Abstract
We have isolated new histone H3 mutants in Saccharomyces cerevisiae that confer phenotypes indicative of transcriptional defects. Here we describe the characterization of one such mutant, encoded by the hht2-11 allele, which contains the single amino acid change L61W in the globular domain of H3. Whole-genome expression analyses show that the hht2-11 mutation confers pleiotropic transcriptional defects and that many of the genes it affects are normally controlled by the Swi/Snf chromatin remodeling complex. Furthermore, we show that Swi/Snf occupancy at two promoters, PHO84 and SER3, is reduced in hht2-11 mutants. Detailed studies of the PHO84 promoter suggest that the hht2-11 mutation impairs Swi/Snf association with chromatin in a direct fashion. Taken together, our results strongly suggest that the integrity of the globular domain of histone H3 is an important determinant in the ability of Swi/Snf to associate with chromatin.
In eukaryotic cells, nucleosomes pose a structural barrier to factors that use chromatin as the substrate for various cellular functions, including transcription, replication, recombination, and repair. The four core histones—H2A, H2B, H3, and H4—are the major protein components of chromatin, and it has become increasingly clear that they play crucial and active roles in the regulation of these essential functions. In particular, our understanding of the dynamic function of chromatin has significantly advanced as a result of studies of its role in the regulation of transcription (33).
One approach to defining the interplay between chromatin and transcription has been to study the protein complexes that act upon chromatin and alter its properties. One class of such complexes functions by covalently modifying specific residues within histones (6, 24). Most complexes identified thus far direct their activities toward residues located in the tails, particularly the N-terminal tails, of the core histones (65). Modifications, including acetylation, methylation, phosphorylation, and ubiquitylation, have been associated with specific chromatin effects that ultimately affect transcription. The combination of posttranslational modifications present on a particular histone is thought to constitute a “histone code” that dictates the property of that particular nucleosome (17, 52, 58).
Members of a second class of complexes, referred to as chromatin-remodeling complexes, utilize the energy from ATP hydrolysis to remodel nucleosomes in order to facilitate subsequent binding of proteins to a particular site on DNA (33, 54, 60). The yeast Swi/Snf complex is the founding member of this family, and both it and other members of this class function as activators as well as repressors of transcription (31). Genetic and biochemical experiments have shown that Swi/Snf is recruited to gene promoters by transcription factors and to subsequently alter chromatin structure (40). However, despite our basic understanding of the function of this complex, relatively little is known regarding the specific interactions that occur between Swi/Snf and chromatin.
An alternative strategy to defining the role of chromatin in transcriptional regulation has been to study the effects of histone mutations on gene expression (49). The N-terminal tails of histones H3 and H4 have been the focus of extensive mutational analyses. For example, genetic experiments in yeast have implicated the tails of H3 and H4 in maintaining silencing at the silent mating-type loci and at telomeres (20, 56). Recent whole-genome transcriptional studies have also shown that the N-terminal tail of histone H3 plays a general role in transcriptional repression (44). A number of experiments have also started to elucidate the function of the globular domains of the histone proteins in gene expression. Histone mutations have been isolated that bypass the requirement of Swi/Snf in transcriptional activation (22, 25, 41, 45, 61). Specific mutations within the globular domain of histone H2A have also been shown to affect transcription (21). Finally, recent genetic experiments have identified a nucleosomal surface that is required to maintain proper transcriptional silencing (38), as well as additional residues in the histone H3 and H4 globular domains that are important for normal levels of silencing (48, 57). In particular, K79 in histone H3, which is important for transcriptional silencing, has been shown to be the target of methylation by the Dot1 protein (9, 27, 36, 59).
The experiments presented in the present study provide new insights into the relationship between the Swi/Snf complex and chromatin. Specifically, we describe the isolation of a novel histone H3-globular domain mutant in Saccharomyces cerevisiae that appears to directly impair the association of Swi/Snf with chromatin. Combined with previous studies, our experiments strongly suggest that not only can Swi/Snf regulate chromatin structure but that the nature of the chromatin environment itself can influence how efficiently Swi/Snf is able to associate with specific promoters.
MATERIALS AND METHODS
Yeast strains, genetic methods, and media.
All S. cerevisiae strains used in the present study (listed in Table 1) are GAL2+ derivatives of the S288C strain background (64). Replacements of the HHT1-HHF1 locus with LEU2 and the HHT2-HHF2 locus with HIS3 were achieved, respectively, by transformations with pUK192 digested with HindIII and pUK431 digested with EcoRI and HincII (plasmids are gifts from M. Grunstein [29]). The (hht1-hhf1)Δ::HIS3 alleles were generated by replacing the LEU2 gene in (hht1-hhf1)Δ::LEU2 cells with the HIS3 gene amplified from pRS413 (8). The hht2Δ::URA3 alleles were constructed by replacing the HHT2 gene with the URA3 gene from pRS426 (11). The Ty912Δ35-lacZ::his4 reporter gene is a derivative of the Ty912Δ44-lacZ::his4 allele previously described (14) containing an extra 225 bp of the ɛ region. The SNF5::C18Myc::kanMX4 constructs were obtained by replacing K. lactis TRP1 within the SNF5::C18Myc::kTRP1 fusion (30) with PCR-amplified kanMX4 from pRS400 (8). Construction of snf2Δ::LEU2 alleles has been described previously (10). Generation of the SER33Δ::kanMX4 alleles has been described previously (30). Strain FY2235 (pho4Δ::KanMX4) was obtained through crosses with a commercially available deletion strain (Research Genetics). Deletion of the ARG82 gene was achieved by replacing the open reading frame (ORF) with the PCR-amplified kanMX4 gene from plasmid pRS400. Integration of the hht2-11 allele in the genome was performed by first replacing the HHT2 gene with a URA3-TRP1 cassette and then cotransforming the resulting strain with a BglII-SmaI fragment derived from pAAD11 and pRS413. His+ transformants were then selected and then screened for Ura− and Trp− phenotypes. Correct integration was confirmed by PCR. Cells containing the integrated version of hht2-11 as the sole source of histone H3 display phenotypes very similar to cells expressing the mutant histone protein from a plasmid as the only source of H3 (data not shown).
TABLE 1.
S. cerevisiae strains
| Strain | Genotype |
|---|---|
| FY84 | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δ |
| FY2160 | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δ hht2-11 |
| FY2161 | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δhht2Δ::URA3 |
| FY2162 | MATahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 Ty912Δ35-lacZ::his4 <pDM9> |
| FY2163 | MATahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 Ty912Δ35-lacZ::his4 <pDM18> |
| FY2164 | MATahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 Ty912Δ35-lacZ::his4 <pAAD11> |
| FY2165 | MATahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 Ty912Δ35-lacZ::his4 <pAAD180> |
| FY2166 | MATα his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 Ty912Δ35-lacZ::his4 met15Δ0 SNF5- C18myc::KanMX4 <pDM18> |
| FY2167 | MATα his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 Ty912Δ35-lacZ::his4 met15Δ0 SNF5-C18myc::KanMX4 <pAAD11> |
| FY2168 | MATα his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 Ty912Δ35-lacZ::his4 <pDM18> |
| FY2169 | MATα his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ::HIS3 (hht2-hhf2)Δ::HIS3 snf2Δ::LEU2 <pDM18> |
| FY2170 | MATahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ::HIS3 (hht2-hhf2)Δ::HIS3 snf2Δ::LEU2 <pDM18> |
| FY2171 | MATα his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ::HIS3 (hht2-hhf2)Δ::HIS3 snf2Δ::LEU2 <pAAD11> |
| FY2172 | MATahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ::HIS3 (hht2-hhf2)Δ::HIS3 arg82Δ::KanMX4 <pDM18> |
| FY2201 | MATahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 Ty912Δ35-lacZ::his4 ser33Δ::KanMX4 <pDM18> |
| FY2202 | MATahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 Ty912Δ35-lacZ::his4 ser33Δ::KanMX4 <pAAD11> |
| FY2203 | MATahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ::HIS3 (hht2-hhf2)Δ::HIS3 snf2Δ::LEU2 ser33Δ::KanMX4 <pDM18> |
| FY2234 | MATahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ::HIS3 (hht2-hhf2)Δ::HIS3 snf2Δ::LEU2 <pAAD11> |
| FY2235 | MATα his3Δ200 or his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 SNF5-C18myc::kTRP1 pho4Δ::KanMX4 |
Mating, transformation, sporulation, and tetrad analysis were performed by standard procedures previously described (43). Rich (yeast extract-peptone-dextrose [YPD]), synthetic dextrose (SD), synthetic complete (SC), omission (SC−), 5-fluoroorotic acid (5-FOA), and sporulation media were prepared as previously described (43). Where indicated, drugs were added to the following final concentrations: 150 mM for hydroxyurea, 15 mM for caffeine, and 3% (vol/vol) for formamide. Selection for cells harboring the kanMX4 gene was conducted on YPD medium containing 200 μg of G418 sulfate (Gibco) per ml.
