Abstract
The pathway determining malignant cellular transformation, which depends upon mutation of the BRCA1 tumor suppressor gene, is poorly defined. A growing body of evidence suggests that promotion of DNA double-strand break repair by homologous recombination (HR) may be the means by which BRCA1 maintains genomic stability, while a role of BRCA1 in error-prone nonhomologous recombination (NHR) processes has just begun to be elucidated. The BRCA1 protein becomes phosphorylated in response to DNA damage, but the effects of phosphorylation on recombinational repair are unknown. In this study, we tested the hypothesis that the BRCA1-mediated regulation of recombination requires the Chk2- and ATM-dependent phosphorylation sites. We studied Rad51-dependent HR and random chromosomal integration of linearized plasmid DNA, a subtype of NHR, which we demonstrate to be dependent on the Mre11-Rad50-Nbs1 complex. Prevention of Chk2-mediated phosphorylation via mutation of the serine 988 residue of BRCA1 disrupted both the BRCA1-dependent promotion of HR and the suppression of NHR. Similar results were obtained when endogenous Chk2 kinase activity was inhibited by expression of a dominant-negative Chk2 mutant. Surprisingly, the opposing regulation of HR and NHR did not require the ATM phosphorylation sites on serines 1423 and 1524. Together, these data suggest a functional link between recombination control and breast cancer predisposition in carriers of Chk2 and BRCA1 germ line mutations. We propose a dual regulatory role for BRCA1 in maintaining genome integrity, whereby BRCA1 phosphorylation status controls the selectivity of repair events dictated by HR and error-prone NHR.
The tumor suppressor gene BRCA1 is mutated in up to 50% of cases of familial early-onset breast cancer and in most families with hereditary breast and ovarian cancer (50). BRCA1 is necessary for cellular processes ranging from cell cycle checkpoint control, DNA repair, regulation of transcription, protein ubiquitination, and apoptosis to chromatin remodeling (28, 48, 54, 63). Dysfunction of many or all of these BRCA1 properties may be invoked in cancer development.
Of the many types of DNA damage, DNA double-strand breaks (DSBs) represent a particularly dangerous form of damage. If not properly repaired, a DSB causes genetic changes and/or cell death. DSBs can arise spontaneously or may be induced by exogenous DNA damaging agents. The cell utilizes two principal pathways for the repair of DSBs: homologous recombination (HR) and nonhomologous recombination (NHR) (29, 62). Homology-mediated repair requires an undamaged template molecule that contains a homologous DNA sequence ordinarily on a sister chromatid or a homologous chromosome. HR is mediated through multiple proteins, including the Rad51/Rad52 recombinases and BRCA2. Several lines of evidence have indicated a role of BRCA1 in the HR pathway. BRCA1 colocalizes with BRCA2 and Rad51 (10, 41, 52, 70) and forms ionizing radiation (IR)-induced subnuclear foci (IRIF) containing Rad51 protein. Rad51 IRIF are reduced in BRCA1-null cells (5). BRCA1-deficient cells are hypersensitive to IR and DNA cross-linking agents, and the repair of both classes of lesions involves HR (5, 16, 41, 53). Loss of BRCA1 wild-type function in mouse embryonic stem (ES) cells leads to both reduced homology-mediated chromosomal plasmid integration and to decreased homologous repair of chromosomal DSBs induced by the site-specific I-SceI endonuclease (40, 41, 55). The BRCA1 domains responsible for regulation of HR have not been defined.
In contrast to HR, NHR is typically an error-prone process in which nucleotide alterations are tolerated at the sites of rejoining. Several genetically defined subtypes of NHR with different mutagenic potential may exist in mammalian cells (3, 23, 25, 26). The classical pathway of nonhomologous end joining, which mediates DSB repair as measured by pulsed-field gel electrophoresis (PFGE), and VDJ recombination are dependent upon DNA-dependent protein kinase catalytic subunit (DNA-PKcs), the Ku heterodimers, and the XRCC4-ligase IV complex (23, 24, 64). In contrast to studies of its role in HR, studies on the function of BRCA1 in NHR have yielded conflicting observations ranging from a promotion of NHR using plasmid or retroviral substrates (75) to suppressive effects using random chromosomal plasmid integration (40, 55) to no effect in PFGE (64). These results may at least in part be a reflection of different roles of BRCA1 in various subtypes of NHR.
BRCA1 has been linked to the protein complex consisting of the Nbs1 (gene mutated in Nijmegen breakage syndrome), Mre11 (mutated in AT-like disorder) and Rad50 gene products (MRN). The MRN complex possesses several biochemical activities relevant to DNA repair, but its role in mammalian DSB repair has not yet been defined. In Schizosaccharomyces pombe and DT40 chicken cells, MRN homologues are important to HR (34, 58). In Saccharomyces cerevisiae, the complex is linked to both HR and NHR and has been found to be required for the random chromosomal integration of DNA (2, 13, 21, 51). In vitro, the Mre11/Rad50/Xrs2 proteins specifically promote intermolecular nonhomologous end joining and interact with the DNA ligase IV and Ku homologues (11). In mammalian cells, Nbs1 has been shown to be required for normal PFGE (20) but not for VDJ recombination (22, 73). BRCA1 interacts and colocalizes with the MRN complex (65, 69, 76) and also inhibits the nucleolytic activity of Mre11 in vitro (44), but the role of the interaction in the function of BRCA1 in DNA repair remains to be elucidated.
In response to DNA damage, the BRCA1 protein becomes rapidly hyperphosphorylated at multiple sites by several kinases including ATM (1, 27) and Chk2 (hCds1) (30). ATM is the gene mutated in the ataxia telangiectasia syndrome. The ATM protein is critical for the cellular response to DNA damage by regulating the G1, S, and G2/M cell cycle checkpoints and by the phosphorylation of an array of protein substrates (1). Mutation of the BRCA1 target sites for ATM, serines 1423 and 1524, abolishes the ability of BRCA1 to mediate the G2/M checkpoint, while mutation at serine 1387 disrupts the S-phase checkpoint (71, 72). Mutations of the Chk2 gene have been found in a subset of patients with Li-Fraumeni syndrome (4) and with familial breast cancer (37). The Chk2 protein kinase is ATM dependent and itself governs checkpoint responses (1, 17). BRCA1 is phosphorylated by Chk2 on serine 988, and mutation of this residue prevents the dispersion of BRCA1 from subnuclear foci after IR (30). Epidemiological evidence has implicated Chk2 and BRCA1 in the same breast cancer prevention pathway, but the molecular process controlled by their interaction has not been identified (37).
In this study, we sought to discover the roles of Chk2- and ATM-mediated BRCA1 phosphorylation in the regulation of HR and NHR. Rad51-dependent HR and MRN-dependent NHR were characterized in a defined human cancer cell line. We found that an opposing regulation of HR and NHR by BRCA1 was dependent on the Chk2 phosphorylation site, while the ATM phosphorylation sites 1423 and 1524 did not affect recombination. These results support a dual model of BRCA1, in which promotion of HR and suppression of error-prone NHR processes serve to maintain chromosomal stability in proliferating cells.
MATERIALS AND METHODS
Cell lines, plasmids, and transfections.
Parental HCC1937 cells were obtained from the American Type Culture Collection. All HCC1937 lines were maintained in Iscove's modified Dulbecco's medium with 10% fetal bovine serum, 2 mM glutamine, and 100 U of penicillin and 100 μg of streptomycin per ml of medium (all Sigma). Simian Virus 40 (SV40)-transformed NBS LBi cells and the wild-type Nbs1 expression vector pWU71 were previously described (68, 69). Expression vectors for wild-type BRCA1 and the Chk2 mutant and an empty vector control (pcDNA3-BRCA1, pcDNA3-BRCA1-S988A, and pcDNA3, respectively) were reported previously (30). The ATM mutant expression vector (pcDNA3-BRCA1-S1423A/S1524A) was obtained from Stephen Elledge. pUHD10-3-based constructs for expression of wild-type Chk2 or dominant-negative kinase-dead Chk2 were obtained from Daniel Haber. U2OS clones stably transfected with the tetracycline-repressible expression vector for wild-type Chk2 or kinase-dead Chk2 or a vector control were also obtained and maintained as described previously (31).
