Abstract
Background
The BH3-only members of the Bcl-2 protein family have been proposed to play a key role in the control of apoptosis and in the initiation of the apoptotic pathways. In this study, we evaluated the expression of Bim, Noxa, and Puma in non-small cell lung cancer (NSCLC).
Methods
A total of 135 surgically resected NSCLCs were immunohistochemically assessed for Bim, Noxa, and Puma expression. The immunoscores were determined, and then its correlation with either the clinicopathological variables or the survival outcomes were analyzed.
Results
Immunohistochemical reactivity for Bim, Noxa, and Puma was detected in the cytoplasm of the tumor cells. Bim expression was associated with several clinicopathological factors, including sex (p < 0.001), smoking habit (p = 0.03), pathological histology (p = 0.001), pathological T stage (p = 0.03), pathological disease stage (p = 0.02), and differentiation of tumor (p < 0.001). Multivariate logistic regression analysis showed a significant correlation between low Bim expression and squamous cell carcinoma (p = 0.04), in addition to a correlation between high Bim expression and well differentiated tumors (p = 0.02). Analysis of cellular biological expression demonstrated a link between low Bim expression and high Ki67. While Noxa expression was also shown to be correlated with both smoking habit (p = 0.02) and the pathological histology (p = 0.03), there was no strong association observed between the expression and the clinical features when they were examined by a multivariate logistic regression analysis. No correlations were noted between Puma expression and any of the variables. Our analyses also indicated that the expression levels of the BH3-only proteins were not pertinent to the survival outcome.
Conclusions
The current analyses demonstrated that Bim expression in the NSCLCs was associated with both squamous cell carcinoma histology and tumor proliferation.
Introduction
Lung cancer is the most common cause of malignancy-related death in the world, and despite advances in both detection and treatment, its incidence rate is still increasing. Although the development of several new agents and molecular targeted therapies has led to an increased survival of patients with non-small cell lung cancer (NSCLC), the actual cure rates remain low for advanced NSCLC patients [1,2]. In order to improve the poor prognosis of patients with NSCLC, further studies that specifically examine the clinical features of lung cancer in addition to the development of new therapeutic strategies will be required.
Deregulation of apoptosis has been demonstrated to lead to cancer development, proliferation, and treatment resistance [3,4]. The mitochondria-mediated apoptotic pathway is largely regulated by the Bcl-2 family proteins [5], with members of this family possessing at least one of four conserved motifs that are known as the Bcl-2 homology domains (BH1 to BH4). These domains have been divided into three subfamilies, with the BH3-only proteins, such as Bik, Bid, Bim, Bmf, Hrk, Bad, Noxa, and Puma, exhibiting sequence homology only in BH3 [5-7]. Bim is induced by the withdrawal of growth factors via either of the two major epidermal growth factor receptor (EGFR)-dependent pathways: the Raf/MAPK or the Akt/PI3K [8,9]. Noxa and Puma are transcriptional targets of p53 and have been shown to play an active role in p53-induced apoptosis [10,11]. Recent studies have also demonstrated that aberrations of the BH3-only proteins are linked to tumorigenesis in several cancers [12,13]. Furthermore, there has been an accumulation of evidence showing an association between the frequent loss of BH3-only proteins and a poor cancer prognosis [14-16]. Although previous data have demonstrated the expression of Bcl-2 family proteins in lung cancer [17,18], there has been very little information available on the actual role that these proteins might play in lung cancer, and thus, further studies are warranted.
In the current study, we used immunohistochemistry to examine the expression of Bim, Noxa and Puma in a series of NSCLCs. Subsequently, we attempted to determine if there were any correlations between the expression of these proteins and features such as clinical and clinicopathological parameters, cell biological characteristics, or survival outcomes.
Materials and Methods
Patient and Specimens
This study was approved by the Medical Ethics Committee of Hokkaido University School of Medicine. A total of 135 patients (91 males and 44 females) who underwent radical surgery between 1982 and 1994 at Hokkaido University Medical Hospital were included in the study. Informed consent was obtained from all patients prior to enrollment in the study. Histological diagnoses and grades of differentiation of the primary tumor specimens were determined in accordance with the 1982 World Health Organization criteria. These histological analyses demonstrated that 74 samples were adenocarcinoma (Ad), 54 were squamous cell carcinoma (Sq), 5 were adenosquamous cell carcinoma (AS), and 2 were large cell carcinoma (La). For the statistical analyses, tumor specimens were divided into either a Sq or non-Sq group, which included the Ad, AS, and La. The pathological stage (pStage) was based on the guidelines of the American Joint Committee on Cancer for post-operative tumor-lymph node-metastasis (TNM) classification [19].
