Abstract
Sir1p is one of four SIR (silent information regulator) proteins required for silencing the cryptic mating-type locus HMRa in the budding yeast Saccharomyces cerevisiae. A Sir1p interaction with Orc1p, the largest subunit of the origin recognition complex (ORC), is critical for Sir1p's ability to bind HMRa and function in the formation of silent chromatin. Here we show that a discrete domain within Sir1p, the ORC interaction region (OIR), was necessary and sufficient for a Sir1p-ORC interaction. The OIR contains the originally defined silencer recognition-defective region as well as additional amino acids. In addition, a Sir1p-Sir4p interaction required a larger region of Sir1p that included the OIR. Amino acid substitutions causing defects in either a Sir1p-Orc1p or a Sir1p-Sir4p interaction reduced HMRa silencing and Sir1p binding to HMRa in chromatin. These data support a model in which Sir1p's association with HMRa is mediated by separable Sir1p-ORC and Sir1p-Sir4p interactions requiring a common Sir1p domain, and they indicate that a Sir1p-ORC interaction is restricted to silencers, at least in part, through interactions with Sir4p.
In eukaryotes, transcriptional repression can be mediated through the formation of higher-order chromatin structures called heterochromatin. Heterochromatin requires specific posttranslational modifications of nucleosomes and the binding of specialized nonhistone chromatin proteins to these modified nucleosomes (18, 25, 45, 48). Heterochromatin can form over large chromosomal regions, such as the length of an entire chromosome in mammalian X-chromosome inactivation (4). In addition to repressing transcription, heterochromatin can affect other chromosomal functions, including chromosome segregation, recombination, and replication (10, 50, 52). Defects in heterochromatin are associated with chromosomal instability and inappropriate transcription activation that may contribute to aberrant cell proliferation and human diseases (20, 27, 46, 49, 51). An accurate mechanistic description of heterochromatin requires an understanding of the individual protein-protein interactions necessary for the nucleation and spreading of heterochromatin domains.
Silencing of the cryptic mating-type loci, HMR and HML, in Saccharomyces cerevisiae provides an experimentally tractable paradigm for studying heterochromatin formation (21). Silent chromatin in yeast is functionally analogous to heterochromatin in multicellular eukaryotes, causing heritable, position-dependent transcriptional repression and general inaccessibility of the underlying chromosomal DNA (16, 32, 33, 44). Although there are notable differences in terms of the specific histone modifications and nonhistone chromatin binding proteins that mediate silent and heterochromatin formation (17), there are also obvious similarities. For example, both silent chromatin and heterochromatin are comprised of hypoacetylated nucleosomes and depend on the activity of histone deacetylases for their maintenance. In addition, silent chromatin and some forms of heterochromatin require physical interactions between the conserved origin recognition complex (ORC), the multisubunit protein complex that functions in the initiation of eukaryotic DNA replication, and heterochromatin-specific nonhistone proteins (1, 13, 14, 36, 47).
Silent chromatin assembly at HMRa requires the function of a small DNA sequence element called the HMR-E silencer (33). The ∼150-bp HMR-E silencer is necessary and sufficient for the formation of a silent chromatin domain that encompasses 4 to 5 kbp of chromosomal DNA, including the HMRa locus. HMR-E contains a single binding site each for ORC, Rap1p, and Abf1p. Silencing of HMRa is not an essential process, but ORC is essential for viability because it also functions at chromosomal origins in the initiation of DNA replication (1). Rap1p and Abf1p are abundant nuclear proteins that also have essential nonsilencing functions elsewhere in the genome (12, 26, 39, 43). When bound together at the HMR-E silencer, these three proteins function as a silencer-protein complex that recruits a set of four nonessential silencing proteins called the silent information regulator (SIR) proteins to HMRa (40). The SIRs play direct roles in nucleating and assembling heterochromatin at HMRa (15). A working model for the formation of silent chromatin at HMRa predicts that Sir1p and/or Sir4p binds the silencer binding proteins and then recruits the other Sir proteins, Sir2p and Sir3p, to the silencer (23, 41). Once bound to the silencer, Sir2p, the founding member (7) of a family of NAD+-dependent deacetylases, deacetylates neighboring nucleosomes, which in turn enhances binding of Sir3p to nucleosomes adjacent to HMR-E. Sir3p bound to adjacent nucleosomes then helps recruit additional Sir2/Sir4 complexes and Sir2 deacetylates neighboring nucleosomes, allowing the binding of additional Sir3p molecules (5, 40, 41). This process continues until a silent domain of chromatin encompassing an array of nucleosomes is formed.
Based on this model, a key step in the nucleation of silent chromatin assembly is the binding of Sir1p to the HMR-E silencer. Stable association between Sir1p and HMR-E requires an interaction between Sir1p and the Orc1p subunit of the ORC bound to HMR-E (13, 14, 47). In this study we defined and characterized a minimal protein domain within Sir1p that is necessary and sufficient for direct and specific interactions with ORC in vitro and in vivo. Since ORC binds to hundreds of nonsilencer replication origins distributed throughout the yeast genome but Sir1p binding is confined to silencers (13), an important issue relevant to the nucleation of silencing at HMRa is what confines a Sir1p-ORC interaction to silencers? A larger domain within Sir1p that includes the minimal Sir1p-ORC interaction domain mediated a Sir1p-Sir4 interaction. Importantly, genetic and direct molecular analysis indicated that this Sir1p-Sir4p interaction was necessary for silencing HMRa. Furthermore, individual amino acids within the defined Sir1p-ORC interaction domain of Sir1p were required for a Sir1p-Sir4p interaction but not a Sir1p-ORC interaction. These amino acids were also required for the stable binding of Sir1p to HMRa in chromatin. Therefore, independent interactions between Sir1p and both Sir4p and ORC involve a common Sir1p domain and contribute to Sir1p's stable association with HMRa in vivo.
MATERIALS AND METHODS
Yeast strains and plasmids.
Yeast strains and plasmids used in this study were constructed using standard yeast molecular genetics (19) and recombinant DNA techniques (42). A SIR1::3xHA-kanMX6 strain was generated by synthesizing the appropriate DNA fragment using PCR and the pFA6a-3HA-kanMX6 plasmid cassette as previously described (31). This strain was used as a genomic DNA source to construct isogenic mutant SIR1::3xHA-kanMX6 strains using complementary oligos containing the mutation of interest marked by a unique restriction site (28).
Protein-protein interactions.
Two-hybrid interactions were analyzed using the HIS3 reporter gene in a GAL4-based yeast two-hybrid strain (24) containing a sir2 deletion marked by TRP1 (CFY932). HIS3 expression was examined by patching cells to medium lacking histidine.
Wild-type and mutant glutathione S-transferase (GST)-Sir1pOIR(M473-D611) was produced in Escherichia coli (BL-21)pRIL cells using pGEX-KG. One milliliter of bacterial lysate containing GST-Sir1pOIR(M473-D611), in 1× phosphate-buffered saline, 1% Triton, and 1% β-mercaptoethanol, was incubated with 50 μl of swelled glutathione resin (Sigma) for 30 min at 4°C. The resin was then washed four times with 1× phosphate-buffered saline (140 mM NaCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, 2.5 mM KCl) with 0.05% Tween and two times with 50 mM Tris (pH 8) containing 0.25 M NaCl and incubated overnight at 4°C with cell lysate. Resin was incubated with either 200 μl of nuclear lysate from Sf9 cells expressing various ORC subunits (2) or 500 μl of yeast extract (37) in buffer H (pH 7.5) with 0.1 M KCl (2). Yeast extracts were produced as previously described (37), except that 1 liter of cells was grown to an optical density at 600 nm of ∼2, a density of 2 × 107 cells/ml, and the final pellet was resuspended in 2 ml of buffer H-0.1 M KCl, omitting the dialysis step. After incubation with extract, the resin was washed with 200 μl and then 800 μl of 50 mM Tris with 0.5 M NaCl, resuspended in 50 mM Tris with 0.1 M NaCl, and boiled with an equal volume of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. Protein immunoblot assays with antibodies against Orc1p, Orc2p, and Orc3p were used to determine whether ORC or Orc1p bound GST-Sir1pOIR.
Limited proteolysis.
His-Sir1OIR was overexpressed from a pET28b vector in E. coli (BL21) cells containing pRIL. Bacterial extracts were incubated with 0.5 ml of Ni-agarose (Qiagen) at 4°C for 30 min with rotation. The beads were poured into a 10-ml column and washed with five column volumes of nickel binding buffer (20 mM Tris-HCl [pH 7.2], 0.5 M NaCl, and 10% glycerol). His-Sir1OIR, eluted from the Ni beads with imidazole buffer (nickel binding buffer plus 0.5 M imidazole), was further purified by size-exclusion chromatography using an S-100 column (Pharmacia).