All transfections were performed using Lipofectamine according to the manufacturer's recommendations (Invitrogen). NBS LBi cells, transfected with pWU71 or a control vector, were selected with G418 (Mediatech) at 500 μg/ml. The expression of wild-type Nbs1 protein was confirmed by Western blotting. The parental HCC1937 cells or HCC1937pDT220 cells carrying a single copy of the chromosomally integrated HR substrate were transfected with BRCA1 expression vectors and selected with 300 μg of G418/ml. After 20 to 30 days, viable G418-resistant colonies from HCC1937 transfections or pooled populations from HCC1937pDT220 transfections were expanded and screened for expression of wild-type or mutant BRCA1 protein by immunoprecipitation or Western blotting.
Recombination substrate construction and assays.
Plasmid pDT220 was generated from previously employed vectors (32, 67). The 18-bp I-SceI recognition site was created by annealing two oligonucleotides and inserting them into the unique KpnI restriction site of pSV2gpt, thereby disrupting gpt gene function (Fig. 1A). A second gpt copy, which carries a 475-bp 3′ deletion, was excised from pHWI (67) and inserted into the unique BamHI site. A clone with the downstream gpt gene inserted in inverted orientation was used for all experiments. The puromycin resistance gene (67) was introduced into the EcoRI site to allow for stable chromosomal integration. The resulting plasmid is 7.2 kb in size and is linearized by PvuI prior to transfection. The I-SceI expression and control vectors have been described previously (66).
FIG. 1.
Determination of chromosomal HR in HCC1937 breast cancer cells. (A) The recombination substrate pDT220 contains two inactive copies of the bacterial gpt gene and a puromycin resistance gene. The gpt gene copies are arranged as an inverted tandem repeat (direction of transcription indicated by arrow) and are under the control of an early SV40 promoter (SV40). The upstream copy is inactivated by insertion of an I-SceI recognition sequence into the unique KpnI site, and the downstream copy is inactivated by a deletion of the 3′ gene region. (B) Identification of HCC1937 clones carrying a single copy of pDT220 by Southern blot analysis. Probing of the XGPRT gene after EcoRV digestion yielded two bands, indicating single-copy status. The PvuI site was lost upon integration of the plasmid. HindIII digestion produced three bands, consistent with single-copy status, as the substrate contains two HindIII sites.
For the determination of chromosomal HR, HCC1937 cells were transfected with 1 to 3 μg of linearized pDT220 substrate by using Lipofectamine, and stable integrants were selected with 1.0 μg of puromycin/ml (Sigma) for 3 weeks. Puromycin-resistant colonies were isolated, expanded, and subjected to Southern blot analysis to identify single-copy integrants. Genomic DNA was extracted and subjected to digestion by EcoRV, PvuI, or HindIII endonuclease (Fig. 1B). Clones carrying single-copy substrates were then transfected with BRCA1 expression vectors as described above. Pooled populations were obtained, and stable BRCA1 protein expression was confirmed (Fig. 2A). Then, 1.2 × 106 cells were transfected with 4 μg of pCMV-I-SceI or a control vector. After 48 h, cells were reseeded, and after 24 h, XHATM selection was added (xanthine, hypoxanthine, aminopterin, thymidine, and mycophenolic acid at 250, 13.6, 0.17, 3.87, and 10 μg/ml, respectively; all Sigma). Mycophenolic acid-resistant colonies (>50 cells) were quantified after Giemsa staining. The HR frequency was recorded as the number of resistant colonies per number of cells seeded, corrected for plating efficiency.
FIG. 2.
Determination of HR, radiation sensitivity, and S-phase checkpoint in BRCA1-transfected HCC1937/pDT220 cells. (A) Cells carrying a single-copy integrant of pDT220 were transfected with wild-type (wt) BRCA1, the S988A mutant, the S1423A/S1524A mutant, or an empty vector control. Pooled cell populations exhibiting similar protein expression levels in Western blot analysis were used for further study. Expression levels were compared to endogenous wild-type BRCA1 protein expression in MCF-7 breast cancer cells. (B) Spontaneous and I-SceI-break-induced HR frequencies were determined for the cell populations shown in panel A. HR frequencies without I-SceI expression were undetectable (<107) in cells expressing the S988A mutant. Bars with standard errors (SE) are based on the cumulative colony counts from three or four independent experiments. The relative increase of HR frequencies induced by I-SceI endonuclease compared to spontaneous levels is indicated. (C) Clonogenic cell survival following exposure to IR (shown for 6-Gy data point) for cells transfected with wild-type BRCA1, the BRCA1-S988A mutant, or an empty control. Bars represent means with SE from three independent experiments. (D) Measurement of DNA synthesis 60 min following cellular exposure to 10 Gy of IR. Bars represent means with SE from three independent experiments.
The U2OS clones with wild-type Chk2 or kinase-dead Chk2 or a control were transfected with the HR substrate pDR-GFP (from Maria Jasin) (47) and selected with puromycin at 1.0 μg/ml for stable integration. At least two pDR-GFP-containing clones from each of the three cell lineages were verified for I-Sce-I-inducible HR, which produced green fluorescent protein (GFP)-positive cells. Cells were transfected with pCMV-I-SceI or a control and with or without tetracycline to control for Chk2 expression. Three days after transfection, the cells were trypsinized, suspended in phosphate-buffered saline with 0.5% fetal bovine serum, and subjected to flow cytometric analysis. Two-color fluorescence analysis revealed the percentage of green fluorescent cells relative to the total cell number, as described previously (41, 47). For each analysis, 20,000 to 50,000 cells were processed.
Integration-associated NHR in NBS LBi cells with or without wild-type Nbs1 expression or in HCC1937 cells with different BRCA1 status was determined by the random integration frequency of a linearized plasmid (pcDNA3.1/hyg; Invitrogen). To assess the effect of Chk2 on integration frequencies, the plasmid substrate was cotransfected with the wild-type Chk2 or kinase-dead Chk2 expression vector into 1.2 ×10−6 HCC1937wt-BRCA1 cells or HCC1937S988A cells. Transfected cultures were subjected to 50-μg/ml hygromycin selection (Sigma) to select for random chromosomal integration. After 20 days, colonies were scored as described above, and the NHR frequencies were calculated. Under the transfection conditions used, similar DNA uptake rates were confirmed for all cell clones under comparison, using GFP expression.
Growth and cell cycle analysis.
Growth curves for HCC1937-derived subclones were determined before conducting recombination analysis. The cells were plated at a density of 105 cells in an 80-cm2 flask. At the indicated time, cells were trypsinized and counted with a hemocytometer. Cell cycle distributions were determined using standard ethanol fixation and propidium iodide staining followed by flow cytometry (67). Radioresistant DNA synthesis using standard BrdU labeling was assessed 30 min after exposure to 10 Gy of IR in control cells and in HCC1937 cells transfected with wild-type BRCA1, BRCA1-S988A, or a vector control.
Immunoprecipitation and immunoblotting.
Cells transfected with the BRCA1 expression constructs were expanded, and cellular extracts were prepared by resuspending the cells in radioimmunoprecipitation assay lysis buffer and incubating on ice for 30 min. Supernatants were collected following centrifugation at 14,000 × g for 15 min. Ten micrograms of C-terminal BRCA1 antibody (Ab-1; Oncogene) was added to 300 μg of protein extract and rotated for an additional 2 hours at 4°C. The immune complexes were collected with 100 μl of slurry of protein A-Sepharose CL-4B beads (Amersham Pharmacia Biotech AB). The complexes were washed twice with lysis buffer, fractionated by sodium dodecyl sulfate-5% polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, using a semidry membrane, and immunoblotted with an N-terminal BRCA1 antibody (Ab-1; Oncogene). The films were developed by using enhanced chemiluminescence. Nbs1 protein expression was confirmed using anti-hNbs1/p95 polyclonal rabbit antibody (Novus).
Immunofluorescence.