Plasmid DNA construction.
pDM9 was constructed by amplifying the HHT1-HHF1 locus from plasmid pCC64 (12) and ligating it into pRS416 (8) by using HindIII and XmaI. pDM18 is based on the pRS414 plasmid (8) and contains the wild-type HHT2-HHF2 region except for the following mutations that were created by using a site-directed mutagenesis kit (Stratagene): AflII and RsrII sites were generated 3′ and 5′ of HHT2, respectively; HpaI and BglII sites were created 3′ and 5′ of the HHF2 gene, respectively. Plasmids harboring mutant versions of HHT2 were obtained by using random PCR mutagenesis and gap-repair as described below. pAAD180 was derived by subcloning a PvuII fragment containing the hht2-11-HHF2 region (from pAAD11) into pRS424 (11). The plasmids carrying the different hht2 alleles are named as follows: pAAD3 for hht2-3, pAAD5 for hht2-5, pAAD7 for hht2-7, pAAD11 for hht2-11, pAAD41 for hht2-41, pAAD44 for hht2-44, pAAD48 for hht2-48, pAAD49 for hht2-49, pAAD51 for hht2-51, pAAD56 for hht2-56, and pAAD57 for hht2-57.
Random PCR mutagenesis of the HHT2 gene.
Mutations within the HHT2 ORF were obtained by random PCR mutagenesis and gap repair as described below. PCR mutagenesis was done by a procedure similar to a previously described method (21). Plasmid pDM18 was used as a template for PCRs with the primers FO4 (5′-GGATCCCCCGGGGGTAATATGTAGACAGTGATT-3′) and FO125 (5′-GGGCGTCCTACGGATGGGAGTTGG-3′). This reaction generates a product encompassing the entire HHT2 gene, as well as 263 bp 5′ and 303 bp 3′of the ORF. Several identical pools of product were generated by using 35 cycles of PCR (94°C for 30 s, 65°C for 20 s, and 72°C for 45 s) with standard concentrations of MgCl2 (1.5 mM) and deoxynucleoside triphosphates (0.2 mM for each). After this round of PCR, 1 μl from a particular pool was used as a template for a second round of PCR under the conditions described above. The resulting products were then cotransformed into strain FY2162 with an AflII-RsrII fragment derived from pDM18 which retains the TRP1 and CEN sequences, as well as >150 bp of homology with each end of the PCR products but which lacks the HHT2 ORF. Trp+ transformants were then selected for loss of the wild-type plasmid pDM9 by selection on 5-FOA medium and tested for phenotypes.
Approximately 25,000 transformants were screened for several phenotypes. These phenotypes included inability to grow on media containing galactose or raffinose as the sole carbon source (Gal− and Raf− phenotypes) and on media lacking inositol (Ino− phenotype). We also screened for defects in growth at elevated (37°C, Ts− phenotype) or reduced (14°, Cs− phenotype) temperatures. From these analyses, we focused on 11 alleles that displayed one or more of the above phenotypes. Although some mutants initially appeared to have Gal− or Raf− phenotypes, further analysis revealed that these phenotypes were only observed when the cells were transferred from 5-FOA medium (to select for loss of the wild-type plasmid) directly onto media containing either galactose or raffinose a sole carbon source. This phenomenon was also seen to some degree in cells harboring a wild-type copy of HHT2. We do not know the cause or relevance of this effect.
Whole-genome expression analyses.
Independent isolates of strains with genotypes identical to either FY2163 or FY2164 were grown in YPD medium and collected at a density of 1 × 107 to 3 × 107 cells/ml. For the 14°C microarray experiments, HHT2 and hht2-11 cells were grown at 30°C to early log phase and then shifted to 14°C for ca. 24 h. The viability of the mutant cells was not significantly affected by the 24-h shift to the nonpermissive temperature. At 14°C, the division time for HHT2 cells is ∼10 h. Under the same conditions, hht2-11 mutant cells divide one time and then essentially stop growing. These cells do not appear to stop growing at any particular phase of the cell cycle. Total RNA was isolated by using the hot phenol method (5). The RNA was cleaned up by using the RNeasy Minikit (Qiagen). Slides were prepared by either the Harvard Medical School Biopolymers Facility or by the Whitehead Institute Center for Microarray Technology. Both types of slides are glass arrays spotted with 70-mer oligonucleotides corresponding to 6,388 ORFs (from Qiagen). Sample labeling (performed by reverse transcription-direct incorporation of cyanine dyes), analyses, hybridization, and slide washes were performed as described at the Whitehead Institute Center for Microarray Technology web site (http://www.whitehead.mit.edu/CMT/Microarrayhome.html) with the following modifications. Slides were incubated for 20 min in prehybridization solution (3.5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate, 10 mg of bovine serum albumin/ml) prewarmed to 50°C. A total of 20 pmol of each of the labeled samples was pooled and concentrated in a Speed-Vac (Savant). The samples were then resuspended in ∼130 μl of hybridization solution [3× SSC, 0.1% SDS, 0.1 mg of salmon sperm DNA/ml, 0.2 mg of tRNA/ml, 0.4 mg of poly(A)/ml], heated at 90°C for 5 min, and applied to slides placed in CMT hybridization chambers (Corning). The chambers were then placed in a 50°C water bath for an overnight incubation. Slides were scanned by using a GenePix 4000B scanner (Axon Instruments), and the data were analyzed by using the GenePix Pro 4.0 program.
Results from all microarray experiments, as well as details on the analysis of the data, can be found at http://genetics.med.harvard.edu/%7Ewinston/hht2-11.html. For experiments comparing expression profiles of cells grown at 30°C, three independent samples of cells expressing either HHT2 or hht2-11 as the only source of histone H3 were analyzed. Two independent samples of each wild-type and mutant cells were assayed for the experiments performed at 14°C. To avoid misleading data resulting from biased incorporation of one cyanine dye over the other, HHT2 and hht2-11 samples were reciprocally labeled with Cy3-dUTP and Cy5-dUTP in different experiments (see http://genetics.med.harvard.edu/%7Ewinston/hht2-11.html for details). MIPS functional classification of the affected genes in hht2-11 cells and statistical significance calculations were carried out by using the FunSpec web-based tool described elsewhere (42). The statistical significance of the overlap between affected genes in hht2-11 and snf2Δ cells, as well as between genes affected by hht2-11 at 30 and 14°C, was determined by using a hypergeometric distribution calculation. P values were calculated by using the hypergeometric distribution calculator at http://www.alewand.de/stattab/tabdiske.htm.