Immunohistochemistry
Immunohistochemical analyses were performed for Bim, Noxa, Puma, or Bcl-2 using 4 μm-thick histological sections that were cut from formalin-fixed, paraffin-embedded blocks. After the sections were deparaffinized in xylene and then rehydrated in ethanol, the sections were treated in 10 mM citrate buffer, pH 6.0 for 20 minutes at 121°C in an autoclave in order to retrieve the antigenicity. To block the endogenous peroxidase activity, the sections were subsequently immersed in methanol containing 1.5 % hydrogen peroxide for 20 minutes followed by incubation with normal rabbit serum in order to block the nonspecific antibody binding sites. Sections were incubated at 4°C overnight with either anti-Bim/Bod rabbit polyclonal antibody (Sigma-Aldrich corp. St Louis, MO, USA), diluted 1:300, primary anti-Noxa rabbit polyclonal antibody (Biovision, Inc., Mountain View, CA, USA), diluted 1:200, primary anti-Puma rabbit polyclonal antibody (ab27669, Abcam, Cambridge, MA, USA), diluted 1:50, or primary anti-Bcl-2 mouse monoclonal antibody (clone124, DAKO, Tokyo, JAPAN), diluted 1:40. Immunostaining was performed by using the biotin-streptavidin immunoperoxidase method with 3,3-diaminobenzidine as the chromogen (SAB-PO kit; Nichirei, Tokyo, Japan). Hematoxylin solution was used for counterstaining. For each batch of sample that was stained, one sample that was positive for either Bim, Noxa, Puma, or Bcl-2 was included as a positive external control. Using a BX 40 microscope (Olympus, Tokyo, Japan), the immunohistochemical evaluations were assessed twice by a single investigator (J.S.K.) who was blinded to the status of the other immunohistological and clinical data.
Immunohistochemical Scoring
Bim, Noxa, and Puma staining levels were classified as either high or low, as per a previously described method [17,18]. To be defined as having a high expression of Bim, Noxa or Puma, moderate-to-strong staining intensity had to be observed in more than 50 % of the tumor cells. Samples were scored as low when the expression was present in less than 50 % of the tumor cells. Immunohistochemically staining sections were judged high expression for Bcl-2 expression when more than 10 % of cancer cells showed cytoplasmic staining [20,21].
Western Blot Analysis
Cell lysates of Jurkat cells, the NSCLC cell line (HCC2429), and tumor tissues were prepared by lysing cells and tissues in radioimmune precipitation assay buffer (150 mM NaCl; 1 % Triton X-100; 1 % B deoxycholate; 0.1 % sodium dodecyl sulfate (SDS); 10 mM Tris, pH 7.4) supplemented with 100 μg/mL leupeptin, 100 μg/mL aprotinin, and 10 mM phenylmethylsulfonyl fluoride. Protein samples were resolved using SDS-PAGE gels, transferred onto nitrocellulose membranes (Amersham Biosciences Inc., St. Albans, United Kingdom), and then incubated with the primary antibody overnight. Subsequently, the membrane was incubated with anti-rabbit antibody conjugated with horseradish peroxidase (NA934V, Amersham Biosciences Inc.) and developed using the Amersham ECL system. Cell lysates of the Jurkat cells were used as the positive controls for Bim and Noxa [22]. In a prior study, we demonstrated that there is expression of Puma in the HCC2429 cell line [23].
Statistical Analysis
The association between Bim, Noxa, or Puma and the categorical variables were analyzed by chi-square tests or Fisher's exact test, as appropriate. When assessing the effects of more than one factor on Bim or Noxa expression, we performed a multivariate logistic regression analysis. Survival curves were estimated using the Kaplan-Meier method, with differences in the survival distributions then evaluated using the log-rank test. The level of significance was set at p < 0.05. Statistical analyses were done using StatView 5.0.1 (SAS Institute Inc., Cary, NC, USA).