Cells were seeded on four-well chamber slides for 24 h and irradiated with 2 to 10 Gy (Siemens Stabilipan 2; 250 KVp, 12 mA, 2.08 Gy/min). In HCC1937 cells, Mre11 and Rad51 IRIF were analyzed after 1 and 8 h. U2OS cells with inducible exogenous wild-type or kinase-dead Chk2 were treated with 8 Gy, and Rad51 foci were scored after 8 h. For the analysis of Mre11 IRIF, cells were fixed in ice-cold methanol, permeabilized in ice-cold acetone, and blocked with 10% fetal calf serum. Cells were then incubated initially with human Mre11 antibody (hMre1 antibody Ab-1; Oncogene) at a 1:25 dilution, followed by fluorescein isothiocyanate-conjugated secondary antibody at a 1:200 dilution. For Rad51, cells were fixed in a solution of 3% paraformaldehyde and 2% sucrose, permeabilized in Triton buffer (0.5% Triton X-100 in 20 mM HEPES-50 mM NaCl-3 mM MgCl-300 mM sucrose [pH 7.4]), and blocked in 10% goat serum. Cells were then incubated with human Rad51 antibody (Oncogene) at a 1:200 dilution, followed by fluorochrome-conjugated secondary antibody at a 1:200 dilution. All slides were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) and visualized by fluorescence microscopy (Olympus BX51). Only cells with nuclei containing more than five foci were scored. At least 300 nuclei were examined for each data set.
RESULTS
Study of chromosomal homologous recombination in BRCA1-deficient human breast cancer cells.
We chose to assess chromosomal HR using an established plasmid assay that involves DSB induction by the rare-cutting I-SceI endonuclease (40, 41, 47). The recombination substrate pDT220 carries two mutated copies of the bacterial gpt gene. The upstream copy has been inactivated by insertion of an I-SceI recognition site, whereas the downstream copy harbors a gene-inactivating 3′ deletion in the inverse orientation (Fig. 1A). DSB induction by transient transfection of an I-SceI expression vector can lead to gene conversion, in most cases, reconstituting the gpt gene and conferring resistance to XHATM selection in a colony assay.
The human breast cancer cell line HCC1937 (12, 30, 53, 61) carries a BRCA1-null mutation and the 5382insC allele, producing a truncated protein that is unstable and expressed at a very low level compared to that in cells with wild-type BRCA1 (10). These cells are hypersensitive to IR and show DSB repair defects (53). HCC1937 cells were stably transfected with pDT220, and clonal lines carrying a single-copy plasmid integrant (HCC1937pDT220) were identified by Southern blot analysis (Fig. 1B). To initiate HR in the template, the cells were transfected with pCMV-I-SceI or a control vector. Recombinants were detected via formation of XHATM-resistant colonies. In the absence of DSBs, the spontaneous HR frequency was found to be 0.6 × 10−6. Following DSB generation by the I-SceI endonuclease, the HR frequency was 2.5 × 10−6, which was an approximately fourfold stimulation over baseline.
Promotion of homologous recombination by BRCA1 requires the Chk2-dependent serine 988 phosphorylation site.
Posttranslational modification of BRCA1 may modulate its functions, associations with other proteins, and/or intracellular localization. To understand the pathways involving BRCA1 in homology-mediated recombination, we transfected HCC1937pDT220 cells with a vector control, a wild-type BRCA1 expression vector, or a plasmid coding for either of two classes of phosphorylation site mutants. The serine 988-to-alanine substitution (S988A) eliminates phosphorylation by Chk2 (30), whereas serine 1423-to-alanine and serine 1524-to-alanine mutations (S1423A/S1524A) interfere with ATM-dependent phosphorylation. We selected cell populations expressing wild-type, S988A, and S1423A/S1524A proteins at comparable levels (Fig. 2A). Growth rates and cell cycle profiles of HCC1937 cells expressing the various BRCA1 proteins were virtually identical to the behavior of control cells (data not shown).
Next, we examined HR events in the pDT220 substrate of the BRCA1-expressing HCC1937pDT220 cells and relevant controls. In the presence of wild-type protein, I-SceI expression resulted in more than 10-fold induction of HR at 18.4 × 10−6, which is an almost fourfold increase over the induction rate observed in vector alone-transfected cells (Fig. 2B). Notably, BRCA1 reconstitution of HCC1937 cells also elevated the spontaneous HR frequencies by twofold, which had not been detected in mouse ES cells (40). In contrast, cells with the S988A mutation displayed defective HR, with the basal level of HR undetectable. The endogenous hemizygous mutant BRCA1 allele in HCC1937 cells encodes a truncated protein that is unstable and expressed at very low levels, but it still carries the serine 988 site and can be phosphorylated by Chk2 (42). Thus, the trace amount of endogenous BRCA1 may have a residual activity in recombination, and overexpression of the exogenous S988A mutant might exhibit a dominant-negative effect in this experiment; however, this was not explored further. Importantly, the S988A mutant does not confer an overall null phenotype, which in principle could be the result of an altered tertiary protein structure, because cells expressing BRCA1-S988A demonstrated wild-type function in other assays including increased radiation resistance and correction of radioresistant DNA synthesis (Fig. 2C and D). Figure 2B shows that in contrast to S988A, the S1423A/S1524A mutant mediated spontaneous and I-SceI-induced HR frequencies that were clearly higher than those in BRCA1-deficient control cells and approached the levels achieved with wild-type BRCA1 reconstitution. Of note, in all experiments transfection frequencies were monitored by parallel transfections of a control vector coding for GFP to assure that each of the pooled populations of HCC1937pDT220 cells had equivalent uptake and expression of transfected plasmids.
The BRCA1 serine 988 phosphorylation site regulates Rad51 subnuclear localization.
Since there is an association of BRCA1 with Rad51, promotion of HR by BRCA1 may also control additional aspects of Rad51 function. In fact, the formation of Rad51 subnuclear foci has been reported to be impaired in BRCA1-deficient cells (5). To define the relationship between BRCA1 and Rad51 further, we studied the formation of Rad51 foci (IRIF) 8 h after exposure to 2 Gy of IR (Fig. 3B). While in unirradiated HCC1937 cells the proportion of cells with Rad51 foci was around 10% irrespective of BRCA1 status, the percentage of cells with Rad51 IRIF in vector-transfected HCC1937 cells was 32.1%. By comparison, in the presence of wild-type BRCA1, the proportion of cells with IRIF rose significantly to 66.8%, which corresponded to a more than a sixfold increase over the baseline in unirradiated cells (P = 0.033, compared to vector control cells with IRIF, t test, two sided). Expression of the S1423A/S1524A mutant also yielded a significantly increased number of cells with IRIF, 60.5%, similar to wild-type BRCA1 (P = 0.35). In contrast, cells expressing the S988A mutant were limited to 27.8% IRIF, not statistically different from the observation for parental cells (P = 0.56). At a higher dose of 10 Gy, virtually identical effects of BRCA1 status on Rad51 IRIF were observed (data not shown). The differences in the percentage of cells with Rad51 foci are not explained by different cell cycle distributions, as the fractions of cells in S phase (19.6 to 22.5%) and in G2 phase (27.1 to 32.9%) were virtually identical among all four HCC1937 cell populations under study. Thus, the effects of disrupting the Chk2 phosphorylation site are clearly revealed for both the enhancement of chromosomal HR and the formation of Rad51 IRIF.
FIG. 3.
Formation of Rad51 subnuclear foci in HCC1937 cells with or without exposure to IR. (A) Rad51 foci form only in a subset of cells, with and without exposure to IR. Green and blue images represent immunofluorescence labeling and DAPI nuclear DNA staining, respectively. (B) Quantitative analysis of Rad51 foci in HCC1937 cells transfected with expression vectors as described for Fig. 2, with or without exposure to 2 Gy of IR. Bars represent means with standard errors and are based on three independent experiments.
Nbs1 stimulates random chromosomal plasmid integration.