Northern hybridization analysis.
Cell growth and RNA isolation were performed as described above. Total RNA was separated on a 1% agarose gel and transferred to nylon membrane. The probe specific to the PHO84 mRNA was synthesized by PCR amplification of the first 246 bases of the ORF. The SER3 probe has been described previously (30). The SNR190 probe was obtained through PCR amplification of the entire gene. All probes were radiolabeled with [α-32P]dATP by random priming (5). Quantitation of relative levels of mRNA was performed by using a PhosphorImager (Molecular Dynamics). Based on the Northern blot, microarray, and chromatin immunoprecipitation experiments presented in here (see Results), as well as on previously reported findings (34, 50), growth in YPD medium is a condition that results in a significant level of activation of the PHO84 gene. The expression level of PHO84 in our experimental conditions in wild-type cells is ca. 63% of that seen in cells in which the PHO pathway is constitutively activated through a deletion for the gene encoding Pho80 (data not shown).
Chromatin immunoprecipitation experiments.
Chromatin immunoprecipitation experiments were performed as previously described (30). Pho4 chromatin immunoprecipitation was performed with a rabbit polyclonal antibody specific to the Pho4 protein (a gift from Dennis Wykoff and Erin O'Shea). Quantitative radioactive PCR was performed as previously described (28). Primers used to detect the SER3 promoter have been previously described (30). The PHO84 promoter was amplified with primers spanning a region from positions −377 to −35 (+1 = ATG). Primers to amplify a region within the POL1 gene have been described previously (26).
Indirect end-labeling analysis of SER3 chromatin structure.
Analysis of SER3 chromatin structure was performed as previously described (30). Briefly, logarithmically growing cells were collected and spheroplated. Spheroplasts were then treated with increasing concentrations of micrococcal nuclease (MNase). Naked DNA was also analyzed in these assays to determine preferential cut sites for the MNase. After these treatments, the DNA was isolated, digested with BglII, and resolved on a 1.5% agarose gel. The DNA was then subjected to indirect end-labeling analysis with a SER3-specific probe.
RESULTS
Isolation of novel histone H3 mutants.
To better understand the contribution of histone H3 in transcriptional regulation, we performed a screen for new histone H3 mutants. Transformants containing mutagenized copies of a plasmid-borne HHT2 gene were screened for a variety of phenotypes indicative of transcriptional defects, such as inositol auxotrophy (Ino− phenotype) and for sensitivity to high and low temperatures (Ts− and Cs− phenotypes; see Materials and Methods). Table 2 shows the amino acid changes and phenotypes conferred by 11 alleles isolated in this screen. DNA sequence analysis showed that most encode single-amino-acid changes and that all of the predicted amino acid changes are within the globular domain of the protein (Table 2). The finding that some mutants confer an Ino− phenotype and that most suppress an insertion mutation in the LYS2 gene (Spt− phenotype) suggests that these alleles affect transcription (for a review of these phenotypes, see references 18 and 63). The breadth and strength of the phenotypes conferred by the different hht2 alleles isolated reveal that mutations within the globular domain of histone H3 can cause a wide range of phenotypic defects.
TABLE 2.
Predicted amino acid substitutions and phenotypes conferred by the hht2 alleles
| Allele | Amino acid change(s) | Scorea
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| YPD | YPD at 14°C | YPD at 37°C | YPD + HU | YPD + Form. | YPD + Caff. | SC−Lysb | SD | SD−ino | ||
| HHT2 | 5 | 5 | 5 | 5 | 5 | 5 | 1 | 5 | 5 | |
| hht2-3 | F104S | 4 | 4 | 1 | 3 | 4 | 1 | 1 | 5 | 5 |
| hht2-5 | E50G, G132S | 3 | 1 | 1 | 0 | 0 | 0 | 3 | 4 | 3 |
| hht2-7 | F78Y, R129Stop | 2 | 2 | 0 | 2 | 0 | 0 | 1 | 4 | 4 |
| hht2-11 | L61W | 3 | 0 | 2 | 0 | 0 | 0 | 4 | 3 | 1 |
| hht2-41 | Q68R | 4 | 4 | 3 | 1 | 2 | 2 | 4 | 4 | 4 |
| hht2-44 | Q93R | 4 | 3 | 3 | 2 | 4 | 3 | 4 | 4 | 4 |
| hht2-48 | L60P, F84L | 4 | 4 | 0 | 0 | 0 | 1 | 4 | 5 | 2 |
| hht2-49 | K37E | 4 | 4 | 4 | 2 | 3 | 0 | 0 | 4 | 4 |
| hht2-51 | F54L | 4 | 4 | 2 | 2 | 4 | 3 | 3 | 5 | 5 |
| hht2-56 | Y41C | 3 | 2 | 3 | 2 | 2 | 0 | 0 | 3 | 3 |
| hht2-57 | I112T | 3 | 2 | 2 | 2 | 3 | 1 | 1 | 4 | 4 |
Mutant phenotypes were scored in strains in which the indicated hht2 allele encoded the only source of histone H3 in the cell. Growth was scored on a scale of 5 to 0, where 5 represents wild-type growth, and 0 represents no growth. Media and conditions are as described in the legend to Fig. 2.
Growth on SC-Lys scores for Spt− phenotypes. All strains contain the lys2-128δ mutation, an insertion of a Ty δ element in the ORF of the LYS2 gene (15a). In a wild-type (HHT2) background, this mutation causes a Lys− phenotype (growth score of 1). Some hht2 alleles suppress this defect (growth scores of 2 to 4), whereas some strengthen the defect (growth score of 0). These hht2 alleles are likely to affect transcription at this locus (see the text).
Analysis of the hht2-11 mutant.
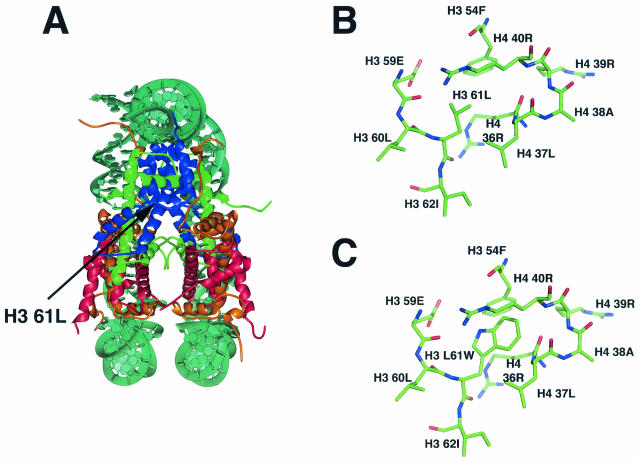
To learn more about the relationship between histone H3 and transcription, we decided to focus our analysis on the hht2-11 allele, since this mutant confers the phenotypes most strongly suggestive of transcriptional defects. Furthermore, the conditional Cs− phenotype allows the analysis of this mutant under conditions that strengthen its effects and might therefore reveal additional characteristics of the mutant. The hht2-11 allele encodes a single amino acid change at position 61 of H3, from a leucine to a tryptophan (H3 L61W). Leucine 61 is located within the globular domain of the protein in the loop region connecting the N-terminal helix and the first helix of the histone fold domain and lies on an interface between histones H3 and H4 (Fig. 1A). The substitution to a tryptophan is not predicted to confer any major structural change in the nucleosome, but the residue does appear to pack comfortably at the H3-H4 interface, possibly increasing hydrophobic interactions between H3 and H4 (Fig. 1B and C). These new interactions would result in increased H3-H4 tetramer stability that might ultimately lead to a stabilization of the H3 N-terminal helix (C. White and K. Luger, unpublished data). This stabilization might pose a barrier to the function of factors that operate by altering nucleosome-DNA interactions to mediate cellular processes, such as chromatin remodeling complexes in transcriptional regulation.
