cassette (pSKmyc68D) was then digested with BamHI and inserted into the BglII site of pUAST.
To generate the catalytically inactive PI3K_68D construct, Asp1457 (GAC) within the kinase domain was mutated to Ala (GC4370C) by using pSKmyc68D as a template and the Chameleon double-stranded site-directed mutagenesis kit (Stratagene). The EcoRV site (GATATC) was also altered (GT3354TATC) to facilitate selection of the mutated plasmid. The BamHI cassette incorporating the mutation was then inserted into the BglII site of pUAST. The same approach was used to introduce mutations (P407A, P410A, and R412Q) in the polyproline motif PPPLPPR to generate pSKmyc68DPII.
An EcoRI cassette incorporating the wild-type sequence was also generated by the PCR overlap extension method (30). Primers were used to mutate the two internal EcoRI sites (A4956G and A4965G) and to introduce an EcoRI site 3′ to both the stop codon and 3′ BamHI site. A 1.4-kb fragment incorporating these changes was amplified, digested with XhoI and Asp718, and exchanged for the Xho-Asp718 fragment in myc-tagged pUAST PI3K_68D.
To generate an amino-terminal fragment of PI3K_68D (N-PI3K_68D) containing the polyproline motifs R123MQPtNP129 and P455PPLPPR461, a 1,446-bp fragment corresponding to the first 470 amino acids of PI3K_68D was amplified from the WT-PI3K_68D construct (described above). A 5′ NdeI site and a 3′ XhoI site were introduced. The fragment was digested with these restriction enzymes and inserted into the same sites in the pET-16α His tag expression vector (Novagen). An amino-terminal fragment, NPII-PI3K_68D, containing mutations in the second polyproline motif PII (P407A, P410A, and R412Q) was generated in the same manner but with pSKmyc68DPII as a template. Mutations in the first polyproline motif, PI (R123Q, P126A, and P129A) were introduced in the N-PI3K_68D fragment by using the PCR overlap extension method to generate NPI-PI3K_68D. A further construct incorporating mutations in both polyproline motifs, NPI; PII-PI3K_68D, was generated by using the PCR overlap extension method with the singly mutated templates.
Generation of recombinant GST-SH3 fusion proteins.
The glutathione S-transferase (GST)-Drk, GST-DrkW36A, and GST-DrkW189A constructs were generously provided by Tony Pawson (46, 50). The GST-DrkW36A;W189A plasmid was produced by the double ligation of a 304-bp BamHI-BglI fragment (from the plasmid for GST-drkW36A) and a 331-bp BglI-EcoRI fragment (from the plasmid κencoding GST-DrkW189A) into the pGEXKT vector (Pharmacia) digested with BamHI and EcoRI. Plasmids encoding GST fusion proteins for the individual SH3 domains of Drk (N-terminal SH3, amino acids 1 to 72; C-terminal SH3, amino acids 154 to 211) were generated by PCR amplification with the GST-Drk construct as a template. The amplified fragments were digested with BamHI and EcoRI and inserted into the same sites in the pGEXKT vector.
The GST-p120 RasGAP SH3 domain was generated by Simon Woodcock from p120 RasGAP (24) and provided by David Hughes.
Plasmids encoding GST fusion proteins for the individual SH3 domains of Src64B (SwissProt accession number P00528; amino acids 96 to 158); Abl (SwissProt accession number P00522; amino acids 196 to 270); DPLCγ/small wing (sl) (Protein Identification Resource accession number A53970; amino acids 824 to 890); α-spectrin (SwissProt accession number P13395; amino acids 970 to 1043), and βH-spectrin/karst (kst) (Protein Identification Resource accession number A37792; amino acids 835 to 916) were generated by PCR amplification. First-strand cDNA, prepared from Drosophila poly(A)+ RNA as described previously (40), was used as a template. Restriction sites were incorporated at the 5′ (EcoRI) and 3′ (XhoI) ends to facilitate subcloning into these sites in the pGEXKG vector (26).
Southern hybridizations.
Genomic DNA was digested with XhoI, fractionated on 0.8% agarose gels, and transferred to Hybond N+ (Amersham). Probes corresponding to the 5′ and 3′ ends of the PI3K_68D coding sequence (nucleotides −26 to +1063 and + 5116 to +5979, respectively, numbered relative to the coding sequence) were generated by PCR and labeled to the same specific radioactivity with [γ-32P]dCTP by using a Multiprime kit (Amersham). Radioactive bands (5.7-kb 5′ fragment and ∼4.5-kb 3′ fragment) were detected and quantified with a PhosphorImager (Molecular Dynamics). Since the deficiency stocks are heterozygous, those which deleted the PI3K_68D gene gave a signal with half the intensity of an Oregon R control. Values were normalized between samples by using a probe for the Dp110 gene, PI3K_92E, which maps to a separate region of the Drosophila chromosome.
Generation of transformants.
Transgenic flies were generated by injecting Qiagen-purified plasmid DNA into yw embryos and selected on the basis of expression of the mini-white gene (7). For upstream activating sequence (UAS) constructs, several independent lines were generated for each plasmid to rule out insertion-specific effects. We found that different insertions for the same construct induced similar phenotypes with different degrees of severity.
Biochemical analyses. (i) Expression of transgenes.
Heat shock Gal4-induced expression of UAS transgenes was achieved by incubating adult flies at 37°C for 1 h and then at 25°C for 4 to 6 h. Flies were lysed in 20 mM HEPES (pH 7.5)-150 mM NaCl-2 mM EGTA-1.5 mM MgCl2-10% (vol/vol) glycerol containing 1% (wt/vol) Triton X-100, 0.1% β-mercaptoethanol, 5 mM benzamidine, 0.2 mM phenylmethylsulfonyl fluoride, and 100 mU of aprotinin (Sigma) ml−1. To maintain the phosphorylation state of the Drosophila EGFR (DER), the protein phosphatase inhibitors sodium orthovanadate (1 mM), β-glycerophosphate (10 mM), molybdate (1 mM), tetrasodium pyrophosphate (1 mM), and sodium fluoride (10 mM) were included. Lysates were clarified by centrifugation.
(ii) Expression of recombinant fusion proteins in bacteria.
Escherichia coli DH5α cells containing plasmids encoding the GST fusion proteins were grown to an optical density at 600 nm of 0.6. Expression of the fusion proteins was induced at 27°C with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h. Bacterial pellets were sonicated in phosphate-buffered saline lysis buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.3]) containing 1% Triton X-100, 2 mM EDTA, 5 mM benzamidine, 0.2 mM phenylmethylsulfonyl fluoride, and 0.1% mercaptoethanol and clarified by centrifugation. The extract was incubated with glutathione-agarose (Pharmacia) beads for 1 h at 4°C with rotation and washed three times with lysis buffer.
Expression of amino-terminal fragments of PI3K_68D was performed in BL21(DH3) cells essentially as for the GST fusion proteins.
(iii) In vitro binding assays.
Clarified extracts from flies and bacteria were incubated with immobilized GST or GST fusion proteins (20 μg) for 2 h at 4°C with rotation. Complexes bound to the beads were recovered by centrifugation, washed three times with lysis buffer, and heated to 95°C for 5 min in sodium dodecyl sulfate (SDS) sample buffer.
(iv) Immunoblotting.
Extracts were resolved on SDS-polyacrylamide gels, transferred to nitrocellulose, and incubated with 9E10 to detect the Myc epitope. For analysis of wild-type and catalytically inactive PI3K_68D, blots were incubated with 9E10 mouse ascites (1/10,000; Developmental Studies Hybridoma Bank) and detected with horseradish peroxidase-linked anti-mouse immunoglobulin G (1/1,000; DAKO) by using ECL (Amersham). For analysis of complexes bound to GST fusion proteins, detection was with horseradish peroxidase-coupled anti-mouse IgG (1/10,000; Pierce) and the SuperSignal West Pico chemiluminescent substrate (Pierce).
(v) Assay of PI3K activity.
PI3K assays were performed as described previously (40), using PI (Sigma) as a substrate, and resolved by thin-layer chromatography in chloroform-methanol-4.9 M ammonium hydroxide (50:39:11). The radioactivity was detected with a PhosphorImager (Molecular Dynamics).
Adult phenotypic analyses.
Wings from female flies were dehydrated in ethanol and mounted in Euparal (Agar Scientific). Images were collected with a Leaf Microlumina color charge-coupled device camera (ISS, Greater Manchester, United Kingdom).
RESULTS
Analysis of class II PI3K function by ectopic expression.
To identify the biological function of the class II PI3K in a multicellular organism, we expressed wild-type and mutated versions of PI3K_68D in different patterns during Drosophila imaginal disk development and looked for phenotypes in the adult cuticle. cDNAs for PI3K_68D were fused to an epitope tag and cloned into P-element expression vectors under the control of yeast Gal4 UASs (9).
Ectopic expression of the class I and class II PI3Ks generates distinct phenotypes.
