Abstract
The Wilms' tumor suppressor protein WT1 is a transcriptional regulator that plays a key role in the development of the kidneys. The transcriptional activation domain of WT1 is subject to regulation by a suppression region within the N terminus of WT1. Using a functional assay, we provide direct evidence that this requires a transcriptional cosuppressor, which we identify as brain acid soluble protein 1 (BASP1). WT1 and BASP1 associate within the nuclei of cells that naturally express both proteins. BASP1 can confer WT1 cosuppressor activity in transfection assays, and elimination of endogenous BASP1 expression augments transcriptional activation by WT1. BASP1 is present in the developing nephron structures of the embryonic kidney and, coincident with that of WT1, its expression is restricted to the highly specialized podocyte cells of the adult kidney. Taken together, our results show that BASP1 is a WT1-associated factor that can regulate WT1 transcriptional activity.
Wilms' tumor, a pediatric malignancy of the kidneys, is the most common solid childhood tumor (reviewed in references 4, 7, 20, 30, and 33). The isolation of genes associated with Wilms' tumor led to the identification of a zinc finger protein, WT1. Subsequently, WT1 was shown to be a transcriptional regulator with putative target genes including those for growth factors and regulators of cell division (5, 6, 18, 21). Approximately 15% of sporadic Wilms' tumors have been found to contain mutations in WT1, while others show aberrant WT1 expression (33).
WT1 knockout mice (homozygous null) do not survive gestation, displaying absence or incorrect development of the kidney, gonads, spleen, heart, diaphragm, and retinal ganglia (12, 14, 35). These findings confirm a major role for WT1 in the formation of the genitourinary system and also a wider role in the development of other tissues.
Alternative splicing, RNA editing, and an alternative translation start codon combine to produce a plethora of WT1 isoforms (reviewed in reference 33). One alternative splice inserts three amino acids (KTS) between zinc fingers three and four, resulting in a form of WT1 that associates with RNA processing factors and localizes to regions of RNA processing in the nucleus (17). Thus, the +KTS and −KTS isoforms of WT1 have been proposed to function in RNA processing and transcription, respectively. These isoforms have both overlapping and distinct roles during development (9, 10). Interestingly, the +KTS isoform of WT1 plays the dominant role in the development of the gonad, while the −KTS isoform has a more extensive function in kidney formation.
The other alternative splice inserts 17 amino acids N terminal to the WT1 zinc fingers and has been shown to have effects on both cell division and cell survival (15, 31, 32). Specific elimination of this isoform of WT1 in mice does not result in any obvious defects in genitourinary development, suggesting that it may be required specifically for a tumor suppressor role or that it performs a subtle function (28).
Several studies have shown that WT1 contains a transcriptional activation domain that is suppressed by an N-terminal region of the protein (33). Suppression of WT1 transcriptional activation also occurs in the context of a GAL4 fusion protein, and the WT1 suppression domain can inhibit the function of other transcriptional activators (19, 22, 24, 36). A 30-amino-acid region of WT1 (residues 71 to 101) is sufficient to confer inhibition of the WT1 transcriptional activation domain. Moreover, this suppression domain is able to inhibit the transcriptional function of the activation domain of SP1 when fused in cis (24).
In this study, we use an in vitro transcription assay to provide direct evidence that the WT1 suppression domain interacts with a transcriptional cosuppressor. We identify brain acid soluble protein 1 (BASP1) as a component of the WT1 cosuppressor. BASP1 associates with WT1 in vivo, and its expression in the developing kidney is coincident with that of WT1. Our data suggest that BASP1 associates with WT1 to regulate its transcription function during development.
MATERIALS AND METHODS
Plasmids and DNA analysis.
The G5E4T, G5E4CAT, G5tkCAT, and W5E4CAT transcription reporter templates have been described previously (24, 32). The Escherichia coli expression vector containing GAL4 amino acids 1 to 93 [GAL4(1-93)] linked in frame to the WT1 suppression domain (amino acids 71 to 101) and the SP1 activation domain has been described before (24). Deletion mutagenesis of the WT1 suppression domain was performed by PCR amplification of the required WT1 suppression domain-encoding DNA and cloning in frame to GAL4(1-93) and the SP1 activation domain. GAL4(1-93)-WT1 (residues 71 to 250 and 99 to 250) and GAL4(1-93)-EVE under the control of a cytomegalovirus (CMV) promoter have been described before (24). Expressed sequence tags (ESTs) containing BASP1 sequence were obtained from the American Type Culture Collection and sequenced. One of the ESTs (accession number 167644) contained the entire coding sequence of BASP1. Full-length BASP1 cDNA was cloned in frame into pGEX2T and used to produce recombinant glutathione S-transferase (GST)-BASP1. BASP1 cDNA was cloned in frame to GAL4(1-93) under the control of a CMV promoter and also in frame to a hemagglutinin (HA) tag under the control of a CMV promoter. A double-stranded oligonucleotide (GATCCCCGGAGGAAGGGGAACCCAAATTCAAGAGATTTGGGTTCCCCTTCCTCCTTTTTGGAAA) was cloned in to the vector pSUPER (3) so as to produce an RNA interference (RNAi) directed against BASP1 (pSUPER-BASP1). The amphiregulin-luciferase reporters were kind gifts of Daniel Haber (18).