Purified Sir1OIR was subjected to proteolysis by trypsin and chymotrypsin (Sigma). Digestions were performed under conditions to limit multiple cleavage events. Trypsin and chymotrypsin were added to 0.75 μg of His6-SirOIR (in imidazole buffer) at room temperature with a 1:300 and a 1:600 protease-to-protein ratio, respectively. Aliquots (1/5) of the reaction mixture were removed after 0, 1, 10, and 30 min of incubation, and digestion was stopped by addition of 4× SDS sample buffer (29). The samples were analyzed on an SDS-15% PAGE gel and visualized by Coomassie staining. Trypsin- and chymotrypsin-digested fragments were analyzed by mass spectrometry (Bruker TOF), and the identity of the protease fragments was confirmed through Edman sequencing.
Screen for Sir1p mutants that fail to interact with Orc1p.
The C terminus of SIR1 (M473 to D678) was PCR mutagenized using a standard Taq PCR and cycle conditions with limiting amounts of either dATP or dGTP (35). SIR1 was mutagenized in the context of the two-hybrid vector (pCF721). PCR fragments were transformed into the yeast two-hybrid strain (24) with a linearized URA3-marked Gal4 binding domain (GBD)-Sir1 plasmid (pCF721) and a LEU2-marked Gal activation domain (GAD)-Orc1p (amino acids L5 to V268) plasmid (pTT50 [47]). Cells containing both plasmids were selected on minimal medium lacking uracil and leucine. Transformants were replica plated to minimal medium lacking uracil, leucine, and histidine. Colonies unable to grow in the absence of histidine contained versions of Sir1p unable to interact with Orc1p in the two-hybrid assay. These mutant clones were isolated and tested for protein expression using protein hybridization blotting with an antibody against the GBD (BAbCO). Clones producing a protein of the correct size were sequenced to determine the location of the mutation(s).
Site-directed mutagenesis of SIR1.
Several amino acid substitutions were engineered in the context of the appropriate coding regions for the two-hybrid GBD-SIR1 clones and/or chromosomal SIR1 locus. Some amino acid substitutions were used to confirm data obtained from the random mutagenesis screen, some were made as part of an alanine-scanning strategy, and others were made to examine the role of conserved amino acids in Sir1p. To make specific mutations in the context of the plasmid copies of GBD-SIR1, smaller fragments of SIR1 containing the region of interest were subcloned into pUC vectors and mutagenized using the QuickChange mutagenesis kit (Stratagene). To make specific mutations in the context of the chromosomal copy of SIR1, fusion PCR with the appropriate oligos and genomic DNA from the SIR1::3xHA-kanMX6 as template was performed as previously described (28), and the resulting mutant SIR1 fragment was integrated into the relevant recipient strain by standard yeast techniques.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were conducted as previously described (13) except that cells were cross-linked for 20 min and protein A-Sepharose resin was blocked by incubating two times for 5 min in 10 mg of bovine serum albumin/ml and two times for 5 min in 1 mg of bovine serum albumin/ml. Blocking steps were done at 4°C. Purified DNA from the immunoprecipitate was resuspended in 25 μl of Tris-EDTA (pH 8). The purified DNA in the starting material (whole-cell extract before immunoprecipitation [IP]) was resuspended in 1.0 ml of Tris-EDTA (pH 8).
The HMR and ACT1 fragments were analyzed by PCR as previously described (13) except that the final volume of all PCR mixtures was 20 μl. The linear range of PCR for comparing wild-type and mutant strains accurately was determined by examining dilutions of the genomic templates under fixed cycle numbers. For each total and IP sample, two directly comparable concentrations of template were used to ensure that the final assay gave linear data points. For examining HMR, the PCR was run for 24 cycles to achieve a signal in the linear range. For ACT1, PCR was run for 26 cycles to ensure the ability to visualize ACT1 and thus normalize the level of the experimental fragment (HMR) in the immunoprecipitate.
RESULTS
A contiguous ∼120-amino-acid region within Sir1p, including the previously defined SRD region, was sufficient for a specific interaction with ORC.
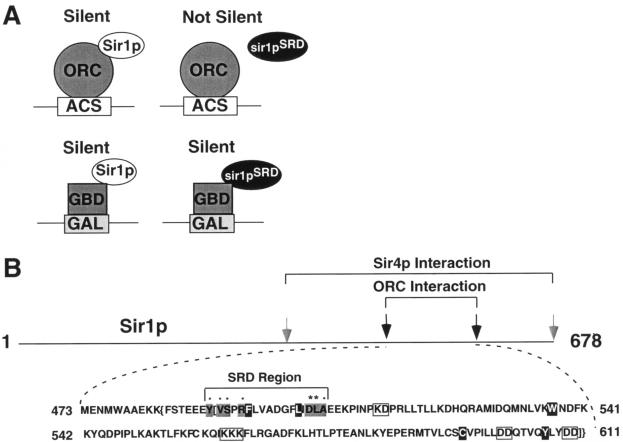
Previously our investigators identified a number of amino acids (Y483, V484, S485, R487, and A505) that are each necessary for Sir1p's ability to recognize the HMR-E silencer in chromatin and to interact with the N terminus of Orc1p in a two-hybrid assay (14) (Fig. 1). Substitution of any one of these amino acids in Sir1p causes a specific silencer recognition-defective (SRD) phenotype, meaning that the mutant proteins are unable to silence HMRa unless they are tethered to the HMR-E silencer via a heterologous DNA binding domain (Fig. 1A). These same substitutions also abolish a two-hybrid interaction between Sir1p and Orc1p and cluster in a 17-amino-acid region of Sir1p that we refer to as the SRD region (14) (Fig. 1B). Though our previous studies indicated that amino acids within this region are necessary for an interaction between Sir1p and the Orc1p N terminus, they did not address whether the SRD region is sufficient to govern this interaction.
FIG. 1.
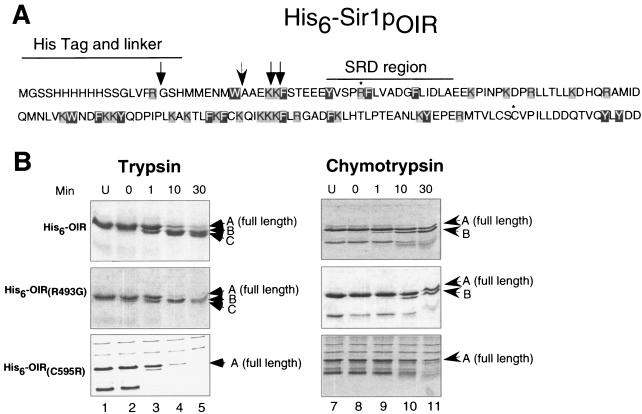
A small region of Sir1p governs a Sir1p-ORC interaction. (A) In a previous study (12), our investigators identified mutant versions of Sir1p incapable of silencing by the natural mechanism but capable of silencing HMR when tethered to the locus by a GBD. (B) Sequence of the OIR. Amino acids causing a SRD phenotype when replaced are shown as black letters on a gray background. Previously identified SRD amino acids are designated by dots (12), and SRD amino acids identified in this study are designated by asterisks. Amino acids important for both Sir1p-ORC and Sir1p-Sir4p interactions are shown as white letters on a black background, and amino acids important for a Sir1p-Sir4p interaction are boxed in gray. The two-hybrid boundaries of the OIR are indicated by brackets, while the protease resistance boundaries are designated by braces.
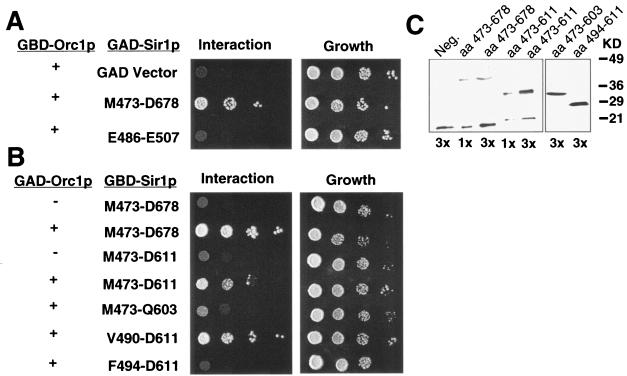
To test whether the SRD region was sufficient for a specific interaction with Orc1p, we asked whether a small region of Sir1p that includes the SRD region, expressed as a GAD-Sir1p(E486-E507) fusion protein, could interact with GBD-Orc1p(L5-V268) in a two-hybrid experiment. We used a GAD-Sir1p fusion because the corresponding GBD-Sir1p fusion activated transcription of the two-hybrid reporter genes on its own (data not shown). The GAD-Sir1p(E486-E507) fusion could not interact with GBD-Orc1p in this assay, although a larger GAD-Sir1p(M473-D678) fusion protein could (Fig. 2A). These data indicated that other amino acids, outside of the previously defined SRD region, were necessary for a Sir1p-Orc1p two-hybrid interaction.