Results
Expression of Bim, Noxa, and Puma
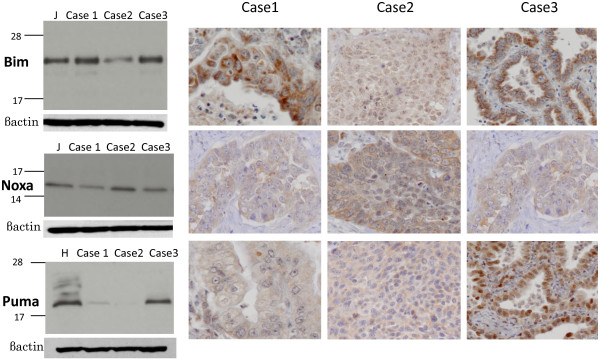
The quality of the antibodies that were used in this study was first tested by Western blot analysis and immunohistochemistry using diverse cell lines and surgical specimens. Cell lysates of Jurkat cells were used as positive controls for Bim and Noxa [22]. In addition, we previously showed that there is expression of Puma in the NSCLC cell line, HCC2429 [23]. The expression of Bim, Noxa and Puma noted during the Western blot and immunohistochemistry analyses is seen in Figure 1. In three surgical specimens that were obtained from the study cohort of NSCLC, Bim was strongly detected in Cases 1 and 3, while Noxa was moderately found in Case 2, and Puma was strongly detected in Case 3. These data were consistent with the results of our immunohistochemical staining, with the staining for all of the antibodies observed within the cytoplasm of the cells (Figures 23 and 4). Bim showed homogenous staining within tumors. Noxa and Puma showed focal and heterogeneous staining within tumors. Normal bronchial epithelial cells, which were used as the internal positive control, showed moderate to strong expression of all BH3-only proteins. No expression of these proteins was noted in the normal alveolar pneumocytes.
Figure 1.
Western blot analysis and immunohistochemistry of the surgical specimens obtained from three patients in the current cohort. In the Western blot analysis, Bim was strongly detected in Cases 1 and 3, while Noxa was moderately found in Case 2, and Puma was strongly detected in Case 3. These findings match the results of the immunohistochemical staining analyses. Cell lysates of the Jurkat cells (J) were used as positive controls for Bim and Noxa. HCC2429 (H) was used as the positive control for Puma.
Figure 2.
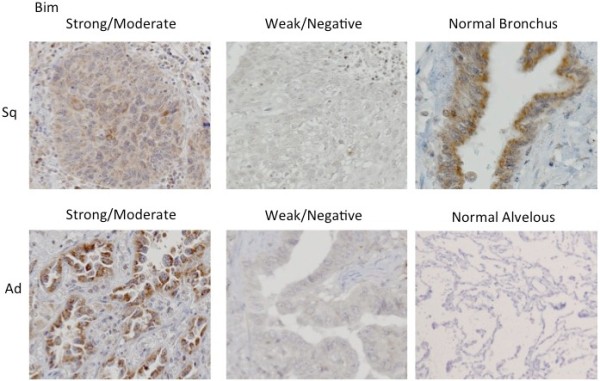
Bim immunohistochemical staining pattern. The immunohistochemical pattern for Bim in both the squamous cell carcinoma (Sq) and adenocarcinoma (Ad) samples.
Figure 3.
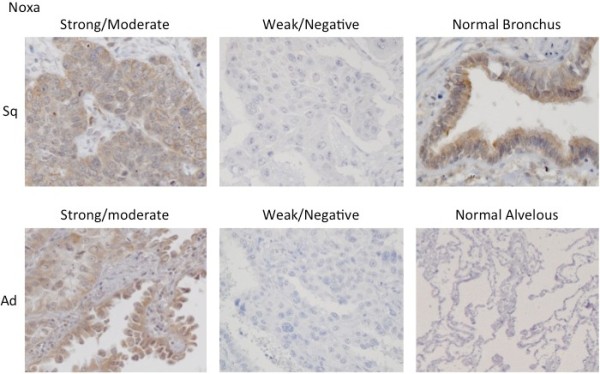
Noxa immunohistochemical staining pattern. The immunohistochemical pattern for Noxa in both the squamous cell carcinoma (Sq) and adenocarcinoma (Ad) samples.
Figure 4.

Puma immunohistochemical staining pattern. The immunohistochemical pattern for Puma in both the squamous cell carcinoma (Sq) and adenocarcinoma (Ad) samples.
Correlation between the expression of the BH3-only proteins and the clinicopathological variables
We evaluated the relationship between Bim, Noxa, or Puma expression and the clinicopathological variables. High Bim expression was observed in 58 patients with NSCLC (43 %). This expression was shown to be associated with sex (p < 0.001), smoking habit (p = 0.03), pathological histology (p = 0.001), pathological T stage (p = 0.03), pathological disease stage (p = 0.02), and differentiation of tumor (Table 1). High Noxa expression was detected in 47 NSCLCs (35 %), with the expression linked to both smoking habit (p = 0.02) and pathological histology (p = 0.03) (Table 2). Although a high Puma expression was seen in 59 patients with NSCLC (44 %), this expression had no influence on any of the variables (Table 3). Multivariate logistic regression analysis used to examine the correlation between Bim expression and the clinicopathological variables demonstrated significant associations between both the low Bim expression and Sq histology (p = 0.04) and the high Bim expression with well differentiated tumors (p = 0.02) (Table 4). No link was observed between Noxa expression and either smoking habit or pathological histology in the multivariate logistic regression analysis (Table 5). When we further analyzed the relationship between BH3-only proteins and patient survival, we found that the expression of these BH3-only proteins was not correlated with the survival outcomes of any of the subgroups analyzed, including both the disease stage and the histological type.