FIG. 1.
Location and nature of the amino acid substitution encoded by the hht2-11 allele. (A) Side view of the yeast nucleosome core particle as solved by White et al. (62). Part of the DNA is removed for clarity. The histone proteins are depicted as follows: H2A is in gold, H2B is in red, H3 is in blue, and H4 is in green. The arrow indicates the location of 61L within one of the two histone H3 proteins. (B and C) Close-up views of the region of interest showing the wild-type structure (B) and the predicted structure modeled with the L61W substitution (C). These figures were kindly provided by Cindy L. White and Karolin Luger.
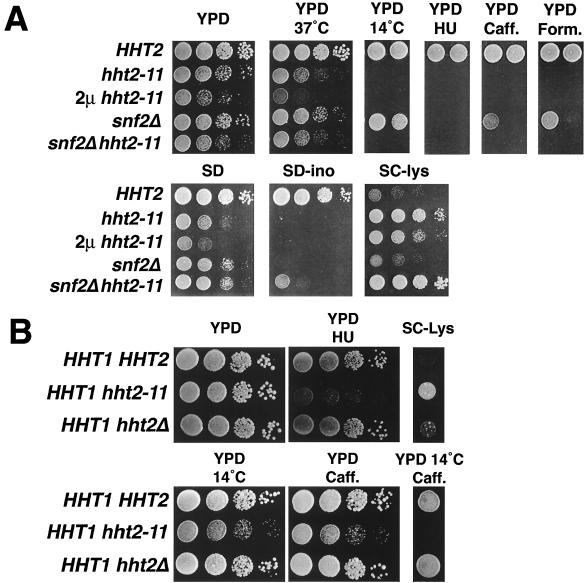
The nature and breadth of the phenotypes conferred by the hht2-11 mutation are shown in Fig. 2A. Interestingly, we found that many of these defects, including the Ino− phenotype, are shared with cells lacking the Swi/Snf chromatin remodeling complex (snf2Δ cells, compare the second and fourth rows in Fig. 2A), thus providing phenotypic evidence that the H3 L61W mutant and Swi/Snf might be functionally related. To examine the relationship between hht2-11 and snf2Δ in greater detail, we also analyzed a snf2Δ hht2-11 double mutant. This analysis (Fig. 2A, bottom row) showed that for most phenotypes examined (sensitivity to caffeine and formamide and the Spt− phenotype) the double-mutant phenotype is the same as the stronger of the two single-mutant phenotypes. For two other phenotypes (inositol auxotrophy and growth on SD medium) the double mutant is less severe (see Discussion).
FIG. 2.
Phenotypic analyses of strains harboring the hht2-11 allele. (A) Phenotypes conferred by hht2-11 when present as the sole source of histone H3. The mutant strains used were as follows: HHT2 (strain FY2163), hht2-11 (FY2164), 2μm hht2-11 (FY2165), snf2Δ (FY2170), and snf2Δ hht2-11 (FY2234). Cells were grown to saturation overnight in YPD medium, washed with H2O, and then spotted in a dilution series from 7 × 106 to 7 × 103 cells/ml on the indicated medium (see Materials and Methods). For some phenotypes, only the two most concentrated spots are shown for simplicity. An Spt− phenotype corresponds to growth on SC−Lys medium (see Table 2). The plates were then incubated at either 30°C or at the indicated temperature for the following times: YPD, 2 days; YPD at 37°C, 2 days; YPD at 14°C, 11 days; YPD + HU, 7 days; YPD + caffeine (Caff.), 5 days; YPD + formamide (Form.), 3 days; SD, 3 days; SD−inositol (ino), 4 days; and SC−Lys, 3 days. (B) Dominant phenotypes conferred by hht2-11. The strains used are as follows: HHT1 HHT2 (strain FY84), HHT1 hht2-11 (strain FY2160), and HHT1 hht2Δ (strain FY2161). Cells were grown and spotted as described above, except that for “SC-Lys” and “YPD 14°C Caff.” the spots shown are at concentrations of 8 × 107 cells/ml. The plates were then incubated as described above, except for the “YPD 14°C Caff. plate,” which was incubated for 30 days.
If H3 L61W alters the property of the nucleosome, for example, by increasing the structural stability as described above, the mutation would be predicted to behave phenotypically as a gain-of-function mutation. Alternatively, if H3 L61W impaired H3 function, for example, by causing reduced levels of H3, it would behave as a loss-of-function mutation. To distinguish between these possibilities, we performed two genetic tests. First, we tested the effect of overexpression of the hht2-11 allele from a multicopy plasmid. These strains displayed all of the hht2-11 mutant phenotypes (Fig. 2A), making it unlikely that the effects seen are a result of reduced levels of H3. Second, we tested whether any of the phenotypes conferred by the hht2-11 mutant are dominant. This was done by constructing strains that contained the hht2-11 allele, as well as a wild-type copy of HHT1, the second histone H3-encoding gene in yeast. As shown in Fig. 2B, the hht2-11 mutant displayed several dominant phenotypes compared to either wild-type or hht2Δ cells. These results strongly suggest that the effects caused by H3 L61W are the result of a mutation that alters the function of histone H3 and are not simply due to reduced H3 function.
Whole-genome expression analysis of hht2-11 cell.
To obtain a comprehensive view of the transcriptional defects conferred by the hht2-11 mutation, we performed whole-genome expression analyses by using microarrays. These experiments were performed on cells grown at both the permissive temperature (30°C) or after a shift to the nonpermissive temperature (14°C). At the permissive temperature, the hht2-11 mutation resulted in increased expression of ∼3% of the genes and decreased expression of ∼0.4% of the genes. Interestingly, whereas the percentages of the genes whose expression is increased were similar whether the cells were grown at 30°C or grown at 14°C, we observed a 10-fold increase in the numbers of genes whose expression was reduced when the cells were shifted to 14°C. Based on the structural information discussed above, the fact that the hht2-11 mutation is less permissive to transcription at 14°C might reflect either an increased stabilization of the H3-H4 tetramer or an increased dependence on normal H3-H4 interactions for transcriptional activation at this temperature. The inability of hht2-11 cells to grow at the nonpermissive temperature might be due in part to the marked increase in abnormally downregulated genes in the mutant. Among the affected genes, we see enrichment of genes involved in several functional categories, including phosphate metabolism, lysosomal and vacuolar degradation, and cell rescue, defense, and virulence (see Tables A1 and A2 at http://genetics.med.harvard.edu/%7Ewinston/hht2-11.html).