BRCA1 binds to double-stranded DNA, interacts with the MRN complex, and inhibits Mre11 exonuclease activity in vitro (44, 65, 76). These findings suggest that BRCA1 regulates the MRN protein complex, but a precise in vivo role for this complex in recombination functions has not yet been established. Therefore, we set out to utilize an NHR assay that is dependent on both BRCA1 and the MRN complex. We sought to characterize the effects of Mre11 and Nbs1 on the random chromosomal integration frequency of a linearized plasmid substrate to measure a specific subtype of NHR (38, 70). The attempts to determine integration proficiency in Mre11-deficient ataxia telangiectasia-like disorder cells (56) were not successful due to the poor clonogenic survival of these cells. In contrast, Nbs1-deficient LBi fibroblast cells were successfully transfected with the linearized pcDNA3.1/hyg substrate, and the integration frequency was determined to be 6.2 × 10−4 (Fig. 4A). A wild-type Nbs1 expression vector was introduced, and protein expression was confirmed (Fig. 4B). The presence of wild-type Nbs1 led to an approximately fivefold-higher integration frequency compared to that of the vector control (Fig. 4A). As MRN complexes fail to form in LBi cells, these observations suggest that the Nbs1, Mre11, and Rad50 proteins constitute a positive mediator of the NHR pathway that is required for random chromosomal DNA integration.
FIG. 4.
Determination of random chromosomal integration-linked NHR in NBS LBi fibroblasts and HCC1937 cells. (A) The random integration frequency of the linear plasmid substrate pcDNA3.1/hyg was determined in Nbs1-deficient cells, with or without exogenous expression of the wild-type Nbs1 protein. (B) Verification of Nbs1 protein expression using pooled populations transfected either with an empty control vector or a wild-type (wt) expression vector. (C) Verification of BRCA1 wild-type and mutant protein expression levels in single-cell-derived clones of HCC1937. (D) Plasmid integration frequency was determined in HCC1937 subclones transfected with BRCA1 expression vectors as described for Fig. 2. The logarithmic means with standard errors based on three independent experiments are shown.
Suppression of random chromosomal plasmid integration by BRCA1 requires the serine 988 phosphorylation site.
Wild-type BRCA1 or a phosphorylation site mutant was stably transfected into clonally derived HCC1937 cells. These single-cell-derived populations were confirmed to express equal levels of BRCA1 proteins (Fig. 4C). Each of these derivatives and the vector alone-transfected control were evaluated for the level of random chromosomal integration of linearized plasmid DNA, as described above. Under the transfection conditions used, we confirmed that the various HCC1937 derivatives had similar plasmid uptake rates, using transient expression of a GFP vector. The integration frequency in BRCA1-deficient HCC1937 cells was found to be 80.7 × 10−5 (Fig. 4D). With reconstitution of wild-type BRCA1, integration frequencies were suppressed more than 10-fold at 7.4 × 10−5. In contrast, expression of the S988A mutant yielded an integration frequency of 98.0 × 10−5, indicative of a loss of the BRCA1-mediated suppression of NHR. The S1423A/S1524A mutant retained the ability to suppress DNA integration, with a mean integration frequency of 8.7 × 10−5. Therefore, the BRCA1-dependent suppression of random plasmid integration is mediated by the serine 988 site and opposite to the effect of Nbs1 in the same recombination assay.
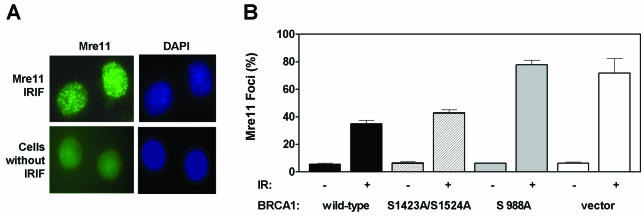
Inhibition of the Mre11-dependent cellular DNA damage response by BRCA1 is also dependent on the S988 site.
MRN complexes appear in subnuclear foci during S phase, and both the number of foci and fraction of cells bearing foci increase following DNA damage (8, 35, 36, 39). Yet, the relationship of MRN foci to BRCA1 function is uncertain (65, 69, 76). To test the hypothesis that MRN function is inhibited by BRCA1 in vivo, we chose to study the effect of cellular BRCA1 status on the subnuclear localization of the MRN protein complex (Fig. 5A). We established clonally derived cell lines stably expressing equal levels of exogenous BRCA1 protein and determined the number of Mre11 IRIF as a function of BRCA1 phosphorylation status. In the absence of IR exposure, the percentage of Mre11 cells containing foci was approximately 6% in all cell lines studied (Fig. 5B). In vector-transfected HCC1937 cells, the proportion of cells with Mre11 IRIF 1 h after exposure to 2 Gy was 71.9%. In contrast, with expression of wild-type BRCA1, cells with Mre11 IRIF were limited to 35.1% (P = 0.038). Introduction of the S988A mutation eliminated the suppressive effect of BRCA1, with 77.9% of the cells showing Mre11 IRIF (compared to wild-type BRCA1, P = 0.016). When disruption of the ATM phosphorylation sites was examined, the proportion of Mre11 IRIF in cells expressing the S1423A/S1524A mutant was comparable to that in cells expressing the wild-type BRCA1. When looking at the effect of BRCA1 status on Mre11 IRIF 8 h after irradiation with 2 Gy, comparable results were obtained (data not shown).
FIG. 5.
Formation of Mre11 subnuclear foci in HCC1937 cells with or without exposure to IR. (A) Mre11 foci form only in a subset of cells, with and without exposure to IR. Green and blue images represent immunofluorescence labeling and DAPI nuclear DNA staining, respectively. (B) Quantitative analysis of Mre11 foci done as described for Fig. 3B except that IRIF were assessed 1 h after exposure to 2 Gy of IR. Bars represent means with standard errors and are based on three independent experiments.
Previous studies have reported independence of Mre11 IRIF from BRCA1 status (65, 69). However, we considered the possibility that the use of heterogeneous pooled cell populations in these studies might have masked any effect on Mre11 in the subfraction of cells expressing functional exogenous BRCA1. Indeed, when similarly measuring Mre11 IRIF in pooled cell populations following BRCA1 transfection, we too could not detect any effect (data not shown).
Inhibition of Chk2 kinase activity mirrors the effects of the BRCA1 S988A mutation.
To test whether the mutation of the serine 988 site was specific for the function of the Chk2 kinase, we employed an expression vector for kinase-dead Chk2 or wild-type Chk2 as a control, as described by Lee et al. (31). The kinase-dead vector exhibits a dominant-negative effect on endogenous wild-type Chk2 kinase activity. Moreover, it has been shown that expression of kinase-dead Chk2 blocks the phosphorylation of BRCA1 at Ser988 in vivo (30). For the study of NHR, a linearized reporter plasmid was cotransfected with either wild-type or kinase-dead Chk2 constructs into HCC1937BRCA1-wt or HCC1937BRCA1-S988A cells, analogously to the experiments for which results are shown in Fig. 4D. The expression of exogenous wild-type Chk2 did not show a significant effect on random integration frequencies compared to the control line, either in the presence of wild-type BRCA1 or with the BRCA1-S988A mutant (Fig. 6A). In comparison, expressing kinase-dead Chk2 in a wild-type BRCA1 background led to a 2- to 3.6-fold increase in integration frequencies in all replicates; however, in the cells expressing mutant S988A, kinase-dead Chk2 failed to cause a further increase of the already elevated NHR levels (P = 0.02, t test, double sided). As perhaps expected, the NHR levels in the cells with wild-type BRCA1 that were transiently transfected with mutant Chk2 did not reach the levels seen in the clones with stable expression of S988A. Together, these results indicate a role of Chk2 phosphorylation in the suppression of integration-associated NHR and the requirement of the BRCA1 serine 988 site to dictate this function.
FIG. 6.
Influence of Chk2 kinase activity on NHR and HR. (A) Assessment of random chromosomal integration of the pcDNA3.1/hyg substrate as described for Fig. 4. HCC1937 cells with wild-type (wt) BRCA1 or mutant BRCA1-S988A were transfected with a wt or a dominant-negative kinase-dead (kd) Chk2 expression vector in the absence of tetracycline. Bars represent the logarithmic means of three independent experiments with standard errors (SE). (B) Assessment of HR using the pDR-GFP substrate. U2OS clones 1 and 6 carry the tetracycline (Tet)-repressible expression vector for wt Ck2 and for kd Chk2, respectively. Cells were transfected with an I-SceI expression vector or a control, with or without tetracycline. Bars represent the results of a comprehensive representative experiment. HR frequencies for kd Chk2 were normalized to the level of spontaneous HR in cells with tetracycline-repressed wt Chk2 expression (i.e., endogenous Chk2 only). (C) Assessment of Rad51 foci in U2OS cells with or without exposure to IR. Bars represent means with SE and are based on four independent experiments.