FIG. 2.
Minimal region of Sir1p required for a two-hybrid Sir1p-Orc1p interaction. (A) The SRD region fused to the GAD was not able to interact with Orc1p in the two-hybrid assay; however, a larger GAD-Sir1(M473-D68) fusion did interact with Orc1p. Cells were plated in 10-fold serial dilutions, starting with 107 cells/ml, on medium lacking histidine to select for an interaction and on complete medium to determine plating efficiency. The James et al. two-hybrid strain (22) containing a SIR2 deletion (CFY932) was used in all two-hybrid experiments. (B) Two-hybrid analysis of GBD-Sir1p fusion proteins defined the minimal OIR of Sir1p. Amino acid boundaries of Sir1p in each experiment are indicated. (C) The GBD-Sir1p fusion proteins examined in panel B were expressed as determined by protein immunoblotting using an anti-GBD antibody (BAbCO).
We used two complementary approaches to identify additional amino acids within Sir1p required for a Sir1p-Orc1p interaction. First, we defined the N-terminal and C-terminal boundaries of an Orc1p-interaction region (OIR) by constructing deletions of the Sir1p coding region within GBD-Sir1p. These experiments indicated that the C-terminal boundary of the OIR was between amino acids Q603 and D611 and the N-terminal boundary was between V490 and F494 (Fig. 2B). Truncated proteins were stable as determined by immunoblotting with the GBD antibody (Fig. 2C and data not shown). Interestingly, although Y489 was defined as necessary for interactions with Orc1p in the context of full-length GBD-Sir1p (14), it could be deleted without affecting a Sir1p-Orc1p interaction in the context of the smaller GBD-Sir1p(V490-D611) fusion used here. However, we note that in this fusion a phenylalanine encoded by plasmid sequences replaces Y489, effectively creating a Y489F substitution in Sir1p. This change may be conservative enough to allow the two-hybrid interaction. Regardless, deleting even a few amino acids within the originally defined SRD region abolished a Sir1p-ORC interaction. Based on this functional analysis, the minimal OIR within Sir1p was between Y489 and D611 and the SRD region was positioned at the extreme N terminus of the OIR.
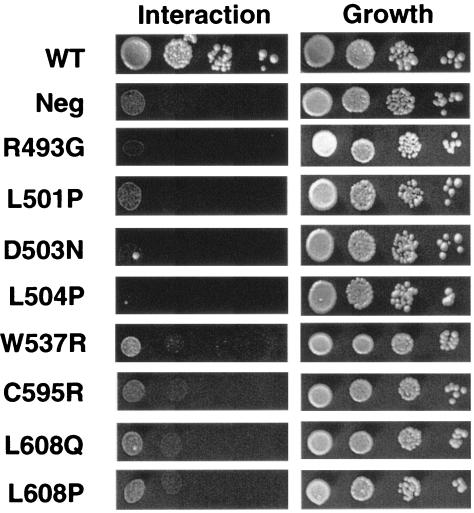
As a second approach to identify amino acids within Sir1p that are required for a Sir1p-Orc1p interaction, we performed PCR-directed random mutagenesis (35) of the Sir1p coding region in the context of GBD-Sir1p(M473-D678). A mutagenized library of plasmids encoding GBD-Sir1p(M473-D678) was transformed into the two-hybrid tester strain harboring the GAD-Orc1p(L5-V268) fusion, and transformants that failed to activate the two-hybrid HIS3 reporter were identified by their inability to grow on medium lacking histidine. Sixty-two independent transformants were identified as harboring potential mutant versions of Sir1p unable to interact with GAD-Orc1p. These were tested for expression of normal levels of GBD-Sir1p(M473-D678) by immunoblot analysis with a GBD antibody (data not shown). GBD-Sir1p(M473-D678)-encoding plasmids expressing normal levels of the GBD-Sir1p fusion (33 transformants) were recovered, retested by transformation into the two-hybrid tester strain, and sequenced. Nine amino acid substitutions that reduced or abolished the two-hybrid Sir1p-ORC interaction were identified in this screen (Table 1). Several of the mutants contained changes in the same amino acids that had been identified in the original SRD screen (14), although in some cases the precise amino acid substitution differed. Four additional amino acids within the SRD region, F494, L501, D503, and L504, were identified as required for a two-hybrid Sir1p-ORC interaction, as were three amino acids C-terminal to the SRD region: W537, C595, and L608 (Fig. 1 and 3; Table 1). These data indicated that amino acids outside the SRD region, including two that were close to the C-terminal boundary of the OIR (C595 and L608), were also required for a two-hybrid Sir1p-ORC interaction. Importantly, as will be demonstrated, substitution of these C-terminal amino acids does not confer an SRD phenotype, suggesting that we have not extended the known SRD region. Instead, we propose that we have defined the boundaries of a larger Sir1p domain required to allow the SRD region to interact with ORC. We refer to this larger domain as the OIR.
TABLE 1.
Sir1p amino acid changes
| Amino acid change(s)a | Orc1p or Sir4p interaction defect(s) |
|---|---|
| E486-488A | Neither |
| V490G* | Orc1p |
| R493G*b | Orc1p |
| F494S* | Orc1p and Sir4p |
| L501P* | Orc1p and Sir4p |
| D503N* | Orc1p |
| L504P* | Orc1p |
| E506-507A | Neither |
| K513A, D514A | Neither |
| K522A, D523A | Neither |
| W537R* | Orc1p and Sir4p |
| K541-542A | Neither |
| C558A | Neither |
| K562-564A | Sir4p |
| C595R* | Orc1p and Sir4p |
| V596G | Neither |
| P597S | Neither |
| D601-602A | Sir4p |
| L608P* | Orc1p and Sir4p |
| L608Q* | Orc1p and Sir4p |
| D610-611A | Sir4p |
*, mutants that were generated through random mutagenesis. All other mutants were generated using site-directed mutagenesis.
This mutation was isolated and described in an earlier study (14).
FIG. 3.
Specific amino acid substitutions abolish a two-hybrid interaction between GBD-Sir1p(M473-D678) and GAD-Orc1p(L5-V268). Experiments were performed as described in the legend for Fig. 2.
The OIR of Sir1p bound both Orc1p and ORC with the appropriate specificities in vitro.
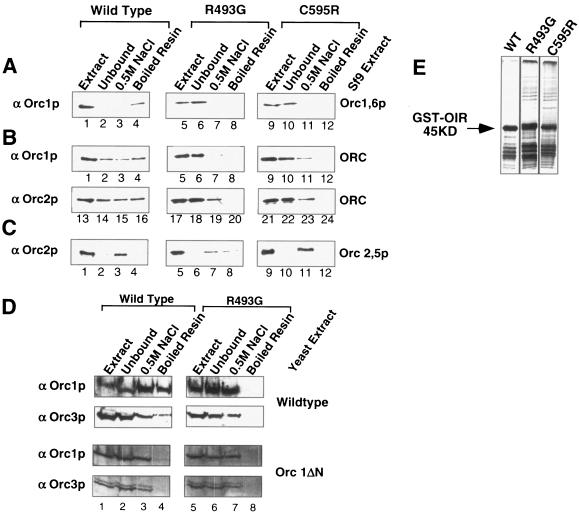
The data presented above provide evidence that Sir1p OIR(V490-D611) is sufficient to interact specifically with the N-terminal region of Orc1p. To extend this conclusion, we tested whether the wild type and two different mutant versions, an original SRD mutant (R493G) (14) and a newly isolated mutant (C595R) (Fig. 3), of GST-Sir1p(M473-D611) could bind full-length Orc1p or the entire ORC in vitro (Fig. 4). Specifically, the GST-Sir1p(M473-D611) fusion, expressed in E. coli, was bound to glutathione resin, washed, and then incubated with nuclear extracts prepared from Sf9 insect cells infected with baculovirus-expressing ORC genes (2). The nuclear extracts were separated into unbound (0.1 M NaCl), high-salt wash (0.5 M NaCl), and bound (resin boiled with SDS-PAGE loading buffer) fractions, and the nuclear extracts and fractions were analyzed by protein immunoblotting with ORC antibodies (Fig. 4).
FIG. 4.