Table 1.
Correlation between Bim expression and the clinicopathological characteristics of 135 resected Non-small cell lung cancers
| |
No. of Patients |
|
|
|---|---|---|---|
| Characteristics | low | high | p |
| Age | |||
| <65 |
38 |
27 |
0.74 |
| ≥65 |
39 |
31 |
|
| Sex | |||
| Men |
61 |
30 |
<0.001 |
| Women |
16 |
28 |
|
| Smoker | |||
| Never |
16 |
22 |
0.03 |
| Smoker |
61 |
36 |
|
| Histology | |||
| Adeno |
32 |
42 |
0.001 |
| Squamous |
39 |
15 |
|
| Other1 |
6 |
1 |
|
| pT2 | |||
| T1 |
16 |
22 |
0.03 |
| T2-4 |
61 |
36 |
|
| pN3 | |||
| N0 |
47 |
44 |
0.09 |
| N1-3 |
30 |
14 |
|
| pStage4 | |||
| I |
41 |
42 |
0.02 |
| II-IV |
36 |
16 |
|
| Differentiation | |||
| Moderate/Poor |
64 |
29 |
<0.001 |
| Well | 13 | 29 | |
1Other: adenosquamous cell carcinoma and large cell carcinoma.
2pT: pathological tumor classifications.
3pN: pathological nodal classifications.
4pStage: pathological disease stage.
Table 2.
Correlation between Noxa expression and the clinicopathological characteristics of 135 resected Non-small cell lung cancers
| |
No. of Patients |
|
|
|---|---|---|---|
| Characteristics | low | high | p |
| Age | |||
| <65 |
39 |
26 |
0.22 |
| ≥65 |
49 |
21 |
|
| Sex | |||
| Men |
63 |
28 |
0.16 |
| Women |
25 |
19 |
|
| Smoker | |||
| Never |
19 |
19 |
0.02 |
| Smoker |
69 |
28 |
|
| Histology | |||
| Adeno |
43 |
31 |
0.03 |
| Squamous |
42 |
12 |
|
| Other1 |
3 |
4 |
|
| pT2 | |||
| T1 |
22 |
16 |
0.28 |
| T2-4 |
66 |
31 |
|
| pN3 | |||
| N0 |
61 |
30 |
0.46 |
| N1-3 |
27 |
17 |
|
| pStage4 | |||
| I |
55 |
28 |
0.74 |
| II-IV |
33 |
19 |
|
| Differentiation | |||
| Moderate/Poor |
64 |
29 |
0.19 |
| Well | 24 | 18 | |
1Other: adenosquamous cell carcinoma and large cell carcinoma.
2pT: pathological tumor classifications.
3pN: pathological nodal classifications.
4pStage: pathological disease stage.
Table 3.
Correlation between Puma expression and the clinicopathologic characteristics of 135 resectedNon-small cell lung cancers
| |
No. of Patients |
|
|
|---|---|---|---|
| Characteristics | low | high | p |
| Age | |||
| <65 |
34 |
31 |
0.81 |
| ≥65 |
42 |
28 |
|
| Sex | |||
| Men |
51 |
40 |
0.93 |
| Women |
25 |
19 |
|
| Smoker | |||
| Never |
21 |
17 |
0.88 |
| Smoker |
55 |
42 |
|
| Histology | |||
| Adeno |
45 |
29 |
0.06 |
| Squamous |
30 |
24 |
|
| Other1 |
1 |
6 |
|
| pT2 | |||
| T1 |
23 |
15 |
0.50 |
| T2-4 |
53 |
44 |
|
| pN3 | |||
| N0 |
49 |
42 |
0.47 |
| N1-3 |
27 |
17 |
|
| pStage4 | |||
| I |
42 |
41 |
0.09 |
| II-IV |
34 |
18 |
|
| Differentiation | |||
| Moderate/Poor |
53 |
40 |
0.81 |
| Well | 23 | 19 | |
1Other: adenosquamous cell carcinoma and large cell carcinoma.