Table 3 lists the genes that are most highly affected by the hht2-11 mutation. Strikingly, among these genes, nearly 50% are affected in a similar fashion by a snf2Δ mutation (53). To further explore a potential correlation between H3 L61W and Swi/Snf functions, we performed a genomewide comparison of the transcriptional defects displayed by the histone mutant with those reported for cells deleted for SNF2 (53). This analysis (Fig. 3) shows that, for all pairwise comparisons (with the possible exception of the comparison shown in the top panel of Fig. 3B), there is a statistically significant overlap between those genes whose expression is similarly affected by both the hht2-11 and the snf2Δ mutations. A comparison of our hht2-11 data with microarray data published for mutations in three other factors that affect chromatin dynamics—Rsc30, Hda1, and Isw2 (3, 7, 16)—showed reduced statistical significance in the overlap in the case of isw2Δ versus hht2-11, and no significant overlap in the cases of rsc30Δ versus hht2-11 and hda1Δ versus hht2-11 (see Fig. A1 to A3 at http://genetics.med.harvard.edu/%7Ewinston/hht2-11.html), possibly suggesting some degree of specificity in the effects of hht2-11 on Swi/Snf. We note, however, that the correlation we observe between hht2-11 and snf2Δ by itself, although suggestive, does not demonstrate that the two mutations affect common functions. For example, unrelated mutations that activate the stress response by different mechanisms might be expected to also show statistically significant correlations in the number of genes they affect. Nevertheless, our microarray data, together with the phenotypic and structural data discussed above, suggested the possibility that nucleosomes containing the H3 L61W mutant protein might in some cases be resistant to the function of the Swi/Snf complex.
TABLE 3.
Genes whose expression is most highly affected by hht2-11
| Temp (°C)b | ORF | Genea | Fold increase (+) or decrease (−) |
|---|---|---|---|
| 30 | YOL052C-A | DDR2 | 10 (+) |
| YER081W | SER3* | 9 (+) | |
| YOL155C | 7 (+) | ||
| YNL160W | YGP1 | 7 (+) | |
| YIL169C | 5 (+) | ||
| YHR136C | SPL2* | 5 (−) | |
| YBR296C | PHO89 | 4 (−) | |
| YML123C | PHO84* | 3 (−) | |
| YDR367W | 3 (−) | ||
| YGR234W | YHB1* | 2 (−) | |
| 14 | YBR040W | FIG1 | 14 (+) |
| YIL082W-A | TyB | 10 (+) | |
| YKL221W | MCH2* | 8 (+) | |
| YFR026C | 8 (+) | ||
| YBR116C | 7 (+) | ||
| YHR215W | PHO12* | 21 (−) | |
| YBR296C | PHO89 | 20 (−) | |
| YBR093C | PHO5* | 18 (−) | |
| YAR071W | PHO11* | 17 (−) | |
| YHR136C | SPL2* | 14 (−) |
Genes affected by the hht2-11 and snf2Δ mutations in a similar fashion are indicated by an asterisk.
There was a highly significant overlap in the number of genes affected by hht2-11 at either temperature (P ≪ 10−9).
FIG. 3.
Overlap in the number of genes whose expression is affected by the hht2-11 and snf2Δ mutations. (A) A Venn diagram showing the number of genes whose expression is either increased or decreased in hht2-11 and snf2Δ cells grown at 30°C and the overlap in the two data sets with the corresponding P value. The data from 3,639 genes were used for this comparison. (B) Same as for panel A, except that the data for the hht2-11 mutant was obtained from experiments in which the cells were shifted to 14°C. The data from 2,955 genes were used for this comparison.
H3 L61W reduces the association of Swi/Snf with the SER3 and PHO84 promoters.
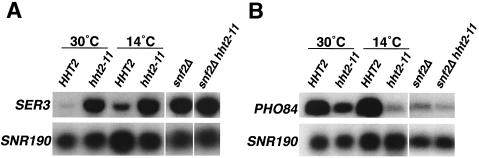
To test the possibility that nucleosomes containing H3 L61W are refractory to the chromatin remodeling activity of the Swi/Snf complex, we focused our analysis on two genes, SER3 and PHO84, whose expression is strongly affected by both hht2-11 and snf2Δ. Previous studies have strongly suggested that Swi/Snf directly represses SER3 (30) and directly activates PHO84 (51). All of the analyses presented in the present study have been carried out on material obtained from cells grown in rich (YPD) media, a condition that results in significant induction of the PHO84 gene (see Materials and Methods and references 34 and 50). Consistent with our microarray results and with previously reported data (30, 51), Northern hybridization experiments showed that both hht2-11 and snf2Δ caused increased levels of SER3 mRNA and decreased levels of PHO84 mRNA (Fig. 4). The finding that at the permissive temperature hht2-11 affects transcription of these genes less than does snf2Δ suggests that H3 L61W mutant decreases but does not eliminate the ability of Swi/Snf to function at these genes. Analysis of snf2Δ hht2-11 double mutants showed that the increase in SER3 expression seen in snf2Δ strains is not further increased by the presence of the hht2-11 mutation (Fig. 4A), a finding consistent with the notion that the two mutations affect SER3 expression through effects on a common pathway. Similarly, we see a lack of additivity of the hht2-11 and snf2Δ mutations in their effects on expression of PHO84 (Fig. 4B), although in this case the results are harder to interpret due to the already very low levels of PHO84 expression in snf2Δ cells.
FIG. 4.
The hht2-11 and snf2Δ mutations affect SER3 and PHO84 expression in a similar manner. (A) Northern analysis of SER3 mRNA levels in HHT2 (strain FY2163), hht2-11 (strain FY2164), snf2Δ (strain FY2169), and snf2Δ hht2-11 (strain FY2171) strains. snf2Δ and snf2Δ hht2-11 cells were grown at 30°C, whereas the HHT2 and hht2-11 cells were either grown at 30°C or shifted to 14°C for 24 h, as indicated. The levels of the SNR190 transcript, used as a loading control, are also shown for each strain. The data shown is representative of at least three independent experiments. (B) Northern analysis of PHO84 mRNA levels was performed with the same strains and conditions as described for panel A. The data shown are representative of at least three independent experiments.
To determine whether the histone mutant affects the association of the Swi/Snf complex with the SER3 and PHO84 promoters, we performed chromatin immunoprecipitation experiments directed against Snf5, a component of Swi/Snf. Our results show that hht2-11 cells have a significantly decreased level of Swi/Snf physically associated with both the SER3 and PHO84 promoters compared to wild-type strains (Fig. 5). The level of Swi/Snf detected at SER3 indicates that Swi/Snf still binds to this promoter to some degree, since previous work has shown that no detectable chromatin immunoprecipitation of Snf5 is observed in cells lacking Snf2, a situation in which the integrity of the Swi/Snf complex is compromised (30). These results are consistent with a model in which the H3 L61W mutant protein alters chromatin structure to reduce binding of Swi/Snf at these promoters.
FIG. 5.
Association of the Swi/Snf complex with the SER3 and PHO84 promoters is perturbed by the hht2-11 mutation. (A) Chromatin immunoprecipitations were performed on HHT2 (strain FY2166) and hht2-11 (strain FY2167) strains expressing the Snf5-Myc protein. A strain lacking the Snf5-Myc fusion (no tag, strain FY2168) was used as negative control. Amplifications of the SER3 promoter and the POL1 region (used as an internal control) were performed on chromatin samples prior to immunoprecipitation (IN, input) and after immunoprecipitation with the A14 anti-Myc antibody (IP, immunoprecipitate). For each sample, two dilutions (2× and 1×) of the chromatin templates were used for the PCR amplification to ensure linearity. The percent immunoprecipitation values (%IPs) of SER3 and POL1 were measured, and the ratios are shown in the bar graph. Each value represents the average ratio of the SER3 %IP to the POL1 %IP with the standard error from three independent experiments. (B) Chromatin immunoprecipitations were performed as described in panel A, except that the PCR amplifications were directed toward the promoter of the PHO84 gene instead of SER3. The bar graph shows the average ratios of the PHO84 %IP to the POL1 %IP with the standard error for each strain measured in three independent experiments.
Analysis of the chromatin structure at SER3 in hht2-11 cells.