Studying the role of Chk2 in HR proved to be more difficult, as the cotransfection of the Chk2 constructs with the I-SceI expression vector into HCC1937pDT220 cells did not yield a sufficiently high number of recombinants for analysis. We therefore employed a panel of U2OS cell clones that had been stably transfected with the inducible tetracycline-based kinase-dead Chk2 expression vector or the corresponding wild-type Chk2 construct (31). Homologous recombination was measured via reconstitution of a GFP reporter gene within a chromosomally integrated plasmid substrate as described previously (41, 47). Removal of tetracycline at the time of I-SceI vector transfection resulted in induced expression of the respective Chk2 protein. In cells with the inducible wild-type Chk2 expression vector, the stimulation of HR by I-SceI breaks was on the order of fivefold with and without tetracycline (Fig. 6B). When dominant-negative kinase-dead Chk2 expression was repressed by tetracycline, we also observed a severalfold stimulation of HR. However, following induced expression of kinase-dead Chk2, break-induced HR frequencies were reduced to the level of spontaneous HR (P = 0.062, paired t test, two sided, n = 4 repeats). Of note, the measured HR frequencies were higher than in HCC1937 cells (Fig. 2B), an observation that likely is cell line or plasmid integration site dependent. To confirm the influence of functional Chk2 status on HR, we assessed Rad51 foci in response to IR, using the same cell system as for Fig. 6B. In the presence of endogenous or exogenous wild-type Chk2, there were no differences in the formation of Rad51 IRIF; i.e., on average between 76 and 83% of the cells scored positive (Fig. 6C). In contrast, upon induced expression of the kinase-dead Chk2 protein we observed a statistically significant reduction to 60.8% (P = 0.01, paired t test, two sided). In summary, these data support the hypothesis that the pathway controlled by Chk2-BRCA1 promotes Rad51-dependent HR while at the same time inhibiting MRN-mediated NHR.
DISCUSSION
The effect of BRCA1 on the suppression of malignant cellular transformation remains a poorly defined pathway. In this paper, we studied the influence of disrupting BRCA1 phosphorylation sites on chromosomal Rad51-dependent HR and MRN-mediated NHR in the well-characterized human breast cancer cell line HCC1937. We provide evidence that the promotion of HR and suppression of NHR are governed by the serine 988 residue of BRCA1, which is phosphorylated by the cell cycle checkpoint kinase Chk2.
Mechanisms of BRCA1-dependent promotion of homologous recombination.
Our observations agree with previous findings obtained with mouse ES cells that chromosomal HR is facilitated by BRCA1 (Fig. 2) (40, 41, 55). The novel finding in this study is that both Rad51-dependent homology-mediated DSB repair and the accumulation of Rad51 IRIF required the BRCA1 serine 988 residue, which is a target for Chk2 phosphorylation, therefore suggesting that Chk2 protein kinase activity on BRCA1 is instrumental in dictating its functions (Fig. 2B and 6B and C). The involvement of a mammalian cell cycle checkpoint protein in HR is perhaps not surprising. It has been demonstrated for Caenorhabditis elegans that Chk2 is required for homologous chromosomal pairing and crossover recombination (33). In S. pombe, Rad3 is necessary for DSB-induced cell cycle delay and efficient HR (49). In another study, hyperphosphorylation of the BRCT domain-containing Crb2 checkpoint protein regulated HR, perhaps similar to the function of BRCA1 (9). Recent epidemiological data place Chk2 and BRCA1 in a common cancer-preventing pathway (37). The 1100delC mutation of Chk2 was linked to an increased incidence of familial breast cancer in noncarriers of BRCA1/2 germ line mutations, while no increased Chk2 mutation frequency was found among individuals with BRCA1/2 mutations. These observations suggest that if the pathway controlled by BRCA1 and Chk2 is already subverted by inactivating BRCA1 mutations, then abolishing Chk2 function may confer no demonstrable additional risk of disease. Intriguingly, the results presented in this paper raise the possibility that the promotion of HR mediated by the Chk2-BRCA1 pathway contributes to the prevention of breast cancer development.
HR requires the involvement of proteins necessary for strand exchange and resolution (i.e., Rad51) but also is likely to be mediated by additional regulatory factors that may influence the timing of the events, the distribution of molecular complexes involved, and integration with other nuclear functions, such as DNA replication. There is no evidence to suggest that BRCA1 plays a direct role in strand exchange in HR per se, but it is likely to play a role in the regulatory events surrounding the integrity of HR. The resolution of the recombination products measured here is a Rad51-dependent event, as neither the pDT220 substrate (Fig. 1A) nor the pDR-GFP plasmid (Fig. 6B) (47) allow for the scoring of Rad51-independent single-strand annealing events. Considering the extensive protein associations within the BASC genome surveillance complex, which contains both Rad51 and BRCA1 (65), and the strength of the BRCA2-Rad51 interaction, it is likely that the effects mediated by BRCA1 are indirectly affecting Rad51.
HR may play a significant role in maintaining chromosome stability by occurring in a controlled manner relative to the activation of cell cycle checkpoints. During S and G2 phases, when chromosomes are partially or nearly completely duplicated, DNA repair by HR rather than NHR may be the preferred pathway (59). However, the disruption of Chk2-mediated phosphorylation of BRCA1 does not cause a defective S-phase checkpoint (Fig. 2D). Recent data on the function of the Fanconi anemia D2 gene product are consistent with this notion (57). Disruption of ATM-dependent phosphorylation of Fanconi anemia D2 resulted in a defective S-phase checkpoint but did not affect cellular resistance to mitomycin C, which is thought to be mediated by an HR process. Thus, regulation of HR can be dissociated from S-phase checkpoint control.
Regulation of BRCA1 function by ATM.
In addition to Chk2, two other regulatory kinases, ATM and ATR, phosphorylate BRCA1 in response to DNA damage and during cell cycle transitions (1, 19, 27, 60). Surprisingly, ATM-mediated phosphorylation of serines 1423 and 1524 was not required for the function of BRCA1 in regulation of either HR or NHR in our study (Fig. 2 to 5). It has been demonstrated that serine 1423 and 1524 are required for the formation of the IR-induced G2/M checkpoint (71, 72). Thus, it is possible that ATM predominantly regulates the involvement of BRCA1 in checkpoint responses. Similar to the dissociation of S-phase control and promotion of HR discussed above, these observations suggest that maintenance of the damage-induced G2/M checkpoint is also not a critical mechanism for the regulation of HR by BRCA1. However, since there is evidence connecting ATM to regulation of HR (32), we cannot exclude the possibility that other ATM-dependent phosphorylation sites of BRCA1 may regulate its function in HR and NHR or that other ATM targets are primarily utilized for these functions of ATM.
Suppression of MRN-dependent recombination by BRCA1.
Previous studies on the role of BRCA1 in NHR have yielded conflicting observations, ranging from a promotion of NHR to suppressive effects to no effect (40, 55, 64, 74, 75). These results may be at least partly explained by the use of various assay types that may measure different subtypes of NHR. The existence of several NHR subpathways has been postulated, but these remain to be defined in mammalian cells (3, 14, 26). It is well established that DNA-PKcs and XRCC4 promote the random chromosomal integration of substrate DNA (18, 26). Here, we report the novel finding that random integration proficiency is also dependent on Nbs1 (Fig. 4A). In addition, preliminary experiments using siRNA against Mre11 have analogously revealed a sixfold reduction of integration frequencies in HeLa cells (J. Zhang, F. Xia, and S. N. Powell, unpublished observations). These data are consistent with yeast studies that have similarly suggested a role of the MRN homologues in the promotion of homology-independent DNA integration frequencies (2, 51). Thus, the MRN complex and the DNA-PK-dependent repair proteins appear to cooperate in some (such as random DNA integration or intermolecular rejoining [11]) but not all types of NHR (such as VDJ recombination [22, 73]).