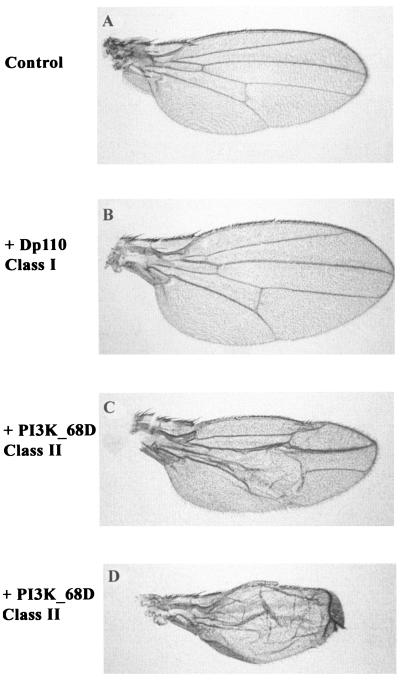
Ectopic expression of the class I PI3K, Dp110, has been shown to enhance organ size by increasing the size of cells without perturbing patterning (37, 72). To evaluate differences between the two classes of PI3Ks, the effects of their expression were directly compared by using a promoter that drives expression at high levels within the prospective wing blade. The wing is a flat structure, and hence changes in size can be easily visualized. The two PI3Ks generated distinct phenotypes (Fig. 1), indicating that class I and class II PI3Ks target distinct pathways. Ectopic expression of the class I PI3K generated a larger wing, as has been previously described (37). In contrast, ectopic expression of the class II PI3K, PI3K_68D, generated very severe developmental defects. When larvae were reared at 18°C rather than at 25°C to induce weaker expression of PI3K_68D, the phenotypic effects obtained were less severe although variable. An example of a weak phenotypic effect is shown in Fig. 1C. Effects on the patterning of the wing veins were observed. In addition, many of the wings were blistered, presumably because the two surfaces of the wing blade failed to adhere properly. In contrast to ectopic expression of the class I PI3K, overgrowth of the wing never occurred with ectopic PI3K_68D expression.
FIG. 1.
Ectopic expression of the class I PI3K affects organ size, while ectopic expression of the class II PI3K affects patterning. Wings of a female control fly (Gal4-MS1096/+) (A) or of female flies expressing wild-type Dp110 (Gal4-MS1096/+; UAS-WT-Dp110/+) (B) or PI3K_68D (Gal4-MS1096/+; UAS-WT-PI3K_68D/+) (C and D) in the precursor to the wing blade are shown. Crosses were performed at 25°C (A, B, and D) or 18°C (C).
While these phenotypes provided a dramatic illustration of the differences in signaling between the class I and class II PI3Ks, those generated by expression of PI3K_68D were too severe and variable to look for genetic interactions with other signaling components. We therefore also generated a version of PI3K_68D that was predicted to be inactive, in order to determine the effects of decreasing the level of class II PI3K signaling. This construct contained a mutation in an aspartate residue (D1457A) that lies within the activation loop of the PI3K catalytic domain and is predicted to contact a metal ion (70). The corresponding residue within protein kinases has been shown to be critical for catalytic activity.
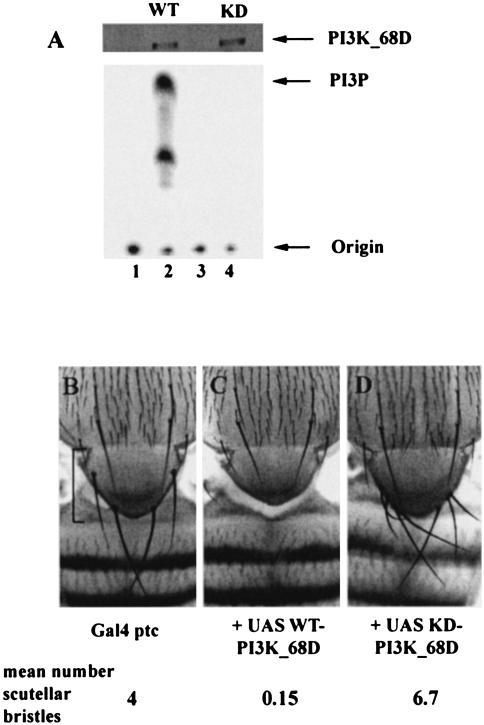
The biochemical properties of these proteins were assessed following heat shock-induced expression in adult flies. The ectopically expressed PI3K was specifically immunoprecipitated from detergent lysates, using an antibody to the epitope tag, and assayed for lipid kinase activity. Figure 2A shows that expression of the wild-type, p210, PI3K (WT-PI3K_68D) was induced following heat shock and that this protein possessed PI3K activity. As anticipated, the construct containing the D1457A mutation lacked detectable PI3K activity when expressed at levels similar to those of the wild-type protein. We have designated this transgene KD-PI3K_68D (for kinase dead). Since the mutant PI3K is unable to generate the downstream lipid signal, it is predicted to have an inhibitory or dominant-negative effect on the function of the endogenous PI3K by competing with the endogenous PI3K for binding sites on interacting molecules. Hence, expression of the wild-type or catalytically inactive PI3K_68D constructs should increase or decrease the levels of the lipid signal, respectively.
FIG. 2.
Lipid-kinase-dependent phenotypic effects generated by expression of PI3K_68D. (A). Mutation of Asp1457 to Ala within the active site of PI3K_68D generates a catalytically inactive protein. Detergent lysates were prepared from adult flies expressing UAS-PI3K_68D constructs under the control of Gal4-heat shock promoters. The extracts were immunoprecipitated with an antibody to the Myc (9E10) epitope and assayed for PI3K activity with PI as a substrate. Equivalent levels of the two constructs were induced as shown by immunodetection with 9E10, but only the wild-type construct possessed PI3K activity (lane 2). Flies were of the following genotypes: lane 1, Gal4-hs/CyO; lane 2, Gal4-hs, UAS-WT-PI3K_68D/CyO; lane 3 Gal4-hs/TM3; lane 4, Gal4-hs, UAS-KD-PI3K_68D/TM3. Arrows mark the positions of the PI3K_68D protein at ∼210 kDa, of PI3P, and of the origin. For each sample the equivalent of 0.5 or 2 female flies were used for the immunoblot (from extracts) and for the PI3K assay (from immunoprecipitations), respectively. Similar results were obtained in three separate experiments. (B to D) Ectopic expression of PI3K_68D affects the number of scutellar sensory bristles in a PI3K-dependent manner. Images of the thoraxes of adult female flies of genotypes Gal4-ptc/+ (B), Gal4-ptc/UAS-WT-PI3K_68D (C), and Gal4-ptc/+; UAS-KD-PI3K_68D/+ (D) are shown. The mean number of scutellar bristles for n = 40 nota is shown. The scutellum is indicated by a bracket. Normally, the four scutellar bristles point distally. In panel D some of these bristles remain curved upwards as in late stages of normal pupae, presumably because they fail to reorient themselves when the flies emerge. Crosses were performed at 25°C.
Ectopic expression of wild-type or catalytically inactive versions of PI3K_68D have opposing effects on bristle number.
In order to generate adult phenotypes, these PI3K_68D transgenic lines were crossed to a number of different enhancer-trap Gal4 lines to induce expression in specific developmental patterns within the larval imaginal disks. We have previously demonstrated that the endogenous enzyme is expressed and possesses lipid kinase activity at this developmental stage (40).
The expression domain of the patched (ptc) Gal4 line (29, 33) includes the scutellum, the shield-shaped structure at the base of the thorax. Expression of the WT- and KD-PI3K_68D transgenes with Gal4-ptc produced opposing and completely penetrant effects on the number of scutellar bristles (Fig. 2B to D). Enhancing the level of the lipid signal with the wild-type transgene resulted in a loss of bristles and was accompanied by a reduction in the size of the scutellum (Fig. 2C), while decreasing the level of the lipid signal with the KD transgene increased the number of the scutellar bristles (Fig. 2D). Hence, expression of catalytically active and inactive versions of PI3K_68D can elicit opposing phenotypic effects. Consistent with the effects of the class I PI3K on growth but not on patterning, expression of either wild-type or catalytically inactive versions of Dp110 with Gal4-ptc had essentially no effect on the number of scutellar bristles (results not shown).
Expression of the class II PI3K affects patterning of the wing imaginal disk.
The Gal4 line 69B induces strong expression within the epidermis. Staining with a β-galactosidase reporter line and with an antibody to the Myc epitope, which identifies expression of the PI3K transgene, showed that Gal4-69B promoted expression within imaginal disks, such as the wing, haltere, and leg disks, and a restricted expression within the eye disks (reference 9 and data not shown). Within the wing disk, expression appeared to be highest in regions that give rise to the wing margin and within parts of the wing blade and hinge. We describe phenotypes generated with Gal4-69B and the PI3K_68D transgenes in the wing imaginal disk that affected the specification of the wing veins, wing margins, and sensory elements and the apposition of the wing surfaces.
The adult wing blade consists of two layers of ectodermal cells (dorsal and ventral) and is divided into regions of vein and intervein tissue by five longitudinal veins composed of more compact, highly pigmented cells (Fig. 3A). The veins provide structural support and form channels for neurons and trachea. The wing is further characterized by sensory organs such as the campaniform sensillae on the wing. These pattern elements, the sensory organs and veins as well as the large sensory bristles (macrochaetae) on the notum, occur in very precise locations (reviewed in reference 61). These are defined within the imaginal disks largely by the spatially restricted domains of expression of the proneural genes, achaete and scute, and the vein-promoting gene, veinlet (also known as Rhomboid-1). veinlet encodes a Golgi protein that regulates cleavage and hence the biological activity of the DER ligand, Spitz (36). In the third-instar wing disk, veinlet promotes the spatially restricted activation of mitogen-activated protein kinase (MAPK) by DER, and this directs wing cells into a vein specification pathway (64). During pupal development MAPK signaling is switched off in the prospective vein cells, allowing their differentiation (41), and the pattern is refined by the Notch and decapentaplegic signaling pathways, which restrict veins to their characteristic widths (18).