To determine the nature of any chromatin changes conferred by H3 L61W, we analyzed the chromatin structure at the SER3 gene by indirect end labeling. The SER3 promoter has been previously shown to have different sensitivities to MNase in snf2Δ mutants that correlate with increased transcription (30). Figure 6 shows results from indirect end-labeling experiments on chromatin that had been treated with MNase from HHT2, hht2-11, and snf2Δ cells. In snf2Δ cells, we observed changes in two regions of the SER3 promoter compared to wild-type cells as previously described (30). In hht2-11 cells, the MNase pattern is similar to that seen in snf2Δ cells, although not quite as extreme, suggesting that the H3 L61W mutant blocks most remodeling of the SER3 promoter by Swi/Snf.
FIG. 6.
H3 L61W causes chromatin changes at the SER3 promoter. Chromatin was isolated from HHT2 (strain FY2201), hht2-11 (strain FY2202), and snf2Δ (strain FY2203) strains and treated with increasing amounts of MNase, as indicated. The resulting material was subjected to indirect end-labeling analysis. All strains were deleted for the SER33 gene to prevent cross-hybridization with the SER3 probe. Part of the SER3 gene and upstream region is depicted on the left. The brackets indicate the chromatin regions most clearly affected by both the hht2-11 and the snf2Δ mutations. Sites that become more sensitive to MNase digestion in either mutant are indicated with closed circles, whereas sites that become more resistant are indicated with open circles. N, naked DNA controls.
Evidence that H3 L61W directly affects the ability of Swi/Snf to associate with the promoter of the PHO84 gene.
The decreased level of Swi/Snf at SER3 and PHO84 might reflect a direct effect of H3 L61W on the ability of Swi/Snf to properly interact with the promoter chromatin of these genes. Alternatively, the histone mutant could act indirectly by controlling earlier events in pathways that would ultimately result in less Swi/Snf association at these promoters. To differentiate between a direct or indirect effect, we analyzed the events that occur at the promoter of the PHO84 gene in greater detail.
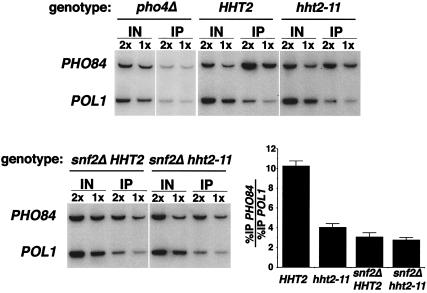
The expression of PHO84 under the conditions used in our assays is dependent upon both Swi/Snf and the Pho4 transcriptional activator (Fig. 4 and data not shown). To determine the relationship between these factors at PHO84, we performed additional chromatin immunoprecipitation experiments. In wild-type cells, we detected strong association of Pho4 at the PHO84 promoter (Fig. 7). The Pho4 activator is required for the recruitment of Swi/Snf to the PHO84 promoter since we observed a loss of Swi/Snf association at PHO84 in a pho4Δ mutant (data not shown). Next, we tested whether Swi/Snf is required for efficient association of the Pho4 protein to the PHO84 promoter. Deletion of SNF2 results in a decrease, but not complete loss, of Pho4 binding, indicating that Swi/Snf is required for full Pho4 promoter occupancy at the PHO84 gene (Fig. 7). This mutual dependence between Pho4 and Swi/Snf for chromatin association is also seen at the promoter of the PHO5 gene (51). At PHO5, Pho4 is able to bind to the accessible UASp1 site in a Swi/Snf-independent manner but requires chromatin remodeling in order to bind to the UAS2p site, which is assembled into a nucleosome (51, 55). Based on our data and the fact that the PHO84 promoter has several Pho4 binding sites (37), it is plausible that a similar situation exists at PHO84 and that Pho4 binds to some sites in a Swi/Snf-independent fashion and to others in a Swi/Snf-dependent fashion.
FIG. 7.
Chromatin association of Pho4 at PHO84 is decreased in hht2-11 and snf2Δ cells, but the effects of the two mutations are not additive. Chromatin immunoprecipitations were performed and are presented as described in Fig. 5B, except that the chromatin samples were immunoprecipitated by using a polyclonal antibody directed against the Pho4 protein. The strains used were as follows: HHT2 (strain FY2166), hht2-11 (strain FY2167), snf2Δ HHT2 (FY2169), snf2Δ hht2-11 (strain FY2171), and pho4Δ (strain FY2235). To ensure that the enrichment of the PHO84 signal detected is specific for the presence of Pho4 at the promoter, we performed control immunoprecipitations with either Pho4 antibody added to pho4Δ cells (first set of experiments shown here) or no Pho4 antibody added to PHO4 cells (data not shown). In both cases, no enrichment for the PHO84 promoter was observed compared to the POL1 internal control. The %IP values for the PHO84 and POL1 regions were measured, and the average values and corresponding standard errors from at least three independent samples for each genotype are shown in the bar graph.
Two predictions can be made if H3 L61W directly impairs Swi/Snf association at the PHO84 promoter. First, the histone mutant should cause a reduced level of Pho4 association at the PHO84 promoter. This effect is predicted to be less than or equal to the defect measured in Swi/Snf-defective cells. As shown in Fig. 7, the hht2-11 mutation does indeed cause reduced association of Pho4 at PHO84 and this defect is less severe than that seen in snf2Δ mutants. A second prediction of the direct model is that the decreased level of association of Pho4 at the PHO84 promoter in snf2Δ cells should not be further reduced by an hht2-11 mutation, since both snf2Δ and hht2-11 are affecting the same process (Swi/Snf function). Mutations that affect Pho4 binding in a Swi/Snf-independent fashion, such as those that affect the PHO pathway or that directly perturb Pho4 binding, would be expected to cause a greater reduction in Pho4 binding in combination with a snf2Δ mutation. Consistent with our prediction, we found that Pho4 binding at the PHO84 promoter in snf2Δ cells is essentially unaffected when combined with the hht2-11 mutation (Fig. 7), indicating that the two mutations affect a common pathway. These results strongly suggest that H3 L61W affects Swi/Snf binding to chromatin at a step subsequent to the initial binding of the Pho4 activator to the PHO84 promoter. Because in vitro experiments have shown that Pho4 can directly interact with the Swi/Snf complex (35), the reduced binding of Swi/Snf in the context of H3 L61W is likely to be a direct effect on Swi/Snf association with chromatin rather than through an intermediate complex connecting Pho4 and Swi/Snf.
H3 L61W perturbs Swi/Snf chromatin association through a mechanism distinct from that involving the production of phosphoinositols.
Recent studies have demonstrated a role for inositol polyphosphates in the regulation of chromatin remodeling (47, 51). Specifically, formation of the small molecules IP4 and IP5 appears to be required for proper transcription and chromatin remodeling at the PHO5 promoter. In these experiments it was shown that the absence of the Arg82 IP3 kinase resulted in decreased association of Swi/Snf at both the PHO5 and PHO84 promoters and that, at least in the case of PHO5, this occurred at a step after Pho4 binding (51). To test whether hht2-11 affects Swi/Snf association at PHO84 through an effect on the production of inositol phosphatases, we compared the extent of the defects in SER3 and PHO84 expression in hht2-11 and arg82Δ cells. As shown in Fig. 8, the hht2-11 mutation conferred stronger transcriptional defects at both SER3 and PHO84 than did an arg82Δ mutation, indicating that at least a component of the mechanism by which the hht2-11 mutation affects the regulation of these genes is distinct from that involving the production of IP4 and IP5. These results support the hypothesis that H3 L61W directly affects Swi/Snf association with chromatin.
FIG. 8.
The hht2-11 mutation affects expression of SER3 and PHO84 to a greater degree than does an arg82Δ mutation. (A) Northern analysis of SER3 levels in HHT2 (strain FY2163), hht2-11 (FY2164), and HHT2 arg82Δ (FY2172) cells. The levels of SNR190 mRNA were measured and used as a normalization control. Shown here is one representative result from three independent experiments. (B) Northern analysis of PHO84 levels was performed as described in panel A.