The OIR of Sir1p bound both Orc1p and ORC with the appropriate specificities in vitro. (A) Wild-type and two mutant versions of GST-OIR were bound to glutathione resin and incubated with extracts from Sf9 cells expressing Orc1p and Orc6p. Starting material (Extract), unbound, 0.5 M NaCl wash, and boiled resin fractions were analyzed by protein immunoblotting with antibodies against Orc1p. (B) Affinity experiments were conducted as described for panel A except that extract from Sf9 cells expressing all six ORC subunits was used. Protein immunoblotting was performed with either antibodies against Orc1p or antibodies against Orc2p, as indicated. (C) Affinity experiments were conducted as described for panel A except that extracts from Sf9 cells expressing only Orc2p and Orc5p were used. (D) Affinity experiments were conducted as described for panel A except that a crude yeast cell extract was used. Extract was produced from either a wild-type strain or a strain expressing an Orc1p lacking its N-terminal 235 amino acids. (E) To ensure that similar amounts of wild-type and mutant versions of GST-OIR were used in affinity experiments, 5 μl of each resin was analyzed by SDS-PAGE. The gel was silver stained to visualize the GST-OIR.
The wild-type GST-Sir1p fusion bound Orc1p more efficiently than either mutant GST-Sir1p fusion (Fig. 4A). In these experiments, Sf9 cells were infected with a baculovirus containing ORC1 and ORC6 (2). Virtually no Orc1p could be detected in the unbound fraction when wild-type GST-Sir1p resin was used, but measurable amounts of Orc1p could be detected by boiling the resin in SDS-PAGE loading buffer (Fig. 4A, lanes 1 to 4). In contrast, when mutant versions of GST-Sir1p were used the majority of Orc1p was recovered in the unbound fraction (Fig. 4A, lanes 5 to 8 and 9 to 12) and no Orc1p could be detected in either the 0.5 M NaCl wash or the boiled resin fractions. For each experiment, similar amounts of wild-type and mutant GST-Sir1p-resins were used (Fig. 4E). In addition, Ponceau S staining of immunoblots indicated that similar amounts of Sf9 extracts were used in each experiment and that the vast majority of proteins in the extract did not bind the GST-Sir1p resin (data not shown).
Since all available evidence indicates that Sir1p binds the Orc1 N terminus in the context of the entire ORC bound to a silencer, we tested whether the GST-Sir1p(M473-D611) fusion could bind ORC with an efficiency and specificity similar to that shown for Orc1p alone (Fig. 4B). In these experiments, nuclear extracts were prepared from Sf9 cells that were coinfected with three different baculoviruses, each containing two of the six ORC genes (ORC1 and -6, ORC2 and -5, and ORC3 and -4) (2). The nuclear extracts were separated over the appropriate GST-Sir1p resins, and binding to ORC was monitored by protein immunoblotting using both Orc1p (α-Orc1p) and Orc2p (α-Orc2p) antibodies. Significantly, the wild-type GST-Sir1p fusion bound ORC more efficiently than either mutant GST-Sir1p fusion (Fig. 4B, compare wild type lanes 1 to 4, α-Orc1p, and lanes 13 to 16, α-Orc2p]) to the R493G mutant (lanes 5 to 8, α-Orc1p, and 17 to 20, α-Orc2p) and the C595R mutant (lanes 9 to 12, α-Orc1p, and 21 to 23, α-Orc2p). To determine whether the GST-Sir1p fusion was enriching for Orc2p as part of the ORC, we also tested whether the GST-Sir1p fusions could bind the Orc2p subunit alone by using nuclear extracts prepared from Sf9 cells infected with only the ORC2,5 baculovirus (Fig. 4C). Although Orc2p bound the GST-Sir1p resin to some degree (eluted at 0.5 M NaCl wash) (Fig. 4C, lane 3), this binding was not reduced by the same amino acid substitutions in Sir1p that reduced binding of Sir1p to Orc1p (Fig. 4A) or ORC (Fig. 4B, lanes 13 to 24, compared to C, lanes 1 to 12). Thus, the major component of Sir1p's ability to interact specifically with ORC in vitro was governed by interactions between Sir1p and Orc1p. Furthermore, a small region of Sir1p that included the OIR was sufficient to bind ORC with the same specificity as Orc1p.
The data described above indicated that the OIR of Sir1p efficiently bound recombinant ORC that was overproduced in Sf9 cells. As a more stringent test of the ability of this region to bind ORC efficiently, we repeated these experiments using crude yeast extracts (37) prepared from yeast cells expressing normal concentrations of ORC. Significantly, the GST-Sir1p(M473-D611) fusion bound a measurable fraction of yeast ORC in these experiments (Fig. 4D). A point mutation, R493G, that abolishes the ability of Sir1p to interact with Orc1p both in vitro and in the two-hybrid assay abolished detectable ORC in the bound fraction (boiled resin) (Fig. 4D, compare wild-type lanes 4 and 8). Similarly, the GST-Sir1p(M473-D611) fusion protein was not able to bind ORC, as assayed by α-Orc1p and α-Orc3p protein immunoblotting, from a strain in which the N terminus of Orc1p had been deleted (Fig. 4D). Importantly, similar amounts of wild-type and mutant GST-Sir1p fusion proteins were used for these affinity experiments (Fig. 4E). Together these data support the view that the OIR of Sir1p can specifically and efficiently bind the yeast ORC through interactions with the N terminus of Orc1p.
The OIR formed a protease-resistant domain.
Two-hybrid and in vitro data indicated that the OIR was the minimal region of Sir1p necessary to interact with ORC with the appropriate specificity. We postulated that the OIR formed a stable domain that positioned the SRD region to promote an ORC interaction. In this view, SRD amino acids interact directly with Orc1p but may not play a major role in stabilizing the structure of the OIR. If this postulate were correct, we would expect that the OIR would be resistant to proteolysis and that substitutions of an SRD amino acid would not affect the gross structure of the OIR as defined by partial proteolysis. To test this hypothesis, we conducted proteolysis experiments with limiting amounts of trypsin or chymotrypsin on His6Sir1p(M473-D611) purified from E. coli. After 30 min of digestion with limiting amounts of trypsin (100:1 His6-OIR-trypsin), His6Sir1p(M473-D611) was cleaved into four distinct protein fragments, suggesting cleavage occurred at only 3 of the 23 possible sites (Fig. 5A and B, lanes 1 to 5). Mass spectrometric analysis and Edman sequencing of the resulting protein fragments indicated that trypsin cut once in the His6 tag and twice in the OIR after K482 and K483. The remainder of the OIR, amino acids 484 to 611, was resistant to trypsin cleavage. Similar results were observed when His6-OIR was digested with chymotrypsin (Fig. 5B, lanes 6 to 10). These data suggest that the OIR forms a folded, protease-resistant domain and that the SRD region itself, containing both trypsin and chymotrypsin cleavage sites, is protected. Importantly, this low-resolution structural analysis supports the functional analysis of the OIR, since the N-terminal region of the OIR that was sensitive to both trypsin and chymotrypsin cleavage in vitro was dispensable for the Sir1p-Orc1p interaction in the two-hybrid assay (Fig. 2B).
FIG. 5.
The OIR formed a protease-resistant domain. (A) The amino acid sequence of His6-Sir1pOIR is shown here with potential trypsin cut sites boxed in gray and potential chymotrypsin cut sites boxed in black. Arrows show sites where the OIR is cleaved by trypsin (normal arrow) and chymotrypsin (barbed arrow). R493G and C595R are marked with an asterisk. (B) Coomassie-stained SDS-PAGE analysis of proteolysis products from limited digestion of wild-type or mutant versions of the OIR with trypsin or chymotrypsin. The full-length OIR is labeled with an A on each gel. Trypsin degradation products are labeled as B and C. Note that C is a doublet produced by cleavage after K481 and K482. Chymotrypsin degradation product is labeled with a B.
Limited proteolysis was also conducted on representative Sir1p mutants containing single amino acid substitutions in the OIR. We first examined the R493G SRD mutant that causes defects in silencer recognition and the Sir1p-Orc1p interaction but does not disrupt other aspects of Sir1p's function (14). Based on this specific phenotype, we predicted that this mutant retained substantial Sir1p function and therefore would not cause a global disruption of the OIR domain structure. Limited proteolysis was consistent with this prediction. There was no discernible difference in trypsin or chymotrypsin digestion patterns between wild-type His6-Sir1p(M473-D611) and mutant OIRs (R493G or A505T [data not shown]) (Fig. 5B, R493G), suggesting that these mutations in the SRD region do not disrupt the global structure of the OIR.