Peptides and antibodies.
Peptide synthesis was performed by Graham Bloomberg (University of Bristol, Bristol, England). The peptide was used to raise antibodies (SAPU, Edinburgh, Scotland), which were purified by peptide affinity chromatography. Antisera were raised against GST-BASP1 by Abcam (Cambridge, England). Antibodies were first purified by absorption to GST-linked beads to remove antibodies that bind GST. Anti-BASP1 antibodies were then purified by affinity chromatography with a column containing GST-BASP1 covalently linked to Sepharose. Anti-WT1 antibodies were the F6 monoclonal or N-180 polyclonal antisera from Santa Cruz. Antitubulin antibodies were from Sigma and anti-TFIIB antibodies have been described before (11).
Cell culture, transfection, and in vitro transcription assays.
Human embryonic kidney (HEK) 293, M15, G-401, HeLa, and Cos-1 cells were cultured as monolayers in Dulbecco's modified Eagle medium containing 10% fetal calf serum, 5 mM l-glutamine, 100 mg of streptomycin per ml, and 100 U of penicillin per ml. Human erythroleukemia cell line K562 was cultured in RPMI 1640 medium with l-glutamine, 10% fetal calf serum, 100 mg of streptomycin per ml, and 100 U of penicillin per ml. 293 cells, M15 cells, and Cos-1 cells were transfected as described previously (11). HeLa cells were transfected by using Lipofectamine 2000 according to the instructions of the manufacturer (Invitrogen). K562 cells were transfected with Lipofectamine 2000 by using a modified protocol for nonadherent cells (2). Chloramphenicol acetyltransferase (CAT) assays were performed and quantified by phosphorimager analysis. Luciferase activity was measured with a Promega kit as described in the manufacturer's instructions. Transfection of the RNAi vectors into HeLa cells was carried out with Lipofectamine 2000 reagent. Cells were plated out into six-well plates on the day prior to the transfection at a density such that they would reach 90 to 95% confluence on the following day. Two micrograms of either pSUPER (3) or pSUPER-BASP1 and 2 μg of CMV-WT1−/−, where appropriate, were transfected into the cells. After 24 h, the cells were split 1:2 and replated. After a further 24 h, 2 μg of either pSUPER or pSUPER-BASP1, 1 μg of pGL2-AR-A (AR) or pGL2-AR-C (ARΔWRE), and 1 μg of CMV-WT1−/−, where appropriate, were transfected. Cells were harvested 48 h later.
HeLa nuclear extracts were purchased from 4C (Mons, Belgium). In vitro transcription assays were performed as described previously (24). Where indicated, antibodies or peptides were added to nuclear extract on ice and left for 30 min prior to the transcription reaction.
Immunofluorescence.
Cells were grown to approximately 50% confluency in eight-well slides, fixed with 1% formaldehyde in phosphate-buffered saline (PBS), and then rinsed in PBS. Cells were incubated with primary antibodies (anti-WT1 F6 and anti-BASP1, both diluted 1:50 in a 500-μg/ml digitonin solution) for 2 h and then washed with PBS containing 1% Tween 20. The cells were then incubated for 30 min with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse (1:100; Dako) or Cy3-congugated goat anti-rabbit (1:500; Jackson Immunological Research) antibodies in PBS and washed with PBS containing 1% Tween 20. Nuclei were counterstained with Hoechst stain (1 μg/ml) in PBS. Samples were mounted with Vectashield (Vector Laboratories, Inc).
Immunohistochemistry.