DISCUSSION
The results presented here provide strong evidence that the histone H3 globular domain plays an important role in ensuring proper association of the Swi/Snf complex with chromatin. Because this effect appears to be direct, these findings suggest that the nature of the chromatin environment at particular chromosomal sites can dictate how efficiently the Swi/Snf complex can be recruited and/or maintained. The broader implication of this idea is that gene-specific regulation by Swi/Snf occurs through two activities: direct recruitment of Swi/Snf by transcription factors and Swi/Snf-nucleosome interactions. This combination of activities could ensure proper transcription levels of Swi/Snf-regulated genes and might allow for differential regulation of genes that use the same factor to recruit Swi/Snf to their promoters. This notion is supported by the finding that the presence of a linker histone, which affects chromatin structure by constraining nucleosomal DNA and by promoting formation of higher order chromatin structure, also results in the decreased ability of chromatin remodeling enzymes to bind and remodel in vitro-reconstituted nucleosomal arrays (23, 39). Similarly, histone variants incorporated into nucleosomes can influence chromatin structure and transcription and in some cases have been shown to functionally interact with remodeling complexes (1, 2, 4, 13, 15, 32). It will be of particular interest to determine which components of chromatin remodeling complexes regulate how efficiently and productively the complex will associate with any given chromatin environment. Recent advances in this regard include the finding that the bromodomain of Snf2 plays a crucial role in retention of Swi/Snf to acetylated nucleosomal templates (19).
What is the mechanism by which H3 L61W reduces the ability of Swi/Snf to associate with chromatin? One possibility is that this amino acid change alters a recognition site on the nucleosome that normally interacts with Swi/Snf. Alternatively, the mutant nucleosome might be more refractory to remodeling, resulting in the disassociation of Swi/Snf from chromatin. The latter model is supported by the prediction that the L61W mutation in histone H3 might result in increased hydrophobic interactions between histones H3 and H4 (White and Luger, unpublished). The effects of substituting other amino acids with different chemical properties at position 61 should lead to a better understanding of the mechanistic nature of H3 L61W. It is also formally possible that the H3 L61W mutant creates a chromatin environment that leads to reduced binding of either a ubiquitous or an activator-recruited factor that in turn affects Swi/Snf association with the promoter. Although in this scenario H3 L61W would not be exerting its effects through direct contact with Swi/Snf, it still implies that the globular domain of histone H3 is responsible for optimal Swi/Snf chromatin association at a step downstream from activator binding in vivo. Whereas the results presented in the present study suggest that the hht2-11 and snf2Δ mutations can affect some functions through effects on common pathways, our studies do not rule out more indirect effects of H3L61W on either expression of factors required for recruitment of Swi/Snf or on components of Swi/Snf itself needed for chromatin association. Biochemical experiments with purified components will need to be conducted to more precisely ascertain the role of the globular domain of H3 in Swi/Snf chromatin association.
One prediction of the proposed model for the effect of the hht2-11 mutation on the Swi/Snf complex might be that all of the genes regulated by Swi/Snf should be affected by the histone mutation. However, whereas we see a significant overlap of the genes affected in the two mutants, there are clearly many Swi/Snf-dependent genes not affected by the hht2-11 mutation and vice versa. This observation can be reconciled with the proposed hypothesis in one of several ways. First, Swi/Snf might function in different ways at different genes and the H3 L61W mutant might only impair some of these activities. In addition, the hht2-11 mutation might confer additional effects that would mask this specific relationship with the Swi/Snf complex. This possibility is supported by our observation that some of the snf2Δ phenotypes are suppressed by the hht2-11 mutation (Gal−, Raf−, and Ino− phenotypes; Fig. 2A and data not shown). The hht2-11 mutation, therefore, might confer two opposing activities: first, in line with the proposed hypothesis, it confers resistance to Swi/Snf activity by decreasing the ability of Swi/Snf to associate with chromatin and second, it can also partially alleviate the requirement for Swi/Snf activity, perhaps by disrupting higher-order chromatin structure or through some other mechanism. As a result, only in cases where the former activity prevails over the latter will a transcriptional correlation with a snf2Δ mutation be observed.
The results presented here set the stage for further analysis of the relationship between nucleosomes and factors that function through interactions with chromatin. Specifically, detailed analyses of the effects of hht2-11 on the functions of other chromatin remodeling complexes, such as the Ino80, RSC, and Isw complexes (46, 60), will be of interest. Furthermore, the isolation of extragenic suppressors of hht2-11 should prove fruitful. For instance, suppressor mutations that alter the Swi/Snf complex or other complexes that interact with chromatin might be expected to be isolated by using such an approach, possibly leading to further insights into the physical and functional interactions that occur within the context of chromatin.
Acknowledgments
We thank Joseph Martens, Jessica Pamment, and Reine Protacio for critical comments on the manuscript and Cindy White and Karolin Luger for providing the illustrations shown in Fig. 1 and for helpful discussions. We are grateful to Delia O'Rourke for providing the pDM9 and pDM18 plasmids and to Dennis Wykoff and Erin O'Shea for the Pho4 antibodies. We thank Wolfram Hörz for reagents and helpful discussions and Tom Volkert, Barak Cohen, Suzanne Komili, Haley Hieronymus, Grant Hartzog, and Todd Burckin for advice on microarray experiments. We are grateful to Natalie Watson for help in constructing the web page containing the supplementary data for this study.
This study was supported by NIH grant GM32967 to F.W. A.A.D. was supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Fellowship (DRG-1502) and by the Charles King Trust Fellowship from The Medical Foundation.
REFERENCES
- 1.Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191-1200. [DOI] [PubMed] [Google Scholar]
- 2.Angelov, D., A. Molla, P. Y. Perche, F. Hans, J. Cote, S. Khochbin, P. Bouvet, and S. Dimitrov. 2003. The histone variant MacroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell 11:1033-1041. [DOI] [PubMed] [Google Scholar]
- 3.Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7:741-751. [DOI] [PubMed] [Google Scholar]
- 4.Ausio, J., and D. W. Abbott. 2002. The many tales of a tail: carboxyl-terminal tail heterogeneity specializes histone H2A variants for defined chromatin function. Biochemistry 41:5945-5949. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology. Greene/Wiley-Interscience, New York, N.Y.