2pT: pathological tumor classifications.
3pN: pathological nodal classifications.
4pStage: pathological disease stage.
Table 4.
Multivariate Logistic Regression Analysis for the correlation between Bim expression and clinical and clinicopathological characteristics
| Variable | Adjusted OR* (95%CI**) | P |
|---|---|---|
| Sex (Men vs Women) |
0.32 (0.09-1.17) |
0.08 |
| Smoker (Smoker vs Never1) |
0.46 (0.30-1.80) |
0.50 |
| Histology (Sq2 vs Non-Sq3) |
0.37 (0.17-0.84) |
0.04 |
| pT4 (pT2-4 vs pT1) |
0.77 (0.34-1.97) |
0.65 |
| pStage5 (II-IV vs I) |
0.47 (0.33-1.83) |
0.12 |
| Differentiation (Well vs Moderate/Poor) | 5.12 (1.16-6.92) | 0.02 |
*OR: odds ratio, **95%CI: 95% confidence interval.
1Never: never smoker.
2Sq: squamous cell carcinoma.
3Non-Sq: Included adenocarcinoma, adenosquamous and large cell carcinoma.
4pT: pathologic tumor classification.
5pStage: pathologic disease stage.
Table 5.
Multivariate Logistic Regression Analysis for the correlation between Noxa expression and clinical and clinicopathological characteristics
| Variable | Adjusted OR* (95%CI**) | P |
|---|---|---|
| Smoker (Smoker vs Never1) |
0.49 (0.21-1.11) |
0.09 |
| Histology (Sq2 vs Non-Sq3) | 0.63 (0.28-1.38) | 0.25 |
*OR: odds ratio, **95%CI: 95% confidence interval.
1Never: never smoker.
2Sq: Squamous cell carcinoma.
3Non-Sq: Included adenocarcinoma, adenosquamous and large cell carcinoma.
Correlation between BH3-only proteins and cellular biological expression
Results of previous studies on the expression of p27, Ki67 and cyclin E have implied that there is a prognostic significance for these expressions [24,25]. Therefore, using the same study cohort, we tested the correlation between the BH3-only proteins and these cellular biological expressions. Our results showed that a high Ki67 expression level was associated with a low Bim expression (p < 0.01), whereas there was no correlation between either the Puma or Noxa expression and any of the tested cellular biological expressions (Table 6). We also examined the correlation between the expression of Bcl-2 protein and BH3-only proteins, because the viability of cells is determined by the balance of expression of pro-apoptotic proteins such as BH3-only proteins and the expression of anti-apoptotic Bcl-2 family members,. However, there was no statistical correlation between Bcl-2 expression and BH3-only proteins. It suggests that other anti-apoptotic Bcl-2 proteins/BH3-only proteins axis may be involved in this study. Moreover, other survival pathway like MAPK or AKT/PI3K pathway may contribute to the cell viability.
Table 6.
Correlation between Bim, Noxa or Puma expression and cellular biological expression
|
Bim |
Noxa |
Puma |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
low |
high |
p |
low |
high |
p |
low |
high |
p |
|
| No. of Patients | No. of Patients | No. of Patients | |||||||
| Ki67 | |||||||||
| High |
56 |
28 |
<0.01 |
59 |
25 |
0.11 |
51 |
33 |
0.18 |
| Low |
21 |
30 |
|
29 |
22 |
|
25 |
26 |
|
| p27 | |||||||||
| High |
71 |
50 |
0.52 |
79 |
42 |
0.95 |
68 |
53 |
0.95 |
| Low |
6 |
8 |
|
9 |
5 |
|
8 |
6 |
|
| CyclinE | |||||||||
| High |
45 |
29 |
0.33 |
49 |
25 |
0.78 |
45 |
29 |
0.24 |
| Low |
32 |
29 |
|
39 |
22 |
|
31 |
30 |
|
| Bcl-2 | |||||||||
| High |
32 |
25 |
0.97 |
37 |
20 |
0.95 |
37 |
20 |
0.51 |
| Low | 45 | 33 | 51 | 27 | 39 | 39 | |||
Discussion
In the current study, we examined the expressions of the BH3-only proteins, Bim, Noxa and Puma, and found that there was a correlation between these expressions and the clinicopathological features. While we also found that low Bim expression was significantly more prevalent in the Sq histology versus non-Sq histologu, this study revealed no significant correlations between any of the clinical variables and the expression of Noxa or Puma. Although there has been one pilot study that used immunohistochemistry to examine the expression of the BH3-only proteins, Bad, Bid and Bim, in NSCLC samples [17], the current study is the first to clarify the frequency of Bim, Noxa and Puma in a large series of NSCLCs.