FIG. 3.
Adult wing phenotypes generated by PI3K_68D transgenes with Gal4-69B. (A) A control wing expressing the Gal4-69B promoter alone generates an essentially wild-type wing characterized by five longitudinal veins (L1 to L5), two cross veins (anterior [acv] and posterior [pcv]), and a number of sensory elements, including the campaniform sensillae (A and G) on the distal portion of L3 (L3-1 to L3-3). (B) Expression of WT-PI3K_68D transgenes produced a small hole (arrow) within the proximal part of the first basal cell in the hinge region. This unusual phenotype was completely penetrant (at 25°C) and was observed with six different insertions. Frequently a hole was also observed in the haltere (not shown). (C to F) Most flies expressing KD-PI3K_68D under control of the Gal4-69B promoter produced mild ectopic wing vein material (C), but in a proportion of flies (D to F) a more severe range of patterning defects was observed. In panel D, a single ectopic cross vein (arrow) is produced between the distal end of L2 and vein L3. In panel E, multiple ectopic cross-veins are formed and there is a thickening of the longitudinal veins in the anterior region. Panels E and F show losses of portions of the wing margin. In panel E, the sensory bristles associated with the anterior wing margin occur at a higher density in the region proximal to the wing margin loss. (G and H) Section of L3 distal to the anterior cross vein which carries the L3-1 to L3-3 campaniform sensillae (see panel A). Flies expressing KD-PI3K_68D show supernumary campaniform sensillae (arrows) (H) compared to controls (Gal4-69B only) (G). The wings shown are from female flies of genotypes Gal4-69B/+ (A and G), WT-PI3K_68D/+; Gal4-69B/+ (B), and Gal4-69B/UAS-KD-PI3K_68D (C to F and H). Anterior is up; proximal is to the left.
Ectopic expression of the WT-PI3K_68D with Gal4-69B principally affected structures derived from the wing imaginal disk, which gives rise to the wing blade, hinge, and notum (dorsal thorax), although effects on the legs and halteres were also observed. The hinge region, which joins the wing blade to the notum, was particularly sensitive to perturbations. WT-PI3K_68D expression resulted in a “held-out” wing phenotype that was accompanied by a small hole in the hinge intervein (Fig. 3B). Although many mutations produce a held-out wing phenotype, this is not accompanied by the small hole characteristic of PI3K_68D overexpression. Holes in the proximal part of the wing have been reported for overexpression of the Rotund RacGap and have been attributed to defects in adhesion (27). Higher levels of expression of WT-PI3K_68D additionally affected patterning of the wing blade and produced blisters, as with Gal4-MS1096 (Fig. 1), and occasionally resulted in complete loss of the wing blade itself. With high levels of WT-PI3K_68D, effects on viability were also observed, presumably because of expression within the embryo.
At 25°C, most flies expressing the catalytically inactive version of PI3K_68D with the Gal4-69B driver showed very minor amounts of additional wing vein material (Fig. 3C). However, a small proportion of the progeny displayed more severe phenotypes affecting the specification of wing veins and wing margins (Fig. 3D to F). The penetrance of these phenotypes was increased when the larvae were reared at 29°C, consistent with the established temperature sensitivity of the Gal4-UAS system (Table 1). This results in higher expression of transgenes with increased temperature. The effects on wing veins were most prominent in anterior regions of the wing and were frequently accompanied by a change in the wing shape resulting from a reduction in the size of anterior intervein regions (see, e.g., Fig. 3E). In some cases only a single ectopic cross vein formed between the distal end of L2, where it meets the anterior margin, and L3 (Fig. 3D); in others more extensive ectopic wing venation occurred (as in Fig. 3E). KD-PI3K_68D expression also affected wing margin formation, resulting in losses of portions of either the posterior wing margin (Fig. 3F) or the anterior wing margin with the concomitant loss of margin bristles (Fig. 3E).
TABLE 1.
Catalytically inactive PI3K_68D wing phenotypes
| Phenotypea | % Penetrance atb:
|
|
|---|---|---|
| 25°C | 29°C | |
| Ectopic cross vein L2-L3 | 5 | 25 |
| Extensive ectopic wing venation | 5 | 20 |
| Wing margin loss | 2 | 20 |
The single cross vein L2-L3 phenotype is as shown in Fig. 3D.
Flies were raised at the indicated temperatures. Values are for 100 flies at 25°C and for 41 flies at 29°C.
Expression of KD-PI3K_68D also affected the external sense organs on the wing and thorax. The campaniform sensillae are dome-shaped sensory structures which are believed to sense strain on the wing cuticle (Fig. 3G). Expression of KD-PI3K_68D increased the number of the campaniform sensilla (Fig. 3H). As with the Gal4-ptc driver (Fig. 2D), Gal4-69B-mediated KD-PI3K_68D expression also generated supernumary bristles on the thorax (results not shown).
Specific mutations in components of the EGFR and Notch signaling pathways also affect wing veins, wing margins, and sensory structures, suggesting a link between these pathways and PI3K_68D signaling.
Class II PI3K interacts genetically with the EGFR and Notch signaling pathways.
The dosage sensitivity of KD-PI3K_68D expressed with the Gal4-69B promoter allowed us to use this combination to look for interactions with genes that give similar mutant overexpression phenotypes. This approach can be used to identify signaling pathways in which the class II PI3Ks might be involved. The observed effects on the specification of wing veins and wing margins suggested links with the EGFR and Notch signaling pathways. The results in Fig. 4 demonstrate that wild-type and catalytically inactive PI3K_68D transgenes can interact genetically, and in an opposing fashion, with components of the EGFR signaling pathway in wing vein development. Furthermore, catalytically inactive PI3K_68D showed interactions with the Notch pathway.
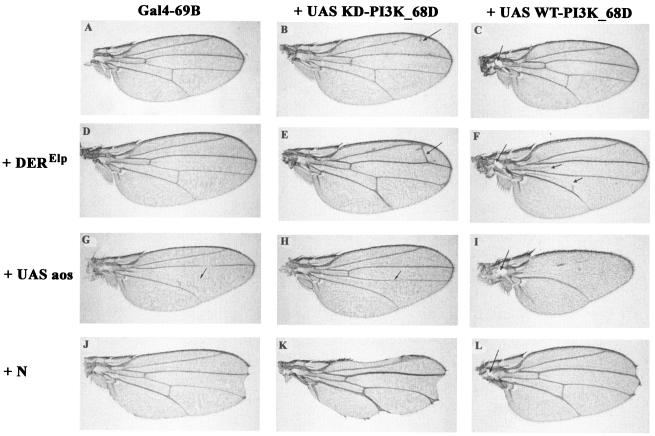
FIG. 4.
Genetic interactions between PI3K_68D transgene phenotypes and components of the Drosophila EGFR and Notch signaling pathways. (A to C) At 25°C, expression of Gal4-69B alone produced an essentially wild-type wing (A), while expression of a KD-PI3K_68D transgene produced only mild ectopic wing vein material (B) and expression of a WT-PI3K_68D transgene produced a small hole within the hinge region (C). (D to F) One copy of an activated form of DER (Elp) alone had little effect on wing venation (D) but enhanced the formation of an ectopic cross vein in flies expressing UAS-KD-PI3K_68D from 5 to 100% at 25°C (for n = 81 flies and for two different KD-PI3K_68D insertions) (E) and the loss of the anterior (and partial loss of the posterior) cross vein in flies expressing UAS-WT-PI3K_68D (F). (G to I) Expression of argos (aos), a secreted inhibitor of DER, with Gal4-69B produced a narrower wing in which L4 and the anterior cross vein were missing (G). Coexpression of KD-PI3K_68D rescued L4 and the anterior cross vein but resulted in the loss of the posterior cross vein (H). Coexpression of WT-PI3K_68D enhanced the loss of wing veins (I). In addition to the loss of L4, the anterior cross vein, and the posterior cross vein, most of L3 and parts of L2 were deleted. Remnants of the latter veins, where present, occupied medial positions. (J to L) Loss-of-function mutations at the Notch locus (such as N264-39) produce a characteristic notching of the wing margin at the anterior-posterior border (J). This was enhanced by expression of the UAS-KD-PI3K_68D transgene (K). In a small proportion of Notch flies, more severe wing notching is obtained. Similarly 2% of flies expressing catalytically inactive PI3K_68D at 25°C display a wing notching phenotype (Fig. 3E and F). Combinations of these more severe phenotypes presumably account for the small proportion of flies showing more severe interactions (not shown). Expression of WT-PI3K-68D did not enhance the loss-of-wing margin or the vein thickening of N264-39 (L). Flies were raised at 25°C. The wings shown are from female flies of genotypes Gal4-69B/+ (A), Gal4-69B/UAS-KD-PI3K_68D (B), UAS-WT-PI3K_68D/+; Gal4-69B/+ (C), Bc, EgfrElpE1/+; Gal4-69B/+ (D), Bc, EgfrElpE1/+; Gal4-69B, UAS-KD-PI3K_68D/+ (E), UAS-WT-PI3K_68D/+; Bc, EgfrElpE1/+; Gal4-69B/+ (F), Gal4-69B/UAS-aos (G), Gal4-69B, UAS-KD-PI3K_68D/UAS-aos (H), UAS-WT-PI3K_68D/+; Gal4-69B/UAS-aos (I), N264-39/+; Gal4-69B/+ (J), N264-39/+; Gal4-69B, UAS-KD-PI3K_68D/+ (K), and N264-39/UAS-WT-PI3K_68D; Gal4-69B/+ (L).