Consistent with previous experiments with mouse ES cells (55), we demonstrated that BRCA1 considerably reduces the levels of NHR as measured by random chromosomal integration of substrate DNA. Because the DNA-PK and XRCC4-ligase IV complexes, but not BRCA1, are required for DNA rejoining in PFGE (64), the function of BRCA1 in NHR may be distinct from the DNA-PK-dependent pathway. Instead, BRCA1 may specifically oppose the activity of Nbs1 or the MRN complex (Fig. 4). It can be assumed that defective phenotypes arising in Nbs1-deficient cells are due to changes in the MRN complex, as the Mre11/Rad50 functions and nuclear localization are disrupted in Nijmegen breakage syndrome cells (8, 15, 58). Also, Nbs1 stimulates Mre11/Rad50 nuclease properties in vitro (45). Further support for an opposing regulation of NHR processes by BRCA1 and the MRN complex was obtained by studying Mre11 IRIF formation in single-clone-derived cell populations that stably express wild-type or mutant BRCA1 (Fig. 5B). Of note, previously discrepant results have been reported regarding the effect of BRCA1 on the appearance of Mre11 IRIF (65, 69, 76), possibly caused by the heterogeneity of the pooled cell populations derived for these studies. We believe that the decrease in Mre11 IRIF seen in the BRCA1-expressing cells may be counterbalanced by the increase in Mre11 IRIF in cells not adequately expressing BRCA1, with the net result being no effect of BRCA1 on Mre11 focus formation. This is an observation we made when we also established pooled cell populations after BRCA1 transfection but not when we used single-clone-derived cells. In conclusion, for all the endpoints studied, BRCA1 opposed the effects attributable to the MRN complex.
An important finding in our study was that the serine 988 site of BRCA1 not only dictated the control of HR but also was critical for the inhibition of the MRN complex in vivo. Interestingly, this site is located within the DNA binding domain of BRCA1, which was recently mapped to amino acids 452 through 1079 (44). As result of this DNA binding, BRCA1 inhibited the exonuclease activity of Mre11 in vitro. It is possible that mutation of serine 988 compromises the function of the DNA binding domain; however, we have not yet been able to obtain direct in vivo evidence for such a relationship (Zhang et al., unpublished). The data presented in Fig. 6A support the hypothesis that Chk2-dependent phosphorylation modulates the function of BRCA1 in NHR, but the potential interplay of this phosphorylation with the DNA binding properties of BRCA1 remains to be uncovered. In this regard, it is interesting that Chk2 is activated in an Nbs1-dependent manner in the DNA damage response as reported by Buscemi et al. (7). Consistent with our data, this activation may result in the modification of BRCA1 and a negative feedback loop to the MRN complex.
Dual regulatory role of BRCA1 in Rad51- and MRN-dependent recombination processes.
It is fascinating that BRCA1's role in regulation of both HR and MRN-dependent NHR is dependent on the Chk2 phosphorylation site. As HR and NHR events are likely to be very different processes, it suggests that BRCA1 exerts regulatory control of these processes at an early stage. Several lines of evidence support the notion that the relationship between BRCA1 and NHR is an active effect of BRCA1. First, there is no significant effect on analogously obtained random integration frequencies in BRCA2- deficient cells exhibiting compromised HR (70). Second, we already observed an inhibition of Mre11 focus formation 1 h after cell exposure to irradiation (Fig. 5B). Third, BRCA1 inhibits the activity of Mre11 in vitro (44). Finally, loss of BRCA1 did not compromise the efficiency of DNA rejoining as determined by PFGE, which measures NHR mediated by the DNA-PK-dependent pathway (62, 64). However, presently we cannot rule out the possibility of a passive competition, which would result in DSB repair becoming more dependent on NHR when BRCA1-controlled HR is diminished. Interestingly, BRCA1 is recruited to sites of γ-H2AX phosphorylation before the MRN complex is seen in association with γ-H2AX (46), so the timing of protein associations after DNA damage is a complex sequence of events. The role of the MRN complex may be to facilitate intermolecular joining in both HR and nonhomologous random integration, since both processes require holding two DNA duplexes in a repair intermediate. Therefore, BRCA1 and the MRN complex may collaborate or compete in HR and NHR processes.
Rad51-dependent HR repairs DSBs with high fidelity, whereas NHR processes are inherently error prone. Precancerous cells harboring defective HR pathways may have to rely more on NHR in the S and G2 phases to repair DSB lesions. Such a shift may ensure repair, but at the cost of accumulating mutations and chromosomal aberrations (59). Thus, in its simplest conception, genomic instability provoked by BRCA1 deficiency is the result of the incorrect routing of DSB processing down nonhomologous and error-prone pathways (63). However, recent reports (74, 75) and the data presented here suggest that BRCA1 also plays an active role in the regulation of at least some aspects of error-prone NHR, including those processes that are controlled by the MRN complex. This notion is consistent with the postulated role of the MRN complex in the processing of DSBs and its associated exonuclease activity. In conclusion, we propose a dual regulatory model of BRCA1 (Fig. 7), in which BRCA1, in conjunction with Chk2, promotes error-free HR while at the same time inhibiting putatively error-prone NHR associated with the MRN complex. This picture of BRCA1 action is distinct from a recent proposal, where BRCA1 acts as a repair switch by binding to DNA at the site of the DSB and recruiting either Rad51 or the MRN complex for homologous or nonhomologous break repair (43). While the model depicted in Fig. 7 likely reflects only one aspect of the functions of Chk2-BRCA1 and MRN in recombinational repair, it represents a very testable hypothesis. Since Nbs1 has been implicated in HR in chicken cells (58), it will be important to determine whether the MRN complex is also involved in the control of BRCA1-mediated HR.
FIG. 7.
Dual-role model of BRCA1 function in regulation of recombinational repair.
BRCA1 is found in large protein complexes such as the BASC complex and a chromatin-remodeling complex (6, 65). These entities likely represent dynamic collections of proteins, both in time and in subnuclear location. We speculate that the composition of these complexes can be modulated in DNA damage responses. Therefore, additional modulators of HR and NHR may well be revealed by analyzing the BRCA1 associations in these complexes. The understanding of these properties of BRCA1 has the potential to generate new targets for therapeutic approaches to breast cancers.
Acknowledgments
We to thank David Cortez, Stephen Elledge, Daniel Haber, and Maria Jasin for their generous contribution of materials.
This work was supported in part by a Breast Cancer Research Grant from the Massachusetts Department of Public Health and an Avon Breast Cancer Research Grant to F.X., as well as a Dana-Farber/Partners Cancer Care Grant and the Harvard Breast Cancer SPORE to S.N.P.