In another study of NSCLCs that reported a similar pattern, the results showed that while a loss of Bim was observed in 2 out of 22 samples in the Sq histology and in 0 out of 19 in the Ad histology, the loss was not statistically significant [17]. Although many promising treatments have been recently developed for adenocarcinoma of the lung, including EGFR-TKI (tyrosine kinase inhibitor) and pemetrexed [1,26], there are few optimal treatments that can be used for Sq. However, our results suggest the possibility that the loss of Bim expression in Sq might be related to resistance to these anticancer drugs.
Bim has previously been shown to have a role in tumor suppression [12,13]. This is supported by our current data that showed a significant association between the loss of Bim expression and a high Ki67 expression, which is a marker of tumor proliferation. In addition, high Bim expression was correlated with well differentiated tumors. Taken together, this suggests that Bim may have an impact on both cell proliferation activity and tumor feature in NSCLC.
Bim has also been shown to be required for initiation of the apoptosis induced by EGFR-TKI in NSCLC cell lines with EGFR-TKI-sensitive mutations [27-29]. Furthermore, inhibition of Bim induction has also been observed in NSCLC with the EGFR-TKI resistance mutation, T790M, which suggests that Bim induction leads to the active response of the EGFR-TKI treatment [28]. Recently, BH3-mimetics have been developed and shown to have cytotoxic effects on many solid tumors [30,31]. The BH3-mimetic, ABT-737, along with its oral analogue, AB7-263, have especially been shown to promote apoptosis and suppress tumor growth in small-cell lung cancer [32]. Although it has been observed that several NSCLC cell lines show relative resistance to ABT-737 monotherapy [33,34], ABT-737 significantly enhanced EGFR-TKI-induced cell killing of NSCLC, and thus, this combined treatment might be a promising new therapy [29]. Although we have not examined the correlation between Bim expression and responses to treatment, our current data suggest that Bim expression might be involved with tumor proliferation and feature, which supports the notion that Bim expression could be a useful therapeutic marker.
Noxa and Puma expression levels have been investigated in various cancers, including colon cancer and ameloblastic tumors, and were shown not to have any clinical significance [35-37]. This is consistent with our current data. To the best of our knowledge, there has been little data reported concerning the relationship between these two BH3-only proteins and the various clinicopathological factors in NSCLC. Unlike Bim expression, these two proteins may have not been implicated in the pathogenesis of NSCLC, which suggests that further investigations of their roles are warranted.
Our findings also did not find any impact of the examined BH3-only proteins on survival outcomes. Likewise, there have been no other reports on any relationships between these proteins and patient prognosis in NSCLC. However, it has been verified that both Bim and Puma expression are independent prognostic factors in colon cancer [14]. Conversely, Bim expression has not been previously linked with patient prognosis in either gastric cancer or malignant mesothelioma [15,38]. Therefore, the origin and stage (early or advanced) of the tumor in addition to the subsequent therapies that have been used against the disease might account for the prognosis discrepancies that have been described. Moreover, these proteins are known to be regulated by many different complex upstream pathways, such as AKT/PI3K, MAPK, or p53 [8-11]. Thus, the various levels of pathway regulation that could potentially exist might also be contributing factors for the differences noted in the prognosis between the different types of malignant diseases.
Conclusions
The current findings demonstrated that the loss of Bim expression was associated with both Sq histology and tumor aggressiveness in NSCLC.
Authors’ contributions
JSK was involved in the design of the study, collected the clinical data, performed the immunohistochemical analysis and drafted the manuscript. SO co-drafted the manuscript. JK, EK and HM collected the clinical data. IK provided general support and helped to draft the manuscript. HDA provided general support and helped to draft the manuscript. MH supervised the study. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Jun Sakakibara-Konishi, Email: konishj@med.hokudai.ac.jp.
Satoshi Oizumi, Email: soizumi@med.hokudai.ac.jp.
Junko Kikuchi, Email: johmuro@med.hokudai.ac.jp.
Eiki Kikuchi, Email: eikik@med.hokudai.ac.jp.
Hidenori Mizugaki, Email: mhide@med.hokudai.ac.jp.
Ichiro Kinoshita, Email: kinoshii@med.hokudai.ac.jp.
Hirotoshi Dosaka-Akita, Email: hdakita@med.hokudai.ac.jp.
Masaharu Nishimura, Email: ma-nishi@med.hokudai.ac.jp.