In contrast, the C595R mutant, located in the C terminus of the OIR, exhibited an altered trypsin digest pattern (Fig. 5B) suggesting that the stability or conformation of the OIR was affected by this amino acid substitution. Thus, the wild-type OIR formed a stable domain as measured by resistance to proteases. An R493G mutation in the SRD region did not affect this domain structure; however, a mutation disrupting Sir1p-Orc1p interactions through a single amino acid substitution outside of the SRD region exhibited altered digest patterns in the presence of trypsin or chymotrypsin.
Conservation of the Sir1p OIR in other yeast species.
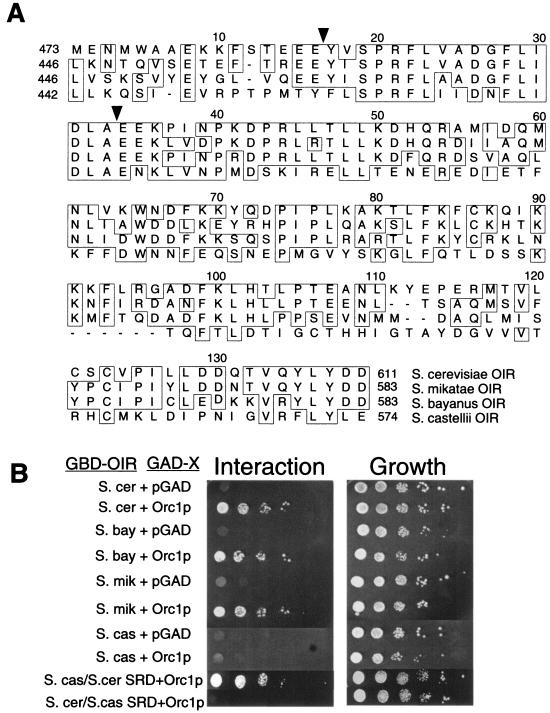
Evolutionarily conserved regions between orthologous proteins in different species can help identify amino acids important for a protein's function. Although Sir1p is not conserved in multicellular organisms, at least at the level of primary amino acid sequence, the availability of genome sequences from other yeast species makes it possible to identify orthologs of Sir1p in related yeasts. We therefore used a comparative approach to gain additional insights into regions and amino acids within the Sir1p OIR that were important for Sir1p's interaction with ORC. Specifically, we examined sequence orthologs of Sir1p in Saccharomyces mikatae, Saccharomyces bayanus, and Saccharomyces castellii, three yeast species closely related to S. cerevisiae (30) (Fig. 6A). The entire OIR, including the SRD region, is extremely well conserved between the S. cerevisiae, S. mikatae, and S. bayanus orthologs. The three proteins share 48% identity over the length of the S. cerevisiae Sir1p, 54% identity over the OIR, and 88% identity over the SRD region. In S. mikatae, there is only a single conservative amino acid change, V490I, in the SRD region. In S. bayanus there is one additional SRD region substitution, V496A. Notably, the three amino acids outside of the SRD region, W537, C595, and L608, that are important for Orc1p binding in the experiments described above are also conserved. Consistent with this high degree of similarity, the OIR of the S. mikatae and S. bayanus Sir1ps interacted with S. cerevisiae GAD-Orc1p(L5-V268) in the two-hybrid assay (Fig. 6B).
FIG. 6.
Conservation of the Sir1p OIR in other yeast species. (A) Alignments of Sir1p orthologs from S. cerevisiae, S. mikatae, S. bayanus, and S. castellii from amino acids M473 to D611, the same region of Sir1p used in biochemical assays. Identical amino acids are boxed, and the boundaries of the SRD region are indicated by black triangles. The OIR extends from the beginning of the SRD region, Y489 (first black triangle) to D611. (B) Two-hybrid examination of Sir1p orthologs. The regions indicated were cloned and tested for the ability to interact with S. cerevisiae Orc1p as described in the legend for Fig. 2A. In the S. castellii-S. cerevisiae SRD swap, S. castellii amino acids 458 to 468 were replaced with S. cerevisiae amino acids 489 to 499. In the reciprocal swap, S. cerevisiae amino acids 489 to 499 were replaced with S. castellii amino acids 458 to 468.
The extensive similarity between the S. cerevisiae OIR and the corresponding region of the S. mikatae and S. bayanus Sir1ps indicates that Sir1p is present in diverged yeast species; however, this high level of similarity makes the identification of the most critical features of the OIR, required for an ORC interaction, difficult. For this purpose, a comparison between S. cerevisiae and S. castellii Sir1p was more useful. Over the length of the S. cerevisiae Sir1p, S. cerevisiae and S. castellii Sir1ps were 19% identical, and throughout the OIR these two orthologs were 30% identical. Interestingly, however, over the length of the SRD region, the two Sir1p orthologs are 70.6% identical, suggesting that this functional domain has been conserved relative to the rest of the protein. We used site-directed mutagenesis to change a number of individual amino acids conserved in all four species of yeast to alanines (Table 2). Three of the five amino acid substitutions reduced S. cerevisiae Sir1p's ability to silence HMRa, indicating that this comparative approach should be useful in structural and functional analysis of Sir1p.
TABLE 2.
Sir1p evolution-based changes
| Amino acid change | Mating phenotypea |
|---|---|
| K508A | None |
| P512A | Reduced 100-fold |
| R526A | None |
| P546A | Reduced 10-fold |
| L554A, F555A | Reduced 100-fold |
Ability to silence HMRa.
The species comparison also indicated that the S. castellii OIR contains five amino acid substitutions in the SRD region compared to the S. cerevisiae OIR, including the nonconservative G504N substitution (Fig. 6A). If the conservation within the SRD region reflects a critical component of Sir1p's ability to interact with Orc1p, then the S. castellii OIR should fail to interact with the S. cerevisiae Orc1p in the two-hybrid assay, and indeed this is what we observed (Fig. 6B). To test whether changes within the SRD region were responsible for the inability of the S. castellii OIR to interact with the S. cerevisiae Orc1p, we created hybrid proteins and tested their abilities to interact with the S. cerevisiae Orc1p in the two-hybrid assay (Fig. 6B). In one hybrid protein, the S. castellii SRD region was replaced with the S. cerevisiae SRD region within the context of the S. castellii OIR. Significantly, this hybrid version of the Sir1p OIR interacted with the S. cerevisiae Orc1p (Fig. 6B). In contrast, the reciprocal hybrid that replaced the S. cerevisiae SRD region with the S. castellii SRD region within the context of the S. cerevisiae OIR did not interact with S. cerevisiae Orc1p (Fig. 6B). Notably, the S. castellii OIR interacted with the S. castellii Orc1p in a two-hybrid assay (data not shown).
Together, these observations indicate that a Sir1p-Orc1p interaction has been conserved in different species of Saccharomyces and underscore the functional role of the SRD region in this interaction.
The Sir1p OIR includes amino acids necessary for a Sir1p-Sir4p interaction and silencing.
Amino acids in the SRD region were identified in a screen in which Sir1p was unable to silence via the natural mechanism but was fully capable of silencing when tethered to the HMR silencer as a Gal4-Sir1p fusion bound to a Gal4p binding site (14) (Fig. 1B). The observations that these mutants cause defects in a Sir1p-ORC interaction and fail to bind the silencer in chromatin (13, 14) support the view that the primary role for ORC in silencing is to recruit Sir1p to the silencer (6, 47). Other amino acids required for an interaction with ORC may not have been revealed in the original screen (14) because they had roles in addition to controlling an interaction with ORC, such as controlling a Sir1p-Sir4p interaction that has been observed in the two-hybrid assay (47). Since Sir4p is required for silencing HMRa regardless of whether Sir1p functions by the natural or the artificially tethered mechanism, it may not have been possible to identify amino acids substitutions that caused defects in the Sir1p-Sir4p interaction in the original SRD screen.
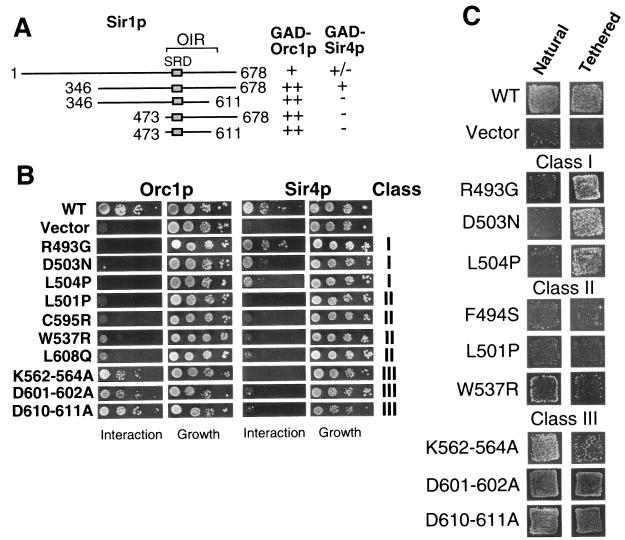
To begin to address these issues, we first determined whether the OIR was necessary or sufficient to interact with Sir4p in the two-hybrid assay by constructing deletion mutants in the context of a GBD-Sir1p fusion protein (Fig. 7A). Full-length GBD-Sir1p or a shorter GBD-Sir1p(346-678) fusion (47) interacted with both Orc1p and Sir4p in a two-hybrid assay. However, deletions of either 127 amino acids N-terminal to the OIR or 67 amino acids C-terminal to the OIR abolished the ability of Sir1p to interact with Sir4p. Thus, the OIR was included in a region of Sir1p that interacted with Sir4p in the two-hybrid assay, but the OIR was not sufficient for a Sir1p-Sir4p interaction.