Mouse embryos of mixed (C57BL/6:CBA) genetic background were recovered 15 days after detection of a copulation plug. Slides from samples fixed in 4% paraformaldehyde were dewaxed and rehydrated, and then antigen retrieval was carried out by boiling in 10 mM sodium citrate buffer (pH 6.0) three times for 5 min each. The slides were washed for 10 min in PBS plus 0.1% Tween 20, and then endogenous peroxidase activity was quenched by incubation in 3% H2O2 in tap water for 5 min, followed by 0.5% H2O2 in methanol for 30 min. Slides were washed three times for 10 min each in PBS plus 0.1% Tween 20 and then blocked and incubated with primary and secondary antibodies. The primary antibody (anti-BASP1 or anti-WT1 [Santa Cruz]) was used at a concentration of 1:500. After the secondary antibody (biotin-conjugated anti-rabbit; Sigma), streptavidin-horseradish peroxidase complex (ABC kit; Vector) and diaminobenzidine (DAB kit; Vector) were used for detection. The slides were then counterstained with Mayer's hematoxylin (Sigma), dehydrated, and mounted.
Paraffin-embedded sections from mouse or human (Novagen) adult kidney were placed in xylene overnight and then washed with ethanol. Endogenous peroxidases were inactivated by treating the samples with 0.3% H2O2 in methanol, and the samples were rehydrated. Target retrieval was achieved by boiling the samples for 10 min in 10 mM Tris-400 μM EGTA (pH 9). Cooled sections were placed in 50 mM NH4Cl in PBS for 30 min and then blocked with 1% bovine serum albumin-0.2% gelatin-0.05% saponin solution in PBS. Sections were incubated with antibodies as described above.
Affinity chromatography and binding assays.
HeLa nuclear extract was dialyzed into buffer D (20 mM HEPES [pH 8], 20% [vol/vol] glycerol, 100 mM KCl, 0.2 mM EDTA, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride). Ten milligrams of HeLa nuclear extract was precleared over a glutathione agarose column and then fractionated over columns containing 1 mg of glutathione-linked GST fusion protein. The columns were washed with 10 volumes of buffer D, and the bound fraction was eluted with buffer D (with 1 M KCl). GST pull-down assays were performed as described before (24). Immunoprecipitation assays were performed with nuclear extracts prepared from M15 cells in buffer D containing 100 mM KCl. Protein A-Sepharose was used to collect the immune complexes, and washes were performed with the same buffer.
Two-dimensional electrophoresis and protein identification.
For protein precipitation, 0.25 volume of trichloroacetic acid solution (100% [wt/vol] trichloroacetic acid, 0.4% [wt/vol] sodium deoxycholate) was added and the proteins were collected by centrifugation for 30 min at 13,000 × g followed by washing with acetone. Two-dimensional electrophoresis was performed with the Pharmacia system, and the second dimension was an 8 to 18% acrylamide gradient gel.
Proteins were identified by mass spectrometry as previously described (34). The unseparated trypsin peptide mixture was analyzed by matrix-assisted laser desorption ionization, using a Bruker Reflex III matrix-assisted laser desorption ionization-time-of-flight mass spectrometer (Bruker Daltronics). Samples for tandem mass spectrometry analysis were prepared essentially as described previously. After in-gel digestion, the supernatant was loaded onto a Poros R2 microcartridge (39) and eluted into nanoelectrospray needles (MDS Proteomics, Odense, Denmark). Nanoelectrospray tandem mass spectrometry analysis was performed on a QSTAR quadrupole time-of-flight mass spectrometer (Applied Biosystems), and fragmentation spectra were obtained for as many peptides as possible. PepSea software (MDS Proteomics) was used to search publicly available sequence databases maintained by the National Center for Biotechnology Information with a list of peptide masses or with peptide sequence tags from fragmentation spectra.
RESULTS
Functional analysis of the WT1 suppression domain.