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Attikum, H. V., P. Bundock, and P. J. J. Hooykaas. 2001. Non-homologous end-joining proteins are required for Agrobacterium T-DNA integration. EMBO J. 20:6550-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann, P., and S. C. West. 1998. DNA end-joining catalyzed by human cell-free extracts. Proc. Natl. Acad. Sci. USA 95:14066-14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, D. W., J. M. Varley, T. E. Szydlo, D. H. Kang, D. C. R. Wahrer, K. E. Shannon, M. Lubratovich, S. J. Verselis, K. J. Isselbacher, J. F. Fraumeni, J. M. Birch, F. P. Li, J. E. Garber, and D. A. Haber. 1999. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science 286:2528-2531. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharyya, A., U. S. Ear, B. H. Koller, R. R. Weichselbaum, and D. K. Bishop. 2000. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 275:23899-23903. [DOI] [PubMed] [Google Scholar]
- 6.Bochar, D. A., L. Wang, H. Beniya, A. Kinev, Y. Xue, W. S. Lane, W. Wang, F. Kashanchi, and R. Shiekhattar. 2000. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell 102:257-265. [DOI] [PubMed] [Google Scholar]
- 7.Buscemi, G., C. Savio, L. Zannini, F. Micciche, D. Masnada, M. Nakanishi, H. Tauchi, K. Komatsu, S. Mizutani, K. Khanna, P. Chen, P. Concannon, L. Chessa, and D. Delia. 2001. Chk2 activation dependence on Nbs1 after DNA damage. Mol. Cell. Biol. 21:5214-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates, 3rd, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477-486. [DOI] [PubMed] [Google Scholar]
- 9.Caspari, T., J. M. Murray, and A. M. Carr. 2002. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 16:1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J., D. P. Silver, D. Walpita, S. B. Cantor, A. F. Gazdar, G. Tomlinson, F. J. Couch, B. L. Weber, T. Ashley, D. M. Livingston, and R. Scully. 1998. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell 2:317-328. [DOI] [PubMed] [Google Scholar]
- 11.Chen, L., K. Trujillo, W. Ramos, P. Sung, and A. E. Tomkinson. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdfl/Hdf2 complexes. Mol. Cell 8:1105-1115. [DOI] [PubMed] [Google Scholar]
- 12.Cortez, D., Y. Wang, J. Qin, and S. J. Elledge. 1999. Requirement of ATM-dependent phosphorylation of BRCA1 in the DNA damage response to double-strand breaks. Science 286:1162-1166. [DOI] [PubMed] [Google Scholar]
- 13.Critchlow, S. E., and S. P. Jackson. 1998. DNA end-joining: from yeast to man. Trends Biochem. Sci. 23:394-398. [DOI] [PubMed] [Google Scholar]
- 14.Dai, Y., B. Kysela, L. A. Hanakahi, K. Manolis, E. Riballo, M. Stumm, T. O. Harville, S. C. West, M. A. Oettinger, and P. A. Jeggo. 2003. Nonhomologous end joining and V(D)J recombination require an additional factor. Proc. Natl. Acad. Sci. USA 100:2462-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desai-Mehta, A., K. M. Cerosaletti, and P. Concannon. 2001. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol. Cell. Biol. 21:2184-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dronkert, M. L., and R. Kanaar. 2001. Repair of DNA interstrand cross-links. Mutat. Res. 486:217-247. [DOI] [PubMed] [Google Scholar]
- 17.Falck, J., J. H. J. Petrini, B. R. Williams, J. Lukas, and J. Bartek. 2002. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat. Genet. 30:290-294. [DOI] [PubMed] [Google Scholar]
- 18.Finnie, N. J., T. M. Gottlieb, T. Blunt, P. A. Jeggo, and S. P. Jackson. 1995. DNA-dependent protein kinase activity is absent in xrs-6 cells: implications for site-specific recombination and DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 92:320-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gatei, M., B. B. Zhou, K. Hobson, S. Scott, D. Young, and K. K. Khanna. 2001. Ataxia telangiectasia mutated (ATM) kinase and ATM and Rad3 related kinase mediate phosphorylation of Brca1 at distinct and overlapping sites. In vivo assessment using phospho-specific antibodies. J. Biol. Chem. 276:17276-17280. [DOI] [PubMed] [Google Scholar]
- 20.Girard, P. M., N. Foray, M. Stumm, A. Waugh, E. Riballo, R. S. Maser, W. P. Phillips, J. Petrini, C. F. Arlett, and P. A. Jeggo. 2000. Radiosensitivity in Nijmegen Breakage syndrome cells is attributable to a repair defect and not cell cycle checkpoint defects. Cancer Res. 60:4881-4888. [PubMed] [Google Scholar]
- 21.Haber, J. E. 1998. The many interfaces of Mre11. Cell 95:583-586. [DOI] [PubMed] [Google Scholar]
- 22.Harfst, E., S. Cooper, S. Neubauer, L. Distel, and U. Grawunder. 2000. Normal V(D)J recombination in cells from patients with Nijmegen breakage syndrome. Mol. Immunol. 37:915-929. [DOI] [PubMed] [Google Scholar]
- 23.Harrington, J., C. L. Hsieh, J. Gerton, G. Bosma, and M. R. Lieber. 1992. Analysis of the defect in DNA end joining in the murine scid mutation. Mol. Cell. Biol. 12:4758-4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeggo, P. A., and J. Smith-Ravin. 1989. Decreased stable transfection frequencies of six X-ray-sensitive CHO strains, all members of the xrs complementation group. Mutat. Res. 218:75-86. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, A. P., and M. P. Fairman. 1996. The identification and characterization of mammalian proteins involved in the rejoining of DNA double-strand breaks in vitro. Mutat. Res. 364:103-116. [DOI] [PubMed] [Google Scholar]
- 26.Kabotyanski, E. B., L. Gomelsky, J. O. Han, T. D. Stamato, and D. B. Roth. 1998. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 26:5333-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kastan, M. B., and D. S. Lim. 2000. The many substrates and functions of ATM. Nat. Rev. Mol. Cell. Biol. 1:179-186. [DOI] [PubMed] [Google Scholar]
- 28.Kerr P, A. A. 2001. New complexities for BRCA1 and BRCA2. Curr. Biol. 11:R668-R676. [DOI] [PubMed] [Google Scholar]
- 29.Khanna, K. K., and S. P. Jackson. 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27:247-254. [DOI] [PubMed] [Google Scholar]
- 30.Lee, J. S., K. M. Collins, A. L. Brown, C. H. Lee, and J. H. Chung. 2000. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature 404:201-204. [DOI] [PubMed] [Google Scholar]
- 31.Lee, S. B., S. H. Kim, D. W. Bell, D. C. Wahrer, T. A. Schiripo, M. M. Jorczak, D. C. Sgroi, J. E. Garber, F. P. Li, K. E. Nichols, J. M. Varley, A. K. Godwin, K. M. Shannon, E. Harlow, and D. A. Haber. 2001. Destabilization of CHK2 by a missense mutation associated with Li-Fraumeni Syndrome. Cancer Res. 61:8062-8067. [PubMed] [Google Scholar]
- 32.Luo, C. M., W. Tang, K. L. Mekeel, J. S. DeFrank, P. R. Anne, and S. N. Powell. 1996. High frequency and error-prone DNA recombination in ataxia telangiectasia cell lines. J. Biol. Chem. 271:4497-4503. [DOI] [PubMed] [Google Scholar]
- 33.MacQueen, A. J., and A. M. Villeneuve. 2001. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 15:1674-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manolis, K. G., E. R. Nimmo, E. Hartsuiker, A. M. Carr, P. A. Jeggo, and R. C. Allshire. 2001. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 20:210-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maser, R. S., O. K. Mirzoeva, J. Wells, H. Olivares, B. R. Williams, R. A. Zinkel, P. J. Farnham, and J. H. J. Petrini. 2001. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 21:6006-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maser, R. S., K. J. Monsen, B. E. Nelms, and J. H. Petrini. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 17:6087-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meijers-Heijboer, H., A. van den Ouweland, J. Klijn, M. Wasielewski, A. de Snoo, R. Oldenburg, A. Hollestelle, M. Houben, E. Crepin, M. van Veghel-Plandsoen, F. Elstrodt, C. van Duijn, C. Bartels, C. Meijers, M. Schutte, L. McGuffog, D. Thompson, D. Easton, N. Sodha, S. Seal, R. Barfoot, J. Mangion, J. Chang-Claude, D. Eccles, R. Eeles, D. G. Evans, R. Houlston, V. Murday, S. Narod, T. Peretz, J. Peto, C. Phelan, H. X. Zhang, C. Szabo, P. Devilee, D. Goldgar, P. A. Futreal, K. L. Nathanson, B. Weber, N. Rahman, and M. R. Stratton. 2002. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat. Genet. 31:55-59. [DOI] [PubMed] [Google Scholar]