Acknowledgements
We would like to thank S. Shoji (First Department of Medicine, Hokkaido University School of Medicine) for special support and experimental assistance provided during this study.
References
- Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I. et al. Gefitinib or Chemotherapy for Non–Small-Cell Lung Cancer with Mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, Lilenbaum R, Johnson DH. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- Hengatner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- Cory S, Huang DC, Adamas JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros AM, Olejniczak ET, Fesik SW. Structural biology of the Bcl-2 family of proteins. Biochim Biophys Acta. 2004;1644:83–94. doi: 10.1016/j.bbamcr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, Ierino H, Lee EF, Fairlie WD, Bouillet P. et al. Apoptosis Initiated When BH3 Ligands Engage Multiple Bcl-2 Homologs, Not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- Deng J, Shimamura T, Perera S, Carlson NE, Cai D, Shapiro GI, Wong KK, Letai A. Proapoptotic BH3-only BCL-2 family protein BIM connects death signaling from epidermal growth factor receptor inhibition to the mitochondrion. Cancer Res. 2007;67:11867–11875. doi: 10.1158/0008-5472.CAN-07-1961. [DOI] [PubMed] [Google Scholar]
- Urbich C, Knau A, Fichtlscherer S, Walter DH, Brühl T, Potente M, Hofmann WK, de Vos S, Zeiher AM, Dimmeler S. FOXO-dependent expression of the proapoptotic protein Bim: pivotal role for apoptosis signaling in endothelial progenitor cells. FASEB J. 2005;19:974–976. doi: 10.1096/fj.04-2727fje. [DOI] [PubMed] [Google Scholar]
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/S1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- Michalak EM, Vandenberg CJ, Delbridge AR, Wu L, Scott CL, Adams JM, Strasser A. Apoptosis-promoted tumorigenesis: gamma-irradiation-induced thymic lymphomagenesis requires Puma-driven leukocyte death. Genes Dev. 2010;24:1608–1613. doi: 10.1101/gad.1940110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T, Dass CR, Choong PF. Bim-targeted cancer therapy: a link between drug action and underlying molecular changes. Mol Cancer Ther. 2009;12:3173–3180. doi: 10.1158/1535-7163.MCT-09-0685. [DOI] [PubMed] [Google Scholar]
- Sinicrope FA, Rego RL, Okumura K, Foster NR, O'Connell MJ, Sargent DJ, Windschitl HE. Prognostic impact of Bim, Puma, and Noxa expression in human colon carcinoma. Clin Cancer Res. 2008;14:5810–5818. doi: 10.1158/1078-0432.CCR-07-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L, Berrieman HK, O'Kane SL, Campbell A, Maraveyas A, Cawkwell L. Immunohistochemical detection of apoptotic markers in gastric cancer. Oncol Res. 2006;15:441–444. doi: 10.3727/096504005776568246. [DOI] [PubMed] [Google Scholar]
- Zantl N, Weirich G, Zall H, Seiffert BM, Fischer SF, Kirschnek S, Hartmann C, Fritsch RM, Gillissen B, Daniel PT. et al. Frequent loss of expression of the pro-apoptotic protein Bim in renal cell carcinoma: evidence for contribution to apoptosis resistance. Oncogene. 2007;26:7038–7048. doi: 10.1038/sj.onc.1210510. [DOI] [PubMed] [Google Scholar]
- Berrieman HK, Smith L, O'Kane SL, Campbell A, Lind MJ, Cawkwell L. The expression of Bcl-2 family proteins differs between nonsmall cell lung carcinoma subtypes. Cancer. 2005;103:1415–1419. doi: 10.1002/cncr.20907. [DOI] [PubMed] [Google Scholar]
- Borner MM, Brousset P, Pfanner-Meyer B, Bacci M, Vonlanthen S, Hotz MA, Altermatt HJ, Schalifer D, Reed JC, Betticher DC. Expression of apoptosis regulatory proteins of the Bcl-2 family and p53 in primary resected non-small-cell lung cancer. Br J Cancer. 1999;79:952–958. doi: 10.1038/sj.bjc.6690152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beahrs oH, Henson DE, Hutter RVP, Kennedy BJ eds. American Joint Committee on Cancer. Lung. In: AJCC Manual for Staging of Cancer. Philadelphia; Lippincott 1992: 115-122.