Ellipse (EgfrElpE1) is a weak hypermorph of DER in which the first amino acid of the kinase domain (A887T) is mutated (38). In transfected cells, this change results in ligand-independent autophosphorylation of the receptor. In the wing this mutation is associated with a weak extra-vein phenotype (6, 38). The ability of WT- and KD-PI3K_68D expression to modify EgfrElpE1-induced phenotypes was assessed. While a single copy of EgfrElpE1 did not affect ectopic cross vein formation (Fig. 4D), when combined with the KD-PI3K_68D transgene it enhanced the penetrance of the formation of an ectopic L2-L3 cross vein from 5% (Fig. 4B) to 100% (Fig. 4E) of the progeny at 25°C. Conversely, one copy of the EgfrElpE1 chromosome in combination with WT-PI3K_68D resulted in the loss of the anterior and posterior cross veins in a significant proportion of flies (Fig. 4F). These results show that the phenotype of a DER gain-of-function mutation can be enhanced by a catalytically inactive PI3K_68D and suppressed by a catalytically active PI3K_68D.
Argos is a secreted inhibitor of DER (reviewed in references 14 and 52). Ectopic expression of argos by using the Gal4-69B driver mimics loss-of-function mutations in DER. In the wing this results in the loss of wing veins, particularly L4 and the anterior cross vein, and a change in shape resulting from a loss of intervein tissue (Fig. 4G) (54). In addition, regions of L3 may be lost, and there is a prominent rough eye phenotype and a loss of the ocelli (simple eyes) and associated bristles on the head (reference 54 and data not shown). Coexpression of KD-PI3K_68D restored L4 to the wild-type pattern (Fig. 4H) without affecting the eye or ocelli phenotypes, while coexpression of the wild-type transgene enhanced the wing vein loss (Fig. 4I) with complete or partial loss of all veins except L5. Flies expressing argos and WT-PI3K_68D retained the prominent eye and ocelli phenotypes from argos expression and the hole in the hinge region generated by WT-PI3K_68D expression. Thus, targeted expression of WT-PI3K_68D enhanced and that of KD-PI3K_68D suppressed the wing vein loss generated by Argos, an inhibitor of the EGFR signaling pathway.
The wing margin forms at the dorsoventral compartment boundary following expression of transcription factors such as vestigial and cut, which are induced in response to Notch and wingless signaling. Consistent with this, specific loss-of-function mutations in Notch result in loss of parts of the wing margin (Fig. 4J). The loss of wing margin resulting from KD-PI3K_68D overexpression was enhanced by loss-of-function mutations in Notch such as N264-39 (Fig. 4K) and notchoid (results not shown), demonstrating that PI3K_68D can also interact genetically with the Notch signaling pathway. The Notch-dependent loss of the wing margin was not enhanced by WT-PI3K_68D (Fig. 4L).
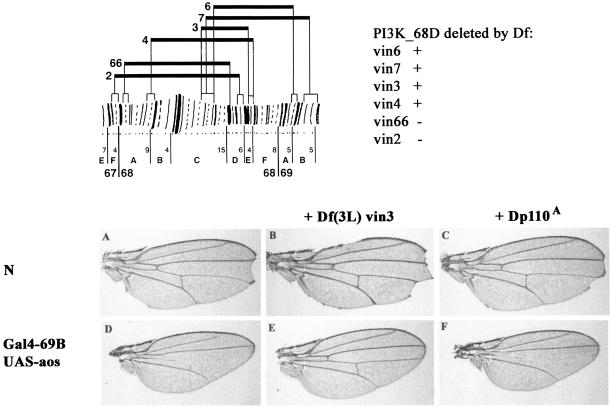
We were unable to identify specific mutations within the PI3K_68D gene among existing mutant stocks that mapped to this region. However, a deficiency which deleted one copy of the gene (Fig. 5), and hence presumably lowered the endogenous levels of the protein, interacted with the DER and Notch signaling pathway phenotypes in a manner similar to that for the catalytically inactive PI3K_68D transgenes. Hence, Df(3L)vin3 enhanced the wing margin loss associated with a Notch mutation and fully rescued the wing vein loss and shape change characteristic of argos overexpression (Fig. 5). In contrast, a deletion at the cytological position 92E, termed Dp110A, which specifically removes the class I PI3K, had no effect on the Notch and argos phenotypes. These results support our assumption that the catalytically inactive PI3K_68D acts in a dominant-negative manner and that the phenotypes it generates reflect a disruption of processes in which the endogenous PI3K_68D is involved. In addition, Df(3L)vin3 suppressed the loss-of-bristle phenotype generated by Gal4-ptc WT-PI3K_68D, consistent with the deficiency decreasing the level of class II PI3K activity (results not shown).
FIG. 5.
Genetic interactions between PI3K_68D deficiencies and components of the Drosophila EGFR and Notch signaling pathways. (Top) Region of chromosome arm containing the PI3K_68D gene. Deficiency mapping by Southern hybridization showed that the PI3K_68D locus is uncovered by the deficiencies Df(3L)vin3, Df(3L)vin4, Df(3L)vin6, and Df(3L)vin7 but excluded from Df(3L)vin2 and Df(3L)vin66, placing it at ∼68D6 to 68E3-4. This region probably contains five lethal complementation groups (31), including the genes cyclin A and brachyenteron. The chromosomal breakpoints of the vin deficiencies are from reference 1. (A to F) Notching of the wing margin by N264-39 (A) is enhanced by a deficiency, Df(3L)vin3, which deletes the class II PI3K, PI3K_68D (B), but not by Dp110A, which removes the class I PI3K, PI3K_92E (C). Similarly, the loss of the L4 wing vein on ectopic expression of argos (D) is rescued by Df(3L)vin3 (E) but unaffected by Dp110A (F). Crosses were performed at 25°C. The wings shown are from female flies of genotypes N264-39/+ (A), N264-39/+; Df(3L)vin3/+ (B), N264-39/+; Dp110A/+ (C), Gal4-69B, UAS-aos/+ (D), Gal4-69B, UAS-aos/Df(3L)vin3 (E), and Gal4-69B, UAS-aos/Dp110A (F).
PI3K_68D can bind the Drk adaptor protein in vitro.
In order to investigate the mechanism by which PI3K_68D can interact genetically with the DER signaling pathway, we initially looked for a direct interaction between PI3K_68D and DER. Following coexpression of PI3K_68D and an activated version of DER in adult flies, we were unable to identify the Myc-tagged PI3K_68D in phosphotyrosine immunoprecipitates of activated DER (results not shown). Analysis of the PI3K_68D sequence had indicated a class II polyproline motif within a region of low complexity towards the N terminus of the molecule. This was predicted to act as a potential binding site for SH3 domains (40). In fact, this sequence, PPPLPPR, is identical to P1, one of the three proline-rich motifs within the C-terminal region of SOS, a guanine nucleotide exchange factor that activates Ras (50). The polyproline motifs within SOS bind the SH3 domains of Drk, the Drosophila homologue of mammalian Grb2. Drk is an adaptor protein that contains a phosphotyrosine binding SH2 domain flanked by two polyproline binding SH3 domains. Hence, Drk can link receptor tyrosine kinases with the activation of Ras and the MAPK pathway. A further sequence within PI3K_68D, RMQPTNP, located at amino acids 123 to 129 contains a basic residue N terminal to the polyproline core and hence conforms to the consensus for class I polyproline motifs (+xxPxxP).
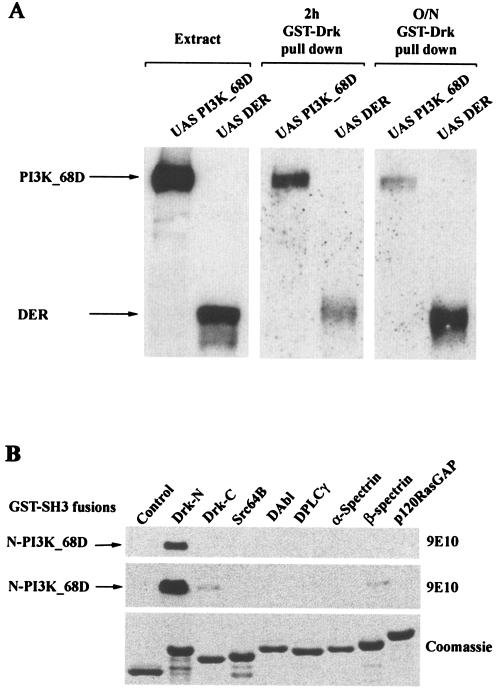
We therefore investigated whether the Myc-tagged wild-type PI3K_68D expressed in adult flies could bind a GST-Drk fusion protein. As a control, an activated version of DER was also used. Both PI3K_68D and DER could associate with Drk (Fig. 6A). The interaction with DER is presumably via an interaction between the Drk SH2 domain and phosphotyrosine residues on DER. For PI3K_68D the interaction is predicted to be via the polyproline motif(s) in the N terminus of PI3K_68D and the SH3 domain(s) of Drk.
FIG. 6.