- 38.Mekeel, K. L., W. Tang, L. A. Kachnic, C. M. Luo, J. S. DeFrank, and S. N. Powell. 1997. Inactivation of p53 results in high rates of homologous recombination. Oncogene 14:1847-1857. [DOI] [PubMed] [Google Scholar]
- 39.Mirzoeva, O. K., and J. H. Petrini. 2001. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moynahan, M. E., J. W. Chiu, B. H. Koller, and M. Jasin. 1999. Brca1 controls homology-directed DNA repair. Mol. Cell 4:511-518. [DOI] [PubMed] [Google Scholar]
- 41.Moynahan, M. E., T. Y. Cui, and M. Jasin. 2001. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 61:4842-4850. [PubMed] [Google Scholar]
- 42.Okada, S., and T. Ouchi. 2003. Cell cycle differences in DNA damage-induced BRCA1 phosphorylation affect its subcellular localization. J. Biol. Chem. 278:2015-2020. [DOI] [PubMed] [Google Scholar]
- 43.Parvin, J. D. 2001. BRCA1 at a branch point. Proc. Natl. Acad. Sci. USA 98:5952-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paull, T. T., D. Cortez, B. Bowers, S. J. Elledge, and M. Gellert. 2001. From the cover: direct DNA binding by Brca1. Proc. Natl. Acad. Sci. USA 98:6086-6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paull, T. T., and M. Gellert. 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13:1276-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 47.Pierce, A. J., R. D. Johnson, L. H. Thompson, and M. Jasin. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13:2633-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierce, A. J., J. M. Stark, F. D. Araujo, M. E. Moynahan, M. Berwick, and M. Jasin. 2001. Double-strand breaks and tumorigenesis. Trends Cell Biol. 11:S52-S59. [DOI] [PubMed] [Google Scholar]
- 49.Prudden, J., J. S. Evans, S. P. Hussey, B. Deans, P. O'Neill, J. Thacker, and T. Humphrey. 2003. Pathway utilization in response to a site-specific DNA double-strand break in fission yeast. EMBO J. 22:1419-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rahman, N., and M. R. Stratton. 1998. The genetics of breast cancer susceptibility. Annu. Rev. Genet. 32:95-121. [DOI] [PubMed] [Google Scholar]
- 51.Schiestl, R. H., J. Zhu, and T. D. Petes. 1994. Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:4493-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scully, R., J. Chen, A. Plug, Y. Xiao, D. Weaver, J. Feunteun, T. Ashley, and D. M. Livingston. 1997. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88:265-275. [DOI] [PubMed] [Google Scholar]
- 53.Scully, R., S. Ganesan, K. Vlasakova, J. Chen, M. Socolovsky, and D. M. Livingston. 1999. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol. Cell 4:1093-1099. [DOI] [PubMed] [Google Scholar]
- 54.Scully, R., and D. M. Livingston. 2000. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408:429-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snouwaert, J. N., L. C. Gowen, A. M. Latour, A. R. Mohn, A. Xiao, L. DiBiase, and B. H. Koller. 1999. BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a Brca1 transgene. Oncogene 18:7900-7907. [DOI] [PubMed] [Google Scholar]
- 56.Stewart, G. S., R. S. Maser, T. Stankovic, D. A. Bressan, M. I. Kaplan, N. G. J. Jaspers, A. Raams, P. J. Byrd, J. H. J. Petrini, and A. M. R. Taylor. 1999. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99:577-587. [DOI] [PubMed] [Google Scholar]
- 57.Taniguchi, T., I. Garcia-Higuera, B. Xu, P. R. Andreassen, R. C. Gregory, S. T. Kim, W. S. Lane, M. B. Kastan, and A. D. D'Andrea. 2002. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109:459-472. [DOI] [PubMed] [Google Scholar]
- 58.Tauchi, H., J. Kobayashi, K. Morishima, D. C. van Gent, T. Shiraishi, N. S. Verkaik, D. vanHeems, E. Ito, A. Nakamura, E. Sonoda, M. Takata, S. Takeda, S. Matsuura, and K. Komatsu. 2002. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature 420:93-98. [DOI] [PubMed] [Google Scholar]
- 59.Thompson, L. H., and D. Schild. 2001. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. 477:131-153. [DOI] [PubMed] [Google Scholar]
- 60.Tibbetts, R. S., D. Cortez, K. M. Brumbaugh, R. Scully, D. Livingston, S. J. Elledge, and R. T. Abraham. 2000. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. 14:2989-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tomlinson, G. E., T. T. Chen, V. A. Stastny, A. K. Virmani, M. A. Spillman, V. Tonk, J. L. Blum, N. R. Schneider, Wistuba II, J. W. Shay, J. D. Minna, and A. F. Gazdar. 1998. Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 58:3237-3242. [PubMed] [Google Scholar]
- 62.van Gent, D. C., J. H. Hoeijmakers, and R. Kanaar. 2001. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2:196-206. [DOI] [PubMed] [Google Scholar]
- 63.Venkitaraman, A. R. 2002. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108:171-182. [DOI] [PubMed] [Google Scholar]
- 64.Wang, H., Z. C. Zeng, T. A. Bui, S. J. DiBiase, W. Qin, F. Xia, S. N. Powell, and G. Iliakis. 2001. Nonhomologous end-joining of ionizing radiation-induced DNA double-stranded breaks in human tumor cells deficient in BRCA1 or BRCA2. Cancer Res. 61:270-277. [PubMed] [Google Scholar]
- 65.Wang, Y., D. Cortez, P. Yazdi, N. Neff, S. J. Elledge, and J. Qin. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14:927-939. [PMC free article] [PubMed] [Google Scholar]
- 66.Willers, H., E. E. McCarthy, P. Hubbe, J. Dahm-Daphi, and S. N. Powell. 2001. Homologous recombination in extrachromosomal plasmid substrates is not suppressed by p53. Carcinogenesis 22:1757-1763. [DOI] [PubMed] [Google Scholar]
- 67.Willers, H., E. E. McCarthy, B. Wu, H. Wunsch, W. Tang, D. G. Taghian, F. Xia, and S. N. Powell. 2000. Dissociation of p53-mediated suppression of homologous recombination from G1/S cell cycle checkpoint control. Oncogene 19:632-639. [DOI] [PubMed] [Google Scholar]
- 68.Wu, X., V. Ranganathan, D. S. Weisman, W. F. Heine, D. N. Ciccone, T. B. O'Neill, K. E. Crick, K. A. Pierce, W. S. Lane, G. Rathbum, D. M. Livingston, and D. T. Weaver. 2000. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405:477-482. [DOI] [PubMed] [Google Scholar]
- 69.Wu, X. H., J. H. Petrini, W. F. Heine, D. T. Weaver, D. M. Livingston, and J. J. Chen. 2000. Independence of R/M/N focus formation and the presence of intact BRCA1. Science 289:11a. [DOI] [PubMed] [Google Scholar]
- 70.Xia, F., D. G. Taghian, J. S. DeFrank, Z. C. Zeng, H. Willers, G. Iliakis, and S. N. Powell. 2001. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc. Natl. Acad. Sci. USA 98:8644-8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu, B., S. Kim, and M. B. Kastan. 2001. Involvement of Brca1 in S-phase and G(2)-phase checkpoints after ionizing irradiation. Mol. Cell. Biol. 21:3445-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu, B., S. T. Kim, D. S. Lim, and M. B. Kastan. 2002. Two molecularly distinct G(2)/M checkpoints are induced by ionizing irradiation. Mol. Cell. Biol. 22:1049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yeo, T. C., D. Xia, S. Hassouneh, X. O. Yang, D. E. Sabath, K. Sperling, R. A. Gatti, P. Concannon, and D. M. Willerford. 2000. V(D)J rearrangement in Nijmegen breakage syndrome. Mol. Immunol. 37:1131-1139. [DOI] [PubMed] [Google Scholar]
- 74.Zhong, Q., T. G. Boyer, P. L. Chen, and W. H. Lee. 2002. Deficient nonhomologous end-joining activity in cell-free extracts from Brca1-null fibroblasts. Cancer Res. 62:3966-3970. [PubMed] [Google Scholar]
- 75.Zhong, Q., C. F. Chen, P. L. Chen, and W. H. Lee. 2002. BRCA1 facilitates microhomology-mediated end joining of DNA double strand breaks. J. Biol. Chem. 277:28641-28647. [DOI] [PubMed] [Google Scholar]
- 76.Zhong, Q., C. F. Chen, S. Li, Y. Chen, C. C. Wang, J. Xiao, P. L. Chen, Z. D. Sharp, and W. H. Lee. 1999. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science 285:747-750. [DOI] [PubMed] [Google Scholar]