FIG. 7.
The OIR includes amino acids necessary for a Sir1p-Sir4p interaction. (A) The Sir1p-Sir4p interaction region was mapped by examining GBD-Sir1p fusions containing different regions of Sir1p. (B) Sir1p mutants were tested for the ability to interact with Orc1p and Sir4p in the two-hybrid assay as described in the legend for Fig. 2A. Mutants were divided into three distinct classes based on their interaction defects. (C) Representative mutants from each class were tested for silencing function at a natural silencer (Natural) and when tethered to silencer via the GBD (Tethered). Silencing was measured as the ability of these MATα strains to mate with a MATa lawn and form viable diploids on selective medium. The “natural” silencer used here was the synthetic silencer (32). The synthetic silencer is more sensitive to Sir1p function (12). The relevant genotype of the strain used for natural silencing was MATα HMR-SSa (CFY762), and the strain used for tethered silencing was MATα HMR-SS(GAL4)a (CFY770). The chromosomal copy of SIR1 has been deleted in both strains.
To test whether any of the individual amino acid substitutions within the OIR also had roles in the Sir1p-Sir4p interaction, we performed two-hybrid assays with wild-type and several mutant versions of the GBD-Sir1p(346-678) fusion and either GAD-Orc1p or GAD-Sir4p (Fig. 7B). This analysis revealed three classes of amino acids within the OIR. Class I amino acids, which included the original SRD amino acid R493 (14), and newly identified amino acids within the SRD region, such as D503, were required for Sir1p interactions with Orc1p but were only minimally required for Sir1p interactions with Sir4p. Class II amino acids, which included amino acids C-terminal to the SRD region, i.e., C595 and W537, were required for Sir1p interactions with both Orc1p and Sir4p. Interestingly, two amino acids within the SRD region itself, F494 and L501, were class II amino acids by this analysis. Potential class II amino acids have also been identified in other regions of Sir1p (8). Finally, class III amino acids, such as K562-564A, were required for interactions with Sir4p but only minimally required for interactions with Orc1p (Fig. 7B). Consistent with data from the original SRD screen, the SRD region of the OIR contained amino acids dedicated to a Sir1p-Orc1p interaction; however, both the SRD region and the remainder of the OIR also contained amino acids required for Sir1p-Sir4p interactions.
Based on current models for HMRa silencing, Sir1p-ORC interactions should be required for silencing HMRa by the natural mechanism but dispensable for Gal4-Sir1p tethered silencing. In contrast, since Sir4p functions to recruit Sir2p and Sir3p to HMRa, Sir1p-Sir4p interactions may be necessary for both natural and Gal4-Sir1p tethered silencing. Therefore, we engineered representative amino acid substitutions from each class discussed above (Fig. 7B) into the context of full-length Gal4-Sir1p and tested the ability of these mutant versions of Gal4-Sir1p to silence via the natural or tethered mechanism (6, 11, 47) (Fig. 1A and 7C). As expected, amino acid substitutions that caused defects in a Sir1p-Orc1p interaction but had minimal effects on a Sir1p-Sir4p interaction (class I) behaved like the original SRD substitutions. Specifically, these mutant Gal4-Sir1p fusions were unable to silence by the natural mechanism but could silence efficiently via the tethered mechanism (Fig. 7C).
In contrast, substitutions of class II amino acids, such as F494S, L501P, and W537R, that caused defects in both Sir1p-Orc1p and Sir1p-Sir4 interactions caused silencing defects by both the natural and tethered mechanisms. We note that the C595R and L608Q substitutions were able to restore some silencing function when tethered (data not shown), suggesting that defects caused by these substitutions may be bypassed somewhat by tethering Sir1p at the silencer via the high-affinity Gal4p-DNA interaction. Regardless, these data are consistent with the conclusion that at least some aspects of the Sir1p-Sir4p interaction play a mechanistically different role than Sir1p-Orc1p interactions in Sir1p's silencing function.
The role of class III amino acids was more difficult to ascertain using the approach described here. Substitutions of class III amino acids, including K562-564A, D601-602A, and D610-611A, had minimal effects on Sir1p-Orc1p interactions but caused obvious defects in Sir1p-Sir4p interactions. Based on these interaction phenotypes, we predicted that these substitutions would cause defects in Sir1p-directed silencing by both the natural and the tethered mechanisms, but instead we observed close-to-wild-type levels of silencing by both mechanisms, suggesting that these amino acids had no measurable role in silencing. However, it is critical to note that the natural and tethered assays used here (Fig. 7C) required high levels of the Gal4-Sir1p fusion protein. Thus, it was possible that these amino acids caused Sir1p silencing defects that could be overcome by overexpression of Sir1p. Therefore, we engineered the relevant codon changes for these substitutions, as well other amino acid substitutions, into the chromosomal SIR1 locus and assessed the levels of HMRa silencing (Fig. 8A). Significantly, when integrated at the endogenous SIR1 locus, the class III amino acid substitutions caused ∼100-fold reductions in silencing, which were similar to the silencing defect caused by a Sir1pSRD mutant. Together these observations indicated that the OIR contained amino acids required for both Sir1p-ORC and Sir1p-Sir4p interactions and each of these interactions was important for governing Sir1p's ability to silence HMRa.
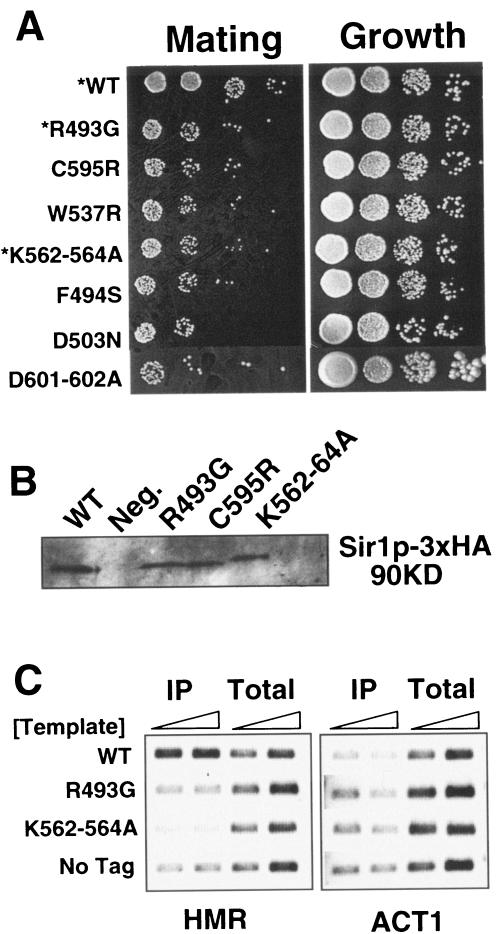
FIG. 8.
Sir1p-ORC and Sir1p-Sir4p interactions each contribute to Sir1p's ability to bind HMRa in chromatin. (A) Amino acid substitutions representing each of the three classes of Sir1p mutants (Fig. 7) were engineered at the chromosomal SIR1 locus (26) in a MATα HMR-SSa strain (CFY345). Strains expressing the mutant Sir1ps were assayed for silencing function by examining 10-fold serial dilutions for the ability to form diploids with a MATa lawn on selective medium. An asterisk indicates a strain used for the ChIP assays shown in panel C. (B) Mutant and wild-type Sir1p-3xHA were expressed at similar levels as measured by protein immunoblotting with anti-HA (by IP followed by immunoblotting). (C) Wild-type and mutant Sir1p binding to HMRa was examined using ChIP assays with anti-HA. Two reaction mixtures, using 2 and 4 μl of template, were set up for each immunoprecipitate (IP) and total sample.
Sir1p-ORC and Sir1p-Sir4p interactions each contributed to Sir1p's ability to bind HMRa in chromatin.