PI3K_68D can associate with the adaptor protein Drk in vitro. (A) Extracts from 25 heat-shocked flies expressing Myc-tagged PI3K_68D or DER were incubated with GST-Drk (20 μg). Bound proteins equivalent to 1.5 flies (extracts) or 25 flies (pull-downs) were resolved by electrophoresis on an SDS-8% polyacrylamide gel, transferred to nitrocellulose, and detected with the 9E10 antibody. (B) Bacterial extracts expressing a Myc-tagged N-terminal fragment of PI3K_68D (20 μg) were incubated with GST alone (control) or with the different GST-SH3 fusion proteins shown (20 μg of each). An amount equivalent to 0.5 or 1% of each of the complexes bound to glutathione-Sepharose beads was resolved on an SDS-12% polyacrylamide gel, transferred to nitrocellulose, and detected with the 9E10 antibody. Upper and middle panels, proteins associated with 100 ng (upper) or 200 ng (middle) of GST-SH3 fusion proteins, respectively; lower panel, Coomassie blue-stained gel of the different fusion proteins (2 μg). Similar results were obtained in three separate experiments.
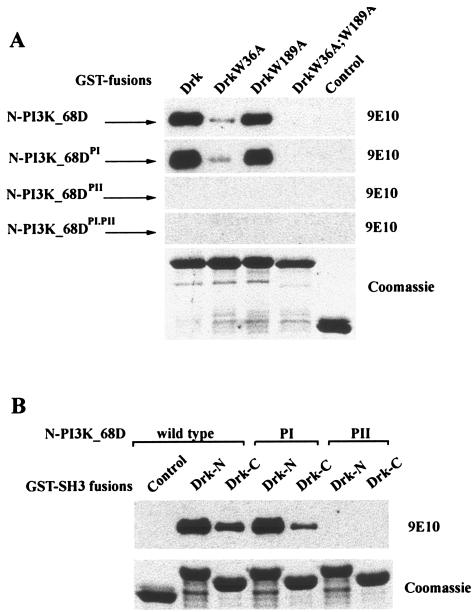
To investigate interactions mediated by these polyproline motifs in PI3K_68D, an N-terminal fragment containing the two polyproline motifs was generated and incubated with a panel of SH3 domains from a number of Drosophila signaling proteins. The N terminus of PI3K_68D showed a strong interaction only with the N-terminal SH3 domain of Drk. Weaker interactions were also observed with both the C-terminal SH3 domain of Drk and the SH3 domain of βH-spectrin (Fig. 6B). No interactions were observed with the SH3 domains from a number of other signaling proteins. These included Drosophila phospholipase Cγ and Src64B. The mammalian homologues of these proteins have been reported to bind the EGFR, and hence it is possible that their SH3 domains may localize interacting molecules such that they can modify EGFR signaling. Similarly, no interaction was detected with Drosophila p120 RasGAP, a negative regulator of Ras; with DAbl, a non-receptor tyrosine kinase; or with α-spectrin, a cytoskeletal protein. Although the SH3 domains tested represented only a small proportion of those present in Drosophila (90 have been identified to date in the PFAM database [http://pfam.wustl.edu/cgi-bin/getdesc?name=SH3]), these results clearly show that there is specificity in the interaction between PI3K_68D and the SH3 domains of Drk. To investigate further the contributions made by the two SH3 domains of Drk in binding to the N terminus of PI3K_68D, we used wild-type Drk and Drk variants containing mutations in the highly conserved tryptophan residues (W36 in N-terminal SH3 and W189 in C-terminal SH3) (50). Binding of Drk to the N terminus of PI3K_68D was almost but not completely abolished by mutation of the N-terminal SH3 domain (Fig. 7A, top panel). In contrast, although binding was largely unaffected by mutation of the C-terminal SH3 domain, a complete loss of binding occurred only when both domains were mutated. These results imply that the N-terminal SH3 domain is primarily responsible for the interaction of Drk with PI3K_68D.
FIG. 7.
A polyproline motif within PI3K_68D mediates binding to the SH3 domains of Drk. Bacterial extracts expressing Myc-tagged wild-type or mutant N-terminal fragments of PI3K_68D were incubated with the indicated GST-Drk fusion proteins (wild type or SH3 domain mutants) (20 μg). An amount equivalent to 0.5% of each of the complexes (100 ng of fusion protein) bound to glutathione-Sepharose beads was resolved on an SDS-12% polyacrylamide gel, transferred to nitrocellulose, and detected with the 9E10 antibody. The lower panel in each section shows a Coomassie blue-stained gel of the different fusion proteins (2.5 μg). Similar results were obtained in three separate experiments.
In order to identify the region within PI3K_68D responsible for the interaction, the binding experiments were repeated with proteins containing mutations in the critical proline and arginine residues within either or both of the identified polyproline motifs (RMQPTNP [PI] and PPPLPPR [PII]). Binding of Drk to the N-terminal fragment of PI3K_68D was largely unaffected by mutation of the first polyproline motif, PI (Fig. 7A, second panel), but the interaction was completely abolished by mutation of the second motif, PII (Fig. 7A, third panel). These results imply that a single polyproline motif within PI3K_68D can mediate the interaction with the adaptor protein Drk in vitro, primarily through binding to the N-terminal SH3 domain of Drk (Fig. 7A). To confirm these results, the wild-type and mutant (PI and PII) N-terminal PI3K_68D fragments were incubated with the isolated N- and C-terminal Drk SH3 domains. Consistent with the results obtained with the full-length Drk protein, binding of PI3K_68D to the N-terminal SH3 domain of Drk was significantly greater than that to the C-terminal SH3 domain and the interaction with both the N- and C-terminal SH3 domains was completely abolished when the PII polyproline motif was mutated (Fig. 7B). However, the results also implied that the C-terminal SH3 domain of Drk when expressed alone could bind to the PI sequence, as binding to the N-terminal fragment of PI3K_68D was consistently reduced when this PI sequence was mutated (Fig. 7B). Binding of the N-terminal fragment of PI3K_68D to the N-terminal SH3 domain was unaffected by mutation of the PI sequence (Fig. 7B), as was shown for the full-length Drk (Fig. 7A).
Genetic interactions between drk and a class II PI3K.
In order to address the in vivo significance of the interaction observed between PI3K_68D and Drk, we looked for a genetic interaction between the PI3K_68D transgenes and the drk mutants. Since Drk acts downstream of the EGFR in wing vein formation (19a), drk loss-of-function alleles would be anticipated to suppress ectopic wing veins. While a single copy of either drk10626 or drke0A alone had no effect on wing vein formation, the drke0A allele suppressed the formation of ectopic veins generated by KD-PI3K_68D (Table 2). In the drke0A allele, His106 in the SH2 domain is mutated to Tyr (46). Since this mutant is probably unable to bind to tyrosine-phosphorylated proteins but may still bind to SOS, it most likely has a dominant-negative effect. Hence, it is likely to behave as a stronger mutant allele than the drk10626 hypomorphic allele, for which we were unable to observe an interaction. No interaction was observed between the drk mutant alleles and the WT-PI3K_68D transgene; this may be because the normal wing veins are less sensitive to perturbations than ectopic venation.
TABLE 2.
Effect of drk alleles on catalytically inactive PI3K_68D wing phenotypes
| drk allele | % Penetrance of phenotypea
|
|
|---|---|---|
| Single cross vein L2-L3 | Multiple ectopic cross veins | |
| − | 22 | 7 |
| drk10626 hypomorph | 25 | 2 |
| drke0A antimorph | 3 | 0 |
The single cross vein L2-L3 phenotype is as shown in Fig. 3D. Flies were raised at 30°C. Values are for 100 flies of each genotype.
DISCUSSION
Ectopic expression of different classes of PI3Ks generates distinct phenotypes.
We embarked on a phenotypic analysis of the class II Drosophila PI3K in order to identify developmental systems and signaling pathways in which this class of PI3K might be involved. In the absence of a mutation in the endogenous gene, we expressed PI3K_68D within Drosophila imaginal disks and looked at the effects on the adult cuticle. Class I and class II PI3Ks differ in their in vitro substrate specificity, and hence their presumed lipid signal, and in their domain structures, suggesting that they interact with different molecules. Consistent with these biochemical and structural properties, expression of these distinct classes of PI3Ks in vivo produced markedly different effects (Fig. 1).
Ectopic expression of the class I PI3K (PI3K_92E/Dp110) within imaginal disks generated effects on growth and final organ size without perturbing patterning. Moreover, this effect was observed in all adult structures derived from regions of Dp110 overexpression in the larval disks (37). In contrast, we found that PI3K_68D affected pattern elements, particularly the wing veins and margins (Fig. 3) and the sensory bristles on the notum and head. Notably, while patterning of the adult structures derived from the wing disk was affected by PI3K_68D expression with Gal4-69B, the eye appeared phenotypically normal. Similarly, the EGFR ligand, vein, affects wing veins but not the eye when expressed with Gal4-69B (reference 54 and data not shown). However, ectopic expression of a number of other molecules with this driver, such as the EGFR DER, argos, and PI3K_92E/Dp110, produced markedly rough or enlarged eyes in addition to the wing effects (reference 54 and unpublished observations). The lack of a detectable phenotype in the eye could be due to a threshold effect. Hence, the apparently ubiquitous effects of Dp110 expression may reflect a more linear response of growth and/or cell size to growth signals, while a threshold of signal may be required to trigger the changes in patterning observed with PI3K_68D.
Genetic interactions between PI3K_68D and the DER signaling pathway.