- Doaka-Akita H, Katabami M, Hommura H, Fujioka Y, Katoh H, Kawakami Y. Bcl-2 expression in non-small cell lung cancers: Higher frequency of expression in squamous cell carcinomas with earlier pT status. Oncology. 1999;56:259–264. doi: 10.1159/000011974. [DOI] [PubMed] [Google Scholar]
- Harada T, Ogura S, Yamazaki K, Kinoshita I, Itoh T, Isobe H, Yamashiro K, Dosaka-Akita H, Nishimura M. Predictive value of expression of P53, Bcl-2 and lung resistance-related protein for response to chemotherapy in non-small cell lung cancers. Cancer Sci. 2003;94:394–399. doi: 10.1111/j.1349-7006.2003.tb01453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High LM, Szymanska B, Wilczynska-Kalak U, Barber N, O'Brien R, Khaw SL, Vikstrom IB, Roberts AW, Lock RB. The Bcl-2 homology domain 3 mimetic ABT-737 targets the apoptotic machinery in acute lymphoblastic leukemia resulting in synergistic in vitro and in vivo interactions with established drugs. Mol Pharmacol. 2010;77:483–494. doi: 10.1124/mol.109.060780. [DOI] [PubMed] [Google Scholar]
- Konishi J, Yi F, Chen X, Vo H, Carbone DP, Dang TP. Notch3 cooperates with the EGFR pathway to modulate apoptosis through the induction of bim. Oncogene. 2010;29:589–596. doi: 10.1038/onc.2009.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommura F, Dosaka-Akita H, Mishina T, Nishi M, Kojima T, Hiroumi H, Ogura S, Shimizu M, Katoh H, Kawakami Y. Prognostic significance of p27KIP1 protein and ki-67 growth fraction in non-small cell lung cancers. Clin Cancer Res. 2000;6:4073–4081. [PubMed] [Google Scholar]
- Mishina T, Dosaka-Akita H, Hommura F, Nishi M, Kojima T, Ogura S, Shimizu M, Katoh H, Kawakami Y. Cyclin E expression, a potential prognostic marker for non-small cell lung cancers. Clin Cancer Res. 2000;6:11–16. [PubMed] [Google Scholar]
- Scagliotti GV, Parikh P, von Pawel J, Biesma B, Vansteenkiste J, Manegold C, Serwatowski P, Gatzemeier U, Digumarti R, Zukin M. et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:3543–3551. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]
- Gong Y, Somwar R, Politi K, Balak M, Chmielecki J, Jiang X, Pao W. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 2007;4:e294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, Tenen DG, Kobayashi S. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4:1669–1679. doi: 10.1371/journal.pmed.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragg MS, Kuroda J, Puthalakath H, Huang DC, Strasser A. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS Med. 2007;4:1681–1689. doi: 10.1371/journal.pmed.0040316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ. et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- Chonghaile TN, Letai A. Mimicking the BH3 domain to kill cancer cells. Oncogene. 2008;1:S149–157. doi: 10.1038/onc.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, Johnson EF, Marsh KC, Mitten MJ, Nimmer P. et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- Wesarg E, Hoffarth S, Wiewrodt R, Kröll M, Biesterfeld S, Huber C, Schuler M. Targeting BCL-2 family proteins to overcome drug resistance in non-small cell lung cancer. Int J Cancer. 2007;121:2387–2394. doi: 10.1002/ijc.22977. [DOI] [PubMed] [Google Scholar]
- Kim KW, Moretti L, Mitchell LR, Jung DK, Lu B. Combined Bcl-2/mammalian target of rapamycin inhibition leads to enhanced radiosensitization via induction of apoptosis and autophagy in non-small cell lung tumor xenograft model. Clin Cancer Res. 2009;15:6096–6105. doi: 10.1158/1078-0432.CCR-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson AK, Emterling AM, Arbman G, Sun XF. Noxa in cololectal cancer: a study on DNA, mRNA and protein expression. Oncogene. 2003;22:4675–4678. doi: 10.1038/sj.onc.1206655. [DOI] [PubMed] [Google Scholar]
- Jansson A, Arbman G, Sun XF. mRNA and protein expression of PUMA in sporadic cololectal cancer. Oncol Rep. 2004;12:1245–1249. [PubMed] [Google Scholar]
- Kumamoto H, Ooya K. Immunohistochemical detection of BH3-only proteins in ameloblastic tumors. Oral Dis. 2008;14:550–555. doi: 10.1111/j.1601-0825.2007.01417.x. [DOI] [PubMed] [Google Scholar]
- O'Kane SL, Pound RJ, Campbell A, Chaudhuri N, Lind MJ, Cawkwell L. Expression of bcl-2 family members in malignant pleural mesothelioma. Acta Oncol. 2006;45:449–453. doi: 10.1080/02841860500468927. [DOI] [PubMed] [Google Scholar]