- 6.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97:13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 9.Briggs, S. D., T. Xiao, Z. W. Sun, J. A. Caldwell, J. Shabanowitz, D. F. Hunt, C. D. Allis, and B. D. Strahl. 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418:498. [DOI] [PubMed] [Google Scholar]
- 10.Cairns, B. R., R. S. Levinson, K. R. Yamamoto, and R. D. Kornberg. 1996. Essential role of Swp73p in the function of yeast Swi/Snf complex. Genes Dev. 10:2131-2144. [DOI] [PubMed] [Google Scholar]
- 11.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 12.Clark-Adams, C. D., D. Norris, M. A. Osley, J. S. Fassler, and F. Winston. 1988. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 2:150-159. [DOI] [PubMed] [Google Scholar]
- 13.Dhillon, N., and R. T. Kamakaka. 2000. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell 6:769-780. [DOI] [PubMed] [Google Scholar]
- 14.Dudley, A. M., L. J. Gansheroff, and F. Winston. 1999. Specific components of the SAGA complex are required for Gcn4- and Gcr1-mediated activation of the his4-912delta promoter in Saccharomyces cerevisiae. Genetics 151:1365-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan, J. Y., F. Gordon, K. Luger, J. C. Hansen, and D. J. Tremethick. 2002. The essential histone variant H2A. Z regulates the equilibrium between different chromatin conformational states. Nat. Struct. Biol. 9:172-176. [DOI] [PubMed] [Google Scholar]
- 15a.Fassler, J. S., and F. Winston. 1988. Isolation and analysis of a novel class of suppressor of Ty insertion mutations in Saccharomyces cerevisiae. Genetics 118:203-212. [DOI] [PMC free article] [PubMed]
- 16.Fazzio, T. G., C. Kooperberg, J. P. Goldmark, C. Neal, R. Basom, J. Delrow, and T. Tsukiyama. 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional represssion. Mol. Cell. Biol. 21:6450-6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischle, W., Y. Wang, and C. D. Allis. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15:172-183. [DOI] [PubMed] [Google Scholar]
- 18.Hampsey, M. 1997. A review of phenotypes in Saccharomyces cerevisiae. Yeast 13:1099-1133. [DOI] [PubMed] [Google Scholar]
- 19.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 20.Hecht, A., T. Laroche, S. Strahl-Bolsinger, S. M. Gasser, and M. Grunstein. 1995. Histone H3 and H4 N termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80:583-592. [DOI] [PubMed] [Google Scholar]
- 21.Hirschhorn, J. N., A. L. Bortvin, S. L. Ricupero-Hovasse, and F. Winston. 1995. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol. Cell. Biol. 15:1999-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirschhorn, J. N., S. A. Brown, C. D. Clark, and F. Winston. 1992. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 6:2288-2298. [DOI] [PubMed] [Google Scholar]
- 23.Horn, P. J., L. M. Carruthers, C. Logie, D. A. Hill, M. J. Solomon, P. A. Wade, A. N. Imbalzano, J. C. Hansen, and C. L. Peterson. 2002. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat. Struct. Biol. 9:263-267. [DOI] [PubMed] [Google Scholar]
- 24.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 25.Kruger, W., C. L. Peterson, A. Sil, C. Coburn, G. Arents, E. N. Moudrianakis, and I. Herskowitz. 1995. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 9:2770-2779. [DOI] [PubMed] [Google Scholar]
- 26.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 27.Lacoste, N., R. T. Utley, J. M. Hunter, G. G. Poirier, and J. Cote. 2002. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 277:30421-30424. [DOI] [PubMed] [Google Scholar]
- 28.Larschan, E., and F. Winston. 2001. The Saccharomyces cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15:1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mann, R. K., and M. Grunstein. 1992. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 11:3297-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martens, J. A., and F. Winston. 2002. Evidence that Swi/Snf directly represses transcription in Saccharomyces cerevisiae. Genes Dev. 16:2231-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13:136-142. [DOI] [PubMed] [Google Scholar]
- 32.Meneghini, M. D., M. Wu, and H. D. Madhani. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112:725-736. [DOI] [PubMed] [Google Scholar]
- 33.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 34.Neef, D. W., and M. P. Kladde. 2003. Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol. Cell. Biol. 23:3788-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neely, K. E., A. H. Hassan, C. E. Brown, L. Howe, and J. L. Workman. 2002. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 22:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng, H. H., Q. Feng, H. Wang, H. Erdjument-Bromage, P. Tempst, Y. Zhang, and K. Struhl. 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogawa, N., H. Saitoh, K. Miura, J. P. Magbanua, M. Bun-ya, S. Harashima, and Y. Oshima. 1995. Structure and distribution of specific cis-elements for transcriptional regulation of PHO84 in Saccharomyces cerevisiae. Mol. Gen. Genet. 249:406-416. [DOI] [PubMed] [Google Scholar]
- 38.Park, J. H., M. S. Cosgrove, E. Youngman, C. Wolberger, and J. D. Boeke. 2002. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 32:273-279. [DOI] [PubMed] [Google Scholar]
- 39.Peterson, C. L. 2002. Chromatin remodeling enzymes: taming the machines. EMBO Rep. 3:319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 41.Recht, J., and M. A. Osley. 1999. Mutations in both the structured domain and N terminus of histone H2B bypass the requirement for Swi-Snf in yeast. EMBO J. 18:229-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson, M. D., J. Grigull, N. Mohammad, and T. R. Hughes. 2002. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Sabet, N., F. Tong, J. P. Madigan, S. Volo, M. M. Smith, and R. H. Morse. 2003. Global and specific transcriptional repression by the histone H3 amino terminus in yeast. Proc. Natl. Acad. Sci. USA 100:4084-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santisteban, M. S., G. Arents, E. N. Moudrianakis, and M. M. Smith. 1997. Histone octamer function in vivo: mutations in the dimer-tetramer interfaces disrupt both gene activation and repression. EMBO J. 16:2493-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen, X., G. Mizuguchi, A. Hamiche, and C. Wu. 2000. A chromatin remodeling complex involved in transcription and DNA processing. Nature 406:541-544. [DOI] [PubMed] [Google Scholar]
- 47.Shen, X., H. Xiao, R. Ranallo, W. H. Wu, and C. Wu. 2003. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science 299:112-114. [DOI] [PubMed] [Google Scholar]
- 48.Smith, C. M., Z. W. Haimberger, C. O. Johnson, A. J. Wolf, P. R. Gafken, Z. Zhang, M. R. Parthun, and D. E. Gottschling. 2002. Heritable chromatin structure: mapping “memory” in histones H3 and H4. Proc. Natl. Acad. Sci. USA 99(Suppl. 4):16454-16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, M. M., and M. S. Santisteban. 1998. Genetic dissection of histone function. Methods 15:269-281. [DOI] [PubMed] [Google Scholar]
- 50.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steger, D. J., E. S. Haswell, A. L. Miller, S. R. Wente, and E. K. O'Shea. 2003. Regulation of chromatin remodeling by inositol polyphosphates. Science 299:114-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 53.Sudarsanam, P., V. R. Iyer, P. O. Brown, and F. Winston. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:3364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sudarsanam, P., and F. Winston. 2000. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 16:345-351. [DOI] [PubMed] [Google Scholar]
- 55.Svaren, J., J. Schmitz, and W. Horz. 1994. The transactivation domain of Pho4 is required for nucleosome disruption at the PHO5 promoter. EMBO J. 13:4856-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson, J. S., X. Ling, and M. Grunstein. 1994. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369:245-247. [DOI] [PubMed] [Google Scholar]
- 57.Thompson, J. S., M. L. Snow, S. Giles, L. E. McPherson, and M. Grunstein. 2003. Identification of a functional domain within the essential core of histone H3 that is required for telomeric and HM silencing in Saccharomyces cerevisiae. Genetics 163:447-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turner, B. M. 2002. Cellular memory and the histone code. Cell 111:285-291. [DOI] [PubMed] [Google Scholar]
- 59.van Leeuwen, F., P. R. Gafken, and D. E. Gottschling. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109:745-756. [DOI] [PubMed] [Google Scholar]
- 60.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wechser, M. A., M. P. Kladde, J. A. Alfieri, and C. L. Peterson. 1997. Effects of Sin− versions of histone H4 on yeast chromatin structure and function. EMBO J. 16:2086-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White, C. L., R. K. Suto, and K. Luger. 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20:5207-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winston, F., and M. Carlson. 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 8:387-391. [DOI] [PubMed] [Google Scholar]
- 64.Winston, F., C. Dollard, and S. L. Ricupero-Hovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53-55. [DOI] [PubMed] [Google Scholar]
- 65.Wolffe, A. P. 2001. Transcriptional regulation in the context of chromatin structure. Essays Biochem. 37:45-57. [DOI] [PubMed] [Google Scholar]