The EGFR performs multiple functions in Drosophila development (reviewed in references 14 and 55). In imaginal disks, these functions include the specification of cell fate and the survival of postmitotic cells (21). Consequently, a variety of patterning defects are observed in the adult cuticle following an earlier reduction in DER function. These include the deletion of certain wing veins (principally L4 and the anterior cross vein), the duplication or elimination of specific sensory bristles, roughened compound eyes, and shrunken or missing ocelli (15). In the context of the wing imaginal disk, DER is locally activated within all vein primordia by veinlet and signals via the Ras-MAPK pathway to specify vein cell fate. Thus, activated (6) or overexpressed DER produces ectopic wing veins. Since expression of the catalytically inactive PI3K_68D generates a similar phenotype, we looked for genetic interactions between these molecules.
A hypermorphic mutation in DER, known as Ellipse, was able to enhance (Fig. 4E), and an antimorphic mutation in drk was able to suppress (Table 2), the penetrance of the ectopic cross vein phenotype produced by expressing the KD-PI3K_68D transgene. Similarly, this transgene suppressed the loss-of-veins phenotype induced by expression of the inhibitory DER ligand, argos (Fig. 4H), while the vein loss was enhanced by active PI3K_68D (Fig. 4I). These results suggest that PI3K_68D may act antagonistically to the Ras-MAPK pathway.
Examination of the sequence of PI3K_68D had previously identified a putative SH3 binding motif (40). In vitro binding studies demonstrated that this motif could interact with both SH3 domains of Drk (Fig. 6 and 7), an adaptor protein that links activated receptor tyrosine kinases such as DER to the Ras-MAPK signaling pathway. While the binding was mediated primarily by the N-terminal SH3 domain, the interaction was completely abolished only by the mutation of both N- and C-terminal SH3 domains. These results are consistent with the conservation in sequence between the class II polyproline motif in PI3K_68D and the P1 sequence in SOS, which interacts primarily but not exclusively with the N-terminal SH3 domain of Drk. In SOS the P1 sequence represents the highest-affinity site for Drk binding, but two further sequences may contribute to this effect.
The EGFR family in mammals has also been shown to recruit PI3Ks, although the mechanism involved is unclear. Indeed, one of the most frequent alterations in the EGFR that has been identified in human tumors (a deletion of exons 2 to 7, termed EGFRvIII, that results in ligand-independent receptor activation) may exert its transforming activity through a PI3K pathway (45). Recent results have indicated that the EGFR may signal through class II, in addition to class I, PI3Ks. First, only one of the four EGFR variants identified in mammals, ErbB3, contains the specific YXXM motifs recognized by the class IA adaptor subunit, yet EGF stimulation of PI3K activity occurs in a number of cell types by an ErbB3-independent mechanism (62). Second, DER, which is equally similar structurally to the four mammalian EGFRs, lacks the YXXM motifs required for association with the class I PI3K. Third, overexpression of the class IA PI3K, Dp110, does not affect wing venation (37) (Fig. 1). Recently the mammalian class II PI3Ks, PI3K-C2α and PI3K-C2β, have been identified in association with the EGFRs ErbB-1/ErbB-2 (2), implicating these mammalian class II PI3Ks in EGFR signaling. Within the N terminus of PI3K-C2β there is a cluster of three class II polyproline motifs analogous to the P1 to P3 sequences in SOS that bind Drk (73). Further analysis of the interaction between PI3K-C2β and the EGFR demonstrated that these sequences were required for the interaction and that they appeared to bind both the N- and C-terminal SH3 domains of the Drk homologue Grb2 (73). However, while PI3K-C2α has been reported to bind the activated EGFR, it lacks class II polyproline motifs. Furthermore, the third mammalian class II PI3K, PI3K-C2γ, also lacks these sequences. These structural differences indicate that the interaction with Grb2 cannot be essential for the function of all class II PI3Ks. The single C. elegans class II PI3K, however, contains multiple class II polyproline motifs within the N terminus which might interact with the Grb2 homologue, SEM-5. Hence, possession of a class II PI3K containing a class II polyproline motif appears to be conserved among multicellular organisms.
The Drk family of adaptor proteins has been shown to interact with a number of different molecules. In addition to interacting with the polyproline motifs in SOS primarily via the N-terminal SH3 domain (50), the C-terminal SH3 domain can bind the receptor substrate DOS via a novel motif (25). Both of these interactions are anticipated to activate the Ras pathway. In addition Drk/Grb2 family members have been reported to interact with a number of molecules that negatively regulate receptor tyrosine kinase signaling. Among these is c-Cbl, an E3 ubiquitin ligase that marks the EGFR for endocytosis and hence degradation (71). In worms, a tyrosine kinase, ARK-1, has also been shown to inhibit EGFR signaling, and it is dependent upon the Grb2 homologue SEM-5 for this function (32).
Our results suggest that PI3K_68D may negatively regulate Ras-MAPK signaling downstream of the EGFR. However, this cannot simply be due to PI3K_68D competing with SOS for Drk and hence reducing signaling through SOS to Ras and the MAPK pathway, because the wild-type and catalytically inactive transgenes exerted opposite effects on modified EGFR signaling (Fig. 4). Hence, the interaction with Drk may be a mechanism for localization that allows the catalytic activity of PI3K_68D to interfere with the Ras-MAPK pathway. The 3-phosphorylated lipids produced by PI3K_68D might induce downregulation of DER-mediated Ras-MAPK signaling via stimulation of, for example, an endocytic pathway. In this situation, expression of the catalytically inactive transgene could displace the endogenous PI3K_68D but be unable to promote endocytosis. This would enhance DER signaling at the plasma membrane and hence wing vein formation. In mammalian cells, receptor-mediated endocytosis has been shown to be important in the downregulation of the ErbB1 EGFR in response to its ligand EGF (reviewed in reference 56). Alternatively, these lipids could interfere with the activation of EGFR either directly or, for example, via the Notch signaling pathway. While receptor-mediated endocytosis downregulates EGF receptor signaling, this process appears to be required for ligand-stimulated Notch signaling (57; reviewed in reference 56).
Clearly, it will be important to mutate the polyproline motif in PI3K_68D and examine the effects in vivo.
Genetic interactions between PI3K_68D and the Notch signaling pathway.
Notch has multiple roles in development, particularly in the control of cell fate (reviewed in references 5 and 10). In mammals, a number of diseases, including T-cell lymphoma and mammary gland tumors, have been linked to mutations in the human Notch genes (5). In Drosophila, loss-of-function phenotypes for Notch include an increase in the number of sensory bristles, a loss of wing margins, and thickened veins. All of these phenotypes were also observed on expression of catalytically inactive PI3K_68D. Furthermore, a deficiency which deletes PI3K_68D (Fig. 5) or the expression of KD-PI3K_68D (Fig. 4K) enhanced the wing notching phenotypes generated by specific Notch mutations (Fig. 4J). Although loss-of-function Notch mutations characteristically produce thicker veins, ectopic cross vein formation as observed for KD-PI3K_68D expression (Fig. 3D) has been induced through overexpression of an inhibitory version of Notch, the Notch extracellular domain (51). Furthermore, activated versions of Notch (the Abruptex mutations) delete portions of the wing veins. Notch frequently acts antagonistically to the EGFR in cell fate specification (6, 19, 49; reviewed in reference 52). Hence, these phenotypes indicate that PI3K_68D may be acting downstream of, or parallel to, Notch in a pathway that negatively regulates EGFR signaling. Alternatively, PI3K_68D may influence Notch signaling through acting as a negative regulator of the EGFR pathway. There are several potential mechanisms for antagonism between these pathways. For example, Notch negatively regulates veinlet transcription in intervein cells and thus restricts veins to their characteristic widths. If PI3K_68D is a component of this pathway, expression of the catalytically inactive transgene may allow ectopic vein formation by relieving the inhibition on veinlet transcription in intervein regions. Veinlet, like PI3K_68D, potently affects EGFR signaling in the wing but not in the eye. Notch signaling can also upregulate yan, an Ets transcriptional repressor of multiple RTK signaling targets (53). In worms, receptor tyrosine kinase signaling can be inhibited through the Notch-dependent induction of LIP-1, a MAPK phosphatase (8). Conversely ebi and strawberry notch, two transcriptional targets of EGFR signaling in flies, can suppress Notch signaling (65).
Conclusions.
The phenotypes that we have identified by ectopic expression demonstrate that class I and class II PI3Ks target distinct biological pathways. Whereas expression of the class I PI3K affects growth, our results show that expression of the class II PI3K perturbs patterning processes and may do so via effects on signaling by EGFR and the Notch receptor. Furthermore, the opposing phenotypes obtained with wild-type and catalytically inactive versions of PI3K_68D indicate a role for the class II PI3K-generated lipid signal in these processes. While it is necessary to be cautious in interpreting overexpression data, our results obtained by using deficiencies are consistent with the catalytically inactive PI3K_68D acting as a dominant-negative and disrupting processes in which the endogenous PI3K_68D is involved.
The adult cuticle phenotypes that we have identified from PI3K_68D expression can additionally be used for structure-function analyses of PI3K_68D and to screen for mutations, both within PI3K_68D itself and in genetically interacting molecules. We anticipate that these might include mutations in the EGFR and Notch signaling pathways. The identification of biological targets specific to class II PI3Ks will greatly facilitate the elucidation of specific roles for this class of PI3Ks in signaling pathways in both flies and mammals.
Acknowledgments
This work was supported by grants from the BBSRC and from the Royal Society and by the Ludwig Institute. E.H. is supported by a grant from the Swiss National Science Foundation.