Available data provide compelling evidence that Sir1p must bind to HMRa to function in HMRa silencing (13, 14, 47). A Sir1p-Orc1p interaction is important for the stable binding of Sir1p to HMR-E (13). Although there is some evidence that Sir1p binding to HMR is enhanced by the presence of the other Sir proteins (13, 41), the role of Sir1p-Sir4p interactions in Sir1p binding to HMRa has not been addressed directly. Therefore, we tested whether defects in a Sir1p-Sir4p interaction, an interaction required for silencing (Fig. 8A), would affect Sir1p's ability to bind HMR-E using the ChIP assay. To perform these experiments, the appropriate codon mutations were engineered in the SIR1 locus along with a C-terminal 3xHA epitope tag such that wild-type and mutant Sir1p were expressed as Sir1p-3xHA fusions. These integrated versions of SIR1 caused defects in silencing but produced similar levels of Sir1p-3xHA (Fig. 8A and B).
ChIPs were performed using four representative isogenic yeast strains differing only in the form of Sir1p-3xHA they expressed: one strain expressed wild-type Sir1p-3xHA, one strain expressed an untagged version of Sir1p, one strain expressed R493G Sir1p-3xHA (a SRD mutant causing a Sir1p-Orc1p interaction defect), and one strain expressed K562-564A Sir1p-3xHA (a mutant causing a Sir1p-Sir4p interaction defect with only minimal effects on a Sir1p-Orc1p interaction). Strikingly, in contrast to wild-type Sir1p-3xHA, both mutant versions of Sir1p failed to bind HMR-E efficiently in vivo (Fig. 8C). These data provide evidence that both Sir1p-ORC and Sir1p-Sir4p interactions contribute to stable binding of HMRa by Sir1p in vivo.
DISCUSSION
The experiments presented in this report identified a discrete 122-amino-acid region of Sir1p (V490 to D611) that was both necessary and sufficient for formation of a specific Sir1p-ORC complex central to the establishment of a silent chromatin domain at HMRa. This region, referred to as the OIR, was also necessary, though not sufficient, for a Sir1p-Sir4p interaction that was important for Sir1p's silencing functions. Both Sir1p-ORC interactions and Sir1p-Sir4p interactions were required for stable binding of Sir1p to HMRa. Together, these observations support a model in which Sir1p's silencing function requires independent Sir1p-ORC and Sir1p-Sir4p interactions mediated through a common and discrete structural domain within Sir1p.
The Sir1p-ORC interaction.
Several independent studies support a model in which an interaction between Sir1p and the ORC is central to Sir1p's role in silencing HMRa (11, 13, 14, 47). The experiments described here support and extend this model in two important ways. First, this study provides evidence for a stable, direct, and specific physical interaction between a defined Sir1p domain (OIR) and the entire ORC in vitro. The OIR-ORC interaction was stable in the presence of high salt concentrations, and a GST-OIR affinity column could bind measurable amounts of ORC expressed at endogenous levels in a crude yeast extract. Second, this physical interaction depended on individual amino acids within the OIR that were also necessary for a Sir1p-Orc1p two-hybrid interaction and Sir1p's ability to bind to and silence the HMRa locus in vivo.
Confining Sir1p-ORC interactions to the silent loci: a role for Sir1p-Sir4p interactions.
Although these data support a central role for a Sir1p-ORC interaction in HMRa silencing, they do not address how this interaction is restricted to only a few loci such as HMRa. In particular, if an interaction between Sir1p and ORC is reasonably stable, what prevents Sir1p from binding ORC at nonsilencer replication origins? The analysis of Sir1p-Sir4p interactions presented here addresses this question. Specifically, amino acids within Sir1p necessary for a Sir1p-Sir4p interaction were also required for Sir1p's ability to silence and, even more significantly, to bind HMRa in chromatin. Indeed, an amino acid substitution within the OIR that abolished a Sir1p-Sir4p interaction but had only minimal effects on a Sir1-Orc1p interaction reduced Sir1p binding to HMRa to the same degree as an amino acid substitution in the SRD region required specifically for a Sir1p-ORC interaction. Together with the Sir1p-ORC analysis, these data indicate that Sir1p must recognize both ORC and Sir4p to obtain a stable association with HMRa in vivo. Recent studies indicate that Sir4p can bind to a silencer, presumably through direct interactions with the silencer binding protein Rap1p (34), in the absence of the other Sir proteins (23, 41). These data are consistent with the idea that Sir4p is localized near the silencer ORC, providing an ORC/Sir4p surface that is recognized most efficiently by Sir1p. Intriguingly, the OIR and Sir4 interaction regions within Sir1p overlap significantly, consistent with the idea that Sir1p simultaneously recognizes closely juxtaposed ORC and Sir4p molecules.
The next level of questions relevant to Sir1p-silencer interactions will require understanding the strength of relevant individual protein-protein interactions and the possible role that protein conformation(s) may play in these interactions. For example, although recombinant ORC could be bound by a GST-Sir1p(I346-D678) fusion protein capable of binding Sir4p and Orc1p in two-hybrid experiments, this same fusion could not bind recombinant Sir4p expressed in a similar extract (M. E. Bose, unpublished observations). This may mean that Sir1p-Sir4p interactions are inherently weaker than Sir1p-ORC interactions, but this explanation is difficult to reconcile with our observation that interfering with either a Sir1p-Sir4p or a Sir1p-ORC interaction with single amino acid substitutions abolished Sir1p's ability to bind HMRa in vivo as measured by ChIPs. An alternative possibility is that Sir1p can bind Sir4p previously bound to the silencer Rap1p significantly better than it can bind free Sir4p because Sir4p-Rap1p interactions promote a Sir4p conformation accessible to Sir1p. Answers to this and similar questions will require additional molecular analysis to further define functional protein domains, coupled with detailed and quantitative biochemical studies of protein-DNA and protein-protein interactions at the silencer.
Sir1p function in the assembly of a domain of silent chromatin.
To date, mutants that disrupt Sir1p's ability to bind its partner proteins and silence HMRa also abolish Sir1p's ability to physically bind this locus in vivo. Perhaps Sir1p has a relatively simple role in silencing: to stabilize proteins at the silencer through individual protein-protein interactions. The role of spreading the silent state to regions distal to the silencer would fall to Sir1-independent mechanisms carried out by the Sir2, -3, and -4 proteins as recently proposed (5, 23, 41). This view of Sir1p is consistent with the ability to detect by ChIPs Sir1p at HMR-E but not at regions, such as HMR-I, distant from the silencer (41). It is also consistent with the ability of the three other Sir proteins to assemble a relatively stable silent chromatin state under several conditions in the absence of Sir1p (22, 38). However, since the contribution(s) of higher-order folded chromatin structures to protein-chromatin associations is difficult to distinguish from spreading, it is worth considering that Sir1p may bind simultaneously to proteins at the silencer, such as ORC and Sir4p, and to proteins more distal to the silencer, such as Sir4p in a nucleosomal Sir2/Sir3/Sir4 complex. In this manner, Sir1p may contribute directly to stabilizing a higher-order chromatin structure at HMRa. Although this latter view still requires only that Sir1p bind chromatin at a defined location, it involves this protein in structural aspects of silent chromatin at HMRa more than models that confine Sir1p to the HMR-E silencer.
Even if Sir1p binding and function is confined to the HMR-E silencer through interactions with ORC and Sir4p, additional information about its structure, alone and in combination with partner proteins, will be useful in understanding features fundamental to silent chromatin at HMRa. In particular, the silencer-protein complex has a distinct orientation: the silent state spreads more efficiently in one direction (3, 9, 32). Sir1p's orientation and structure upon binding to the silencer, via interactions with ORC and Sir4p, may play an important role in both the specificity and directionality of the silent chromatin state at HMRa.
Finally, a role for the N-terminal region of Orc1p in silencing is conserved in Drosophila melanogaster, where it is important for ORC's interaction with the metazoan heterochromatin protein HP1 (36). Although Sir1p and HP1 share no similarity at the primary sequence level, they may share a structural feature important for interactions with ORC. Such information should contribute to a clearer picture of ORC's role in heterochromatin and perhaps provide another tool for identifying new proteins that interface with ORC and function in chromosome structure and organization.
Acknowledgments
We are particularly grateful to Paul Kaufman and Judith Sharp for helpful discussions and generous technical advice and Paul Clifton and Mark Johnston for sharing their data prior to publication. We also thank Michael Sheets and members of the Fox laboratory. We thank Bruce Stillman for ORC baculoviruses and Rolf Sternglanz for providing ORC1 two-hybrid plasmids, and we appreciate advice from a particularly thorough reviewer.