We thank Kathy Matthews and the Bloomington Stock Center, Mandy Simcox, and Christian Lehner for the fly stocks used in this study; the Developmental Studies Hybridoma Bank for the 9E10 supernatant; Tony Pawson for the GST-Drk, GST-DrkW36A, and GST-DrkW189A constructs; Simon Woodcock and David Hughes for the SH3 domain of p120 RasGAP; Brenda Catelani, Alan Entwhistle, Krishna Pitrola, and Heather Phillips for technical assistance; Juan Riesgo-Escovar and Christoph Hugentobler for help in generating transgenic lines; Jenny Higgs for advice on imaging thoraxes; Carmen Coelho, María Domínguez, Enrique Martín-Blanco, and members of the Hafen lab, past and present, for useful discussions; and David Hughes for constructive criticism of the manuscript.
REFERENCES
- 1.Akam, M. E., D. B. Roberts, G. P. Richards, and M. Ashburner. 1978. Drosophila: the genetics of two major larval proteins. Cell 13:215-225. [DOI] [PubMed] [Google Scholar]
- 2.Arcaro, A., S. Volinia, M. J. Zvelebil, R. Stein, S. J. Watton, M. J. Layton, I. Gout, K. Ahmadi, J. Downward, and M. D. Waterfield. 1998. Human phosphoinositide 3-kinase C2_β—the role of calcium and the C2 domain in enzyme activity. J. Biol. Chem. 273:33082-33090. [DOI] [PubMed] [Google Scholar]
- 3.Arcaro, A., M. J. Zvelebil, C. Wallasch, A. Ullrich, M. D. Waterfield, and J. Domin. 2000. Class II phosphoinositide 3-kinases are downstream targets of activated polypeptide growth factor receptors. Mol. Cell. Biol. 20:3817-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arcaro, A., U. K. Khanzada, B. Vanhaesebroeck, T. D. Tetley, M. D. Waterfield, and M. J. Seckl. 2002. Two distinct phosphoinositide 3-kinases mediate polypeptide growth factor-stimulated PKB activation. EMBO J. 21:5097-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signalling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 6.Baker, N. E., and G. M. Rubin. 1992. Ellipse mutations in the Drosophila homologue of the EGF receptor affect pattern formation, cell division and cell death in eye imaginal discs. Dev. Biol. 150:381-396. [DOI] [PubMed] [Google Scholar]
- 7.Basler, K., B. Christen, and E. Hafen. 1991. Ligand-independent activation of the Sevenless receptor tyrosine kinase changes the fate of cells in the developing Drosophila eye. Cell 64:1069-1081. [DOI] [PubMed] [Google Scholar]
- 8.Berset, T., E. F. Hoier, G. Battu, S. Canevascini, and A. Hajnal. 2001. Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science 291:1055-1058. [DOI] [PubMed] [Google Scholar]
- 9.Brand, A. H., and N. Perrimon. 1993. Targetted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 10.Bray, S. 1998. Notch signalling in Drosophila: three ways to use a pathway. Semin. Cell Dev. Biol. 9:591-597. [DOI] [PubMed] [Google Scholar]
- 11.Brown, R., L. Ho, S. Weber-Hall, J. M. Shipley, and M. J. Fry. 1997. Identification and cDNA cloning of a novel mammalian C2 domain-containing phosphoinositide 3-kinase, HsC2-PI3K. Biochem. Biophys. Res. Commun. 233:537-544. [DOI] [PubMed] [Google Scholar]
- 12.Brown, R., J. Domin, A. Arcaro, M. D. Waterfield, and P. R. Shepherd. 1999. Insulin activates the α isoform of class II phosphoinositide 3-kinase. J. Biol. Chem. 274:14529-14532. [DOI] [PubMed] [Google Scholar]
- 13.Capdevila, J., and I. Guerrero. 1994. Targetted expression of the signalling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J. 13:4459-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casci, T., and M. Freeman. 1999. Control of EGF receptor signalling: lessons from fruitflies. Cancer Metast. Rev. 18:181-201. [DOI] [PubMed] [Google Scholar]
- 15.Clifford, R. J., and T. Shübach. 1989. Coordinately and differentially mutable activities of torpedo, the Drosophila melanogaster homologue of the vertebrate EGF receptor gene. Genetics 123:771-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen, S. M. 1993. Imaginal disc development, p. 747-842. In M. Bate and A. Martinez-Arias (ed.), The development of Drosophila melanogaster, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Crosby, M. A., and E. M. Meyerowitz. 1986. Lethal mutations flanking the 68C glue gene cluster on chromosome 3 of Drosophila melanogaster. Genetics 112:785-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Celis, J. F., S. Bray, and A. Garcia-Bellido. 1997. Notch signalling regulates Veinlet expression and establishes boundaries between veins and interveins in the Drosophila wing. Development 124:1919-1928. [DOI] [PubMed] [Google Scholar]
- 19.Diaz-Benjumea, F. J., and A. Garcia-Bellido. 1990. Behaviour of cells mutant for an EGF receptor homologue of Drosophila in genetic mosaics. Proc. R. Soc. London Biol. 242:36-44. [DOI] [PubMed] [Google Scholar]
- 19a.Diaz-Benjumea, F. J., and E. Hafen. 1994. The sevenless signalling cassette mediates Drosophila EGF receptor function during epidermal development. Development 120:569-578. [DOI] [PubMed] [Google Scholar]
- 20.Domin, J., F. Pages, S. Volinia, S. E. Rittenhouse, M. J. Zvelebil, R. C. Stein, and M. D. Waterfield. 1997. Cloning of a human phosphoinositide 3-kinase with a C2 domain that displays reduced sensitivity to the inhibitor wortmannin. Biochem. J. 326:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domínguez, M., J. D. Wasserman, and M. Freeman. 1998. Multiple functions of the EGF receptor in Drosophila eye development. Curr. Biol. 8:1039-1048. [DOI] [PubMed] [Google Scholar]
- 22.Dove, S. P., F. T. Cooke, M. R. Douglas, L. G. Sayers, P. J. Parker, and R. H. Michell. 1997. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature 390:187-192. [DOI] [PubMed] [Google Scholar]
- 23.Evan, G. I., G. K. Lewis, G. Ramsay, and G. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5:3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldmann, P., E. N. Eischer, S. J. Leevers, E. Hafen, and D. A. Hughes. 1999. Control of growth and differentiation by Drosophila RasGAP, a homolog of p120 Ras-GTPase-activating protein. Mol. Cell. Biol. 19:1928-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feller, S. M., H. Wecklein, M. Lewitzky, E. Kibler, and T. Raabe. 2002. SH3 domain-mediated binding of the Drk protein to Dos is an important step in signalling of Drosophila receptor tyrosine kinases. Mech. Dev. 116:129-139. [DOI] [PubMed] [Google Scholar]
- 26.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 27.Guichard, A., E. Bergeret, and R. Griffin-Shea. 1997. Overexpression of Rn RacGAP in Drosophila melanogaster deregulates cytoskeletal organisation in cellular embryos and induces discrete imaginal phenotypes. Mech. Dev. 61:49-62. [DOI] [PubMed] [Google Scholar]
- 28.Herman, P. K., and S. D. Emr. 1990. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:6742-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinz, U., B. Giebel, and J. A. Campos-Ortega. 1994. The basic-helix-loop-helix domain of Drosophila lethal of scute protein is sufficient for proneural function and activates neurogenic genes. Cell 76:77-87. [DOI] [PubMed] [Google Scholar]
- 30.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 31.Hoogwerf, A. M., M. Akam, and D. Roberts. 1988. A genetic analysis of the rose-gespleten region (68C8-69B5) of Drosophila melanogaster. Genetics 118:665-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hopper, N. A., J. Lee, and P. W. Sternberg. 2000. Ark-1 inhibits EGFR signalling in C. elegans. Mol. Cell 6:65-75. [PubMed] [Google Scholar]
- 33.Johnson, R. L., J. K. Grenier, and M. P. Scott. 1995. Patched overexpression alters wing disc size and pattern: transcriptional and post-transcriptonal effects on Hedgehog targets. Development 121:4161-4170. [DOI] [PubMed] [Google Scholar]
- 34.Kay, B. R., M. P. Williamson, and M. Sudol. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14:231-241. [PubMed] [Google Scholar]
- 35.Kispert, A., B. G. Herrmann, M. Leptin, and R. Reuter. 1994. Homologs of the mouse Brachyury gene are involved in the specification of posterior terminal structures in Drosophila, Tribolium, and Locusta. Genes Dev. 8:2137-2150. [DOI] [PubMed] [Google Scholar]
- 36.Lee, J. R., S. Urban, C. F. Garvey, and M. Freeman. 2001. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell 107:161-171. [DOI] [PubMed] [Google Scholar]
- 37.Leevers, S. J., D. Weinkove, L. K. MacDougall, E. Hafen, and M. D. Waterfield. 1996. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 15:6584-6594. [PMC free article] [PubMed] [Google Scholar]
- 38.Lesokhin, A. M., S. Y. Yu, J. Katz, and N. E. Baker. 1999. Several levels of EGF receptor signalling during photoreceptor specification in wild-type, Ellipse, and null mutant Drosophila. Dev. Biol. 205:129-144. [DOI] [PubMed] [Google Scholar]