M.E.B. was supported by a training grant to the Laboratory of Genetics at the University of Wisconsin—Madison. This work was supported by a grant from the National Institutes of Health (GM056890) to C.A.F.
REFERENCES
- 1.Bell, S. P. 2002. The origin recognition complex: from simple origins to complex functions. Genes Dev. 16:659-672. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S. P., J. Mitchell, J. Leber, R. Kobayashi, and B. Stillman. 1995. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83:563-568. [DOI] [PubMed] [Google Scholar]
- 3.Bi, X., M. Braunstein, G. Shei, and J. Broach. 1999. The yeast HML I silencer defines a heterochromatin domain boundary by directional establishment of silencing. Proc. Natl. Acad. Sci. USA 96:11934-11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boumil, R. M., and J. T. Lee. 2001. Forty years of decoding the silence in X-chromosome inactivation. Hum. Mol. Genet. 10:2225-2232. [DOI] [PubMed] [Google Scholar]
- 5.Carmen, A. A., L. Milne, and M. Grunstein. 2002. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein Sir3p. J. Biol. Chem. 277:4778-4781. [DOI] [PubMed] [Google Scholar]
- 6.Chien, C., S. Buck, R. Sternglanz, and D. Shore. 1993. Targeting of Sir1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75:531-541. [DOI] [PubMed] [Google Scholar]
- 7.Denu, J. M. 2003. Linking chromatin function with metabolic networks: Sir2 family of NAD+-dependent deacetylases. Trends Biochem. Sci. 28:41-48. [DOI] [PubMed] [Google Scholar]
- 8.Dhillon, N., and R. T. Kamakaka. 2000. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell 6:769-780. [DOI] [PubMed] [Google Scholar]
- 9.Donze, D., C. Adams, J. Rine, and R. Kamakaka. 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13:698-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flanagan, J. F., and C. L. Peterson. 1999. A role for the yeast SWI/SNF complex in DNA replication. Nucleic Acids Res. 27:2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox, C. A., A. E. Ehrenhofer-Murray, S. Loo, and J. Rine. 1997. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science 276:1547-1551. [DOI] [PubMed] [Google Scholar]
- 12.Gailus-Durner, V., J. Xie, C. Chintamaneni, and A. Vershon. 1996. Participation of the yeast activator Abf1p in meiosis-specific expression of the HOP1 gene. Mol. Cell. Biol. 16:2777-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner, K., and C. Fox. 2001. The Sir1 protein's association with a silenced chromosome domain. Genes Dev. 15:147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner, K., J. Rine, and C. Fox. 1999. A region of the Sir1 protein dedicated to recognition of a silencer and required for interaction with the Orc1 protein in Saccharomyces cerevisiae. Genetics 151:31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasser, S., and M. Cockell. 2001. The molecular biology of the Sir proteins. Gene 279:1-16. [DOI] [PubMed] [Google Scholar]
- 16.Gottschling, D. 1992. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc. Natl. Acad. Sci. USA 89:4062-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grewal, S. 2000. Transcriptional silencing in fission yeast. J. Cell. Physiol. 184:311-318. [DOI] [PubMed] [Google Scholar]
- 18.Grewal, S. I., and S. C. Elgin. 2002. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12:178-187. [DOI] [PubMed] [Google Scholar]
- 19.Guthrie, C., and G. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, Calif.
- 20.Henikoff, S. 2000. Heterochromatin function in complex genomes. Biochim. Biophys. Acta 1470:O1-O8. [DOI] [PubMed] [Google Scholar]
- 21.Herskowitz, I. 1992. Fungal physiology. Yeast branches out. Nature 357:190-191. [DOI] [PubMed] [Google Scholar]
- 22.Hollenhorst, P., M. Bose, M. Mielke, U. Muller, and C. A. Fox. 2000. Forkhead genes in transcriptional silencing, cell morphology and the cell cycle: overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics 154:1533-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoppe, G., J. Tanny, A. Rudner, S. Gerber, S. Danaie, S. Gygi, and D. Moazed. 2002. Steps in assembly of silent chromatin in yeast: Sir3p-independent binding of a Sir2p/Sir4p complex to silencers and role for Sir2p-dependent deacetylation. Mol. Cell. Biol. 22:4167-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James, P., J. Halladay, and E. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenuwein, T., and C. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 26.Kang, J., T. Yokoi, and M. Holland. 1995. Binding sites for abundant nuclear factors modulate RNA polymerase I-dependent enhancer function in Saccharomyces cerevisiae. J. Biol. Chem. 270:28723-28732. [DOI] [PubMed] [Google Scholar]
- 27.Kirschmann, D., R. Lininger, L. Gardner, E. Seftor, V. Odero, A. Ainsztein, W. Earnshaw, L. Wallrath, and M. Hendrix. 2000. Down-regulation of HP1Hsα expression is associated with the metastatic phenotype in breast cancer. Cancer Res. 60:3359-3363. [PubMed] [Google Scholar]
- 28.Kitazono, A. A., B. T. Tobe, H. Kalton, N. Diamant, and S. J. Kron. 2002. Marker-fusion PCR for one-step mutagenesis of essential genes in yeast. Yeast 19:141-149. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Langkjaer, R. B., P. F. Cliften, M. Johnston, and J. Piskur. 2003. Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature 421:848-852. [DOI] [PubMed] [Google Scholar]
- 31.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 32.Loo, S., and J. Rine. 1994. Silencers and domains of generalized repression. Science 264:1768-1771. [DOI] [PubMed] [Google Scholar]
- 33.Loo, S., and J. Rine. 1995. Silencing and heritable domains of gene expression. Annu. Rev. Cell. Dev. Biol. 11:519-548. [DOI] [PubMed] [Google Scholar]
- 34.Moretti, P., K. Freeman, L. Coodly, and D. Shore. 1994. Evidence that a complex of Sir proteins interacts with the silencer and telomere-binding protein Rap1p. Genes Dev. 8:2257-2269. [DOI] [PubMed] [Google Scholar]
- 35.Muhlrad, D., R. Hunter, and R. Parker. 1992. A rapid method for localized mutagenesis of yeast genes. Yeast 8:79-82. [DOI] [PubMed] [Google Scholar]
- 36.Pak, D., M. Pflumm, I. Chesnokov, D. Huang, R. Kellum, J. Marr, P. Romanowski, and M. Botchan. 1997. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91:311-323. [DOI] [PubMed] [Google Scholar]
- 37.Parviz, F., D. D. Hall, D. D. Markwardt, and W. Heideman. 1998. Transcriptional regulation of CLN3 expression by glucose in Saccharomyces cerevisiae. J. Bacteriol. 180:4508-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pillus, L., and J. Rine. 1989. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell 59:637-647. [DOI] [PubMed] [Google Scholar]
- 39.Rolfes, R., F. Zhang, and A. Hinnebusch. 1997. The transcriptional activators Bas1p, Bas2p, and Abf1p bind positive regulatory sites as the critical elements for adenine regulation of ADE5,7. J. Biol. Chem. 272:13343-13354. [DOI] [PubMed] [Google Scholar]
- 40.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481-516. [DOI] [PubMed] [Google Scholar]
- 41.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2207-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Shore, D. 1994. RAP1: a protein regulator in yeast. Trends Genet. 10:408-412. [DOI] [PubMed] [Google Scholar]
- 44.Singh, J., and A. Klar. 1992. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev. 6:186-196. [DOI] [PubMed] [Google Scholar]
- 45.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 46.Tham, W., and V. Zakian. 2002. Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene 21:512-521. [DOI] [PubMed] [Google Scholar]
- 47.Triolo, T., and R. Sternglanz. 1996. Role of interactions between the origin recognition complex and Sir1p in transcriptional silencing. Nature 381:251-253. [DOI] [PubMed] [Google Scholar]
- 48.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 49.Urnov, F. D. 2003. Chromatin as a tool for the study of genome function in cancer. Ann. N. Y. Acad. Sci. 983:5-21. [DOI] [PubMed] [Google Scholar]
- 50.Wolffe, A., and D. Pruss. 1996. Deviant nucleosomes: the functional specialization of chromatin. Trends Genet. 12:58-62. [DOI] [PubMed] [Google Scholar]
- 51.Yasui, W., N. Oue, S. Ono, Y. Mitani, R. Ito, and H. Nakayama. 2003. Histone acetylation and gastrointestinal carcinogenesis. Ann. N. Y. Acad. Sci. 983:220-231. [DOI] [PubMed] [Google Scholar]
- 52.Zappulla, D. C., R. Sternglanz, and J. Leatherwood. 2002. Control of replication timing by a transcriptional silencer. Curr. Biol. 12:869-875. [DOI] [PubMed] [Google Scholar]