- 39.Linassier, C., L. K. MacDougall, J. Domin, and M. D. Waterfield. 1997. Molecular cloning and biochemical characterisation of a Drosophila PI-specific phosphoinositide 3-kinase. Biochem. J. 321:849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacDougall, L. K., J. Domin, and M. D. Waterfield. 1995. A family of phosphoinositide 3-kinases in Drosophila identifies a new mediator of intracellular signalling. Curr. Biol. 5:1404-1415. [DOI] [PubMed] [Google Scholar]
- 41.Martín-Blanco, E., F. Roch, E. Noll, A. Baonza, J. B. Duffy, and N. Perrimon. 1999. A temporal switch in DER signalling controls the specification and differentiation of veins and interveins in the Drosophila wing. Development 126:5739-5747. [DOI] [PubMed] [Google Scholar]
- 42.Milan, M., F. J. Diaz-Benjumea, and S. M. Cohen. 1998. Beadex encodes an LMO protein that regulates Apterous LIM-homeodomain activity in Drosophila wing development: a model for LMO oncogene function. Genes Dev. 12:2912-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Misawa, H., M. Ohtsubo, N. Copeland, D. Gilbert, N. Jenkins, and A. Yoshimura. 1998. Cloning and characterisation of a novel class II phosphoinositide 3-kinase containing a C2 domain. Biochem. Biophys. Res. Commun. 244:531-539. [DOI] [PubMed] [Google Scholar]
- 44.Molz, L., Y. Chen, and L. T. Williams. 1996. Cpk is a novel class of Drosophila PtdIns 3-kinase containing a C2 domain. J. Biol. Chem. 271:13892-13899. [DOI] [PubMed] [Google Scholar]
- 45.Moscatello, D. K., M. Holgado-Madruga, D. R. Emlet, R. B. Montgomery, and A. J. Wong. 1998. Constitutive activation of phosphatidylinositol 3-kinase by a naturally occurring mutant epidermal growth factor receptor. J. Biol. Chem. 273:200-206. [DOI] [PubMed] [Google Scholar]
- 46.Olivier J.-P., T. Raabe, M. Henkemeyer, B. Dickson, G. Mbamalu, B. Margolis, J. Schlessinger, E. Hafen, and T. Pawson. 1993. A Drosophila SH2-SH3 adaptor protein implicated in coupling the sevenless tyrosine kinase to an activator of Ras guanine nucleotide exchange, Sos. Cell 73:179-191. [DOI] [PubMed] [Google Scholar]
- 47.Ono, F., T. Nakagawa, S. Saito, Y. Owada, H. Sakagami, K. Goto, M. Suzuki, S. Matsuno, and H. Kondo. 1998. A novel class II phosphoinositide 3-kinase predominantly expressed in liver and its enhanced expression during liver regeneration. J. Biol. Chem. 273:7731-7736. [DOI] [PubMed] [Google Scholar]
- 48.Ponting, C. P. 1996. Novel domains in NADPH oxidase subunits, sorting nexins, and PtdIns 3-kinases: binding partners of SH3 domains? Protein Sci. 5:2353-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price, J. V., E. D. Savenye, D. Lum, and A. Breitkreutz. 1997. Dominant enhancers of Egfr in Drosophila melanogaster: genetic links between the Notch and EGFR signalling pathways. Genetics 147:1139-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raabe, T., J. P. Olivier, B. Dickson, X. Liu, G. D. Gish, T. Pawson, and E. Hafen. 1995. Biochemical and genetic analysis of the Drk SH2/SH3 adaptor protein of Drosophila. EMBO J. 14:2509-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rebay, I., R. G. Fehon, and S. Artavanis-Tsakonas. 1993. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell 74:319-329. [DOI] [PubMed] [Google Scholar]
- 52.Rebay, I. 2002. Keeping the receptor tyrosine kinase signalling pathway in check: lessons from Drosophila. Dev. Biol. 251:1-17. [DOI] [PubMed] [Google Scholar]
- 53.Rohrbaugh, M., E. Ramos, D. Nguyen, M. Price, Y. Wen, and Z. C. Lai. 2002. Notch activation of yan expression is antagonized by RTK/pointed signaling in the Drosophila eye. Curr. Biol. 12:576-581. [DOI] [PubMed] [Google Scholar]
- 54.Schnepp, B., T. Donaldon, G. Grumbling, S. Ostrowski, R. Schweitzer, B.-Z. Shilo, and A. Simcox. 1998. EGF domain swap converts a Drosophila EGF receptor activator into an inhibitor. Genes Dev. 12:908-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schweitzer, R., and B.-Z. Shilo. 1997. A thousand and one roles for the Drosophila EGF receptor. Trends Genet. 13:191-196. [DOI] [PubMed] [Google Scholar]
- 56.Seto, E. S., H. J. Bellen, and T. E. Lloyd. 2002. When cell biology meets development: endocytic regulation of signalling pathways. Genes Dev. 16:1314-1336. [DOI] [PubMed] [Google Scholar]
- 57.Seugnet, L., P. Simpson, and M. Haenlin. 1997. Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev. Biol. 192:585-598. [DOI] [PubMed] [Google Scholar]
- 58.Shao, X., C. Li, I. Fernandez, X. Zhang, T. C. Südhof, and J. Rizo. 1997. Synaptotagmin-syntaxin interaction: the C2 domain as a Ca2+-dependent electrostatic switch. Neuron 18:133-142. [DOI] [PubMed] [Google Scholar]
- 59.Simon, M. A., D. D. Bowtell, G. S. Dodson, T. R. Laverty, and G. M. Rubin. 1991. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell 67:701-716. [DOI] [PubMed] [Google Scholar]
- 60.Simon, M. A., G. S. Dodson, and G. M. Rubin. 1993. An SH3-SH2-SH3 protein is required for p21Ras1 activation and binds to sevenless and Sos proteins in vitro. Cell 73:169-177. [DOI] [PubMed] [Google Scholar]
- 61.Simpson, P., R. Woehl, and K. Usui. 1999. The development and evolution of bristle patterns in Diptera. Development 126:1349-1364. [DOI] [PubMed] [Google Scholar]
- 62.Soltoff, S. P., S. A. Carraway, S. A. Prigent, W. G. Gullick, and L. C. Cantley. 1994. ErbB3 is involved in activation of phosphatidylinositol 3-kinase by epidermal growth factor. Mol. Cell. Biol. 14:3550-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stephens, L. R., A. Eguinoa, H. Erdjument-Bromage, M. Lui, F. Cooke, J. Coadwell, A. S. Smrcka, M. Thelen, K. Cadwallader, P. Tempst, and P. T. Hawkins. 1997. The Gβγ sensitivity of a PI3K is dependent upon a tightly associated adaptor, p101. Cell 89:105-114. [DOI] [PubMed] [Google Scholar]
- 64.Sturtevant, M. A., M. Roark, and E. Bier. 1993. The Drosophila Rhomboid gene mediates the localised formation of wing veins and interacts genetically with components of the EGF-R signalling pathway. Genes Dev. 7:961-973. [DOI] [PubMed] [Google Scholar]
- 65.Tsuda, L., R. Nagaraj, S. Zipursky, and U. Banerjee. 2002. An EGFR/Ebi/Sno pathway promotes delta expression by inactivating Su(H)/SMRTER repression during inductive Notch signaling. Cell 110:625-637. [DOI] [PubMed] [Google Scholar]
- 66.Turner, S. J., J. Domin, M. D. Waterfield, S. G. Ward, and J. Westwick. 1998. The CC chemokine monocyte chemotactic peptide-1 activates both the class I p85/p110 phosphatidylinositol 3-kinase and the class II PI3K-C2α. J. Biol. Chem. 273:25987-25995. [DOI] [PubMed] [Google Scholar]
- 67.Vanhaesebroeck, B., S. J. Leevers, K. Ahmadi, J. Timms, R. Katso, P. C. Driscoll, R. Woscholski, P. J. Parker, and M. D. Waterfield. 2001. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 70:535-602. [DOI] [PubMed] [Google Scholar]
- 68.Virbasius, J. V., A. Guilherme, and M. P. Czech. 1996. Mouse p170 is a novel phosphatidylinositol 3-kinase containing a C2 domain. J. Biol. Chem. 271:13304-13307. [DOI] [PubMed] [Google Scholar]
- 69.Volinia, S., R. Dhand, B. Vanhaesebroeck, L. K. MacDougall, R. Stein, M. J. Zvelebil, J. Domin, C. Panaretou, and M. D. Waterfield. 1995. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 14:3339-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walker, E. H., O. Perisic, C. Ried, L. Stephens, and R. L. Williams. 1999. Structural insights into phosphoinositide 3-kinase signalling. Nature 402:313-320. [DOI] [PubMed] [Google Scholar]
- 71.Waterman, H., M. Katz, C. Rubin, K. Shtiegman, S. Lavi, A. Elson, T. Jovin, and Y. Yarden. 2002. A mutant EGF-receptor defective in ubiquitylation and endocytosis unveils a role for Grb2 in negative signalling. EMBO J. 21:303-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weinkove, D., T. P. Neufeld, T. Twardzik, M. D. Waterfield, and S. J. Leevers. 1999. Regulation of imaginal disc cell size, cell number and organ size by Drosophila class IA phosphoinositide 3-kinase and its adaptor. Curr. Biol. 9:1019-1029. [DOI] [PubMed] [Google Scholar]
- 73.Wheeler, M., and J. Domin. 2001. Recruitment of the class II phosphoinositide 3-kinase C2β to the epidermal growth factor receptor: role of Grb2. Mol. Cell. Biol. 21:6660-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, J., H. Banfic, F. Straforini, L. Tosi, S. Volinia, and S. Rittenhouse. 1998. A type II phosphoinositide 3-kinase is stimulated via activated integrin in platelets. A source of phosphatidylinositol 3-phosphate. J. Biol. Chem. 273:14081-14084. [DOI] [PubMed] [Google Scholar]