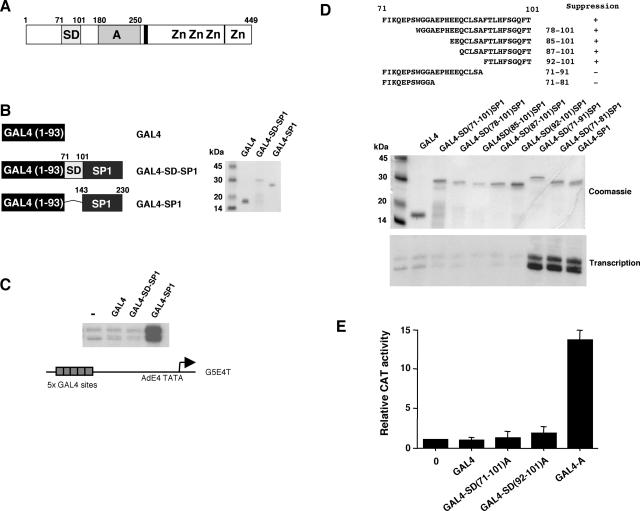
The N terminus of WT1 harbors a suppression domain that can inhibit the function of the transcriptional activation domain (Fig. 1A). We have previously shown that the suppression domain of WT1 (amino acids 71 to 101) can inhibit the stimulation of transcription by the SP1 activation domain when fused in cis in the context of a GAL4 fusion protein (24). We prepared GAL4-SD-SP1, which contains GAL4 (amino acids 1 to 93) linked to the WT1 suppression domain (WT1 amino acids 71 to 101) and the SP1 transcriptional activation domain (residues 143 to 230) (Fig. 1B). In addition, GAL4 and GAL4 linked to the SP1 activation domain (GAL4-SP1) were prepared, and all of the proteins were assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining (Fig. 1B). The GAL4 fusion proteins were then tested in an in vitro transcription assay with HeLa cell nuclear extract and a reporter containing the AdE4 core promoter linked to five GAL4 DNA-binding sites (G5E4T) (Fig. 1C). As we have shown before, GAL4-SP1 activates transcription in a HeLa nuclear extract but GAL4-SD-SP1 does not (24).
FIG. 1.
Dissection of the WT1 transcriptional suppression domain. (A) Schematic of WT1, indicating the transcriptional suppression domain (SD) (amino acids 71 to 101) and the transcriptional activation domain (A). The vertical lines indicate the regions of alternative splicing, with the 17-amino-acid insertion located centrally and the KTS insertion located between the third and fourth zinc fingers (Zn). (B) The indicated GAL4 fusion proteins were prepared, and 1 μg was analyzed by SDS-PAGE and Coomassie blue staining. Molecular mass markers are shown at the left. (C) The GAL4 fusion proteins (100 ng) were tested in an in vitro transcription assay with a HeLa cell nuclear extract and the reporter G5E4T (shown at the bottom). Transcripts were detected by primer extension. (D) The WT1 suppression domain was subjected to deletion and fused in frame to GAL4 and the SP1 activation domain. The amino acid sequences of the intact WT1 suppression domain and derivatives are shown. GAL4-SD-SP1 derivatives were expressed in E. coli, purified, and examined by SDS-PAGE and Coomassie blue staining (1 μg of each protein). Transcription assays were as described for panel C. (E) Expression constructs (1 μg) that produce GAL4(1-93), GAL4-SD (residues 71 to 101)-A (WT1 activation domain residues 180 to 250), GAL4-SD (residues 92 to 101)-A, and GAL4-A were transfected into 293 cells. The reporter is the same as that used in panel C but linked to the CAT gene. The results are presented as mean relative CAT activity in three experiments with standard deviation.
We next tested N- and C-terminal truncations of the suppression domain linked in frame to GAL4 and the SP1 transcriptional activation domain (Fig. 1D). Our analysis showed that just 10 amino acids of WT1 (residues 92 to 101) were sufficient to confer inhibition of the SP1 activation domain. The same 10 amino acids of the WT1 suppression domain were also able to inhibit the WT1 transcriptional activation domain in transient transfection analysis (Fig. 1E; compare GAL4-A with the GAL4-SD-A derivatives).
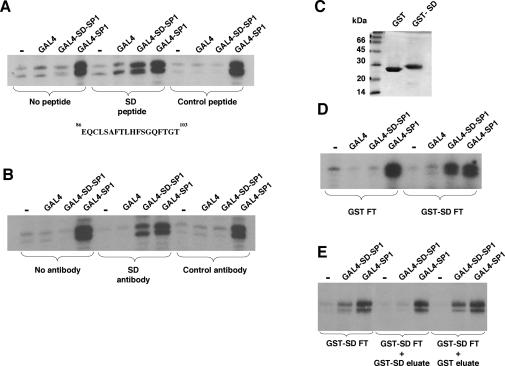
Previous studies have shown that overexpression of an N-terminal fragment of WT1 (containing the suppression domain) causes a normally inactive intact WT1 protein to activate transcription of a promoter containing WT1 DNA-binding sites (37). It was proposed that this manifestation of activator function resulted from titration of a WT1 cosuppressor by the free suppression domain. We sought to determine if this finding could be recapitulated in an in vitro transcription assay. HeLa cell nuclear extract was incubated with a synthetic 18-amino-acid peptide containing the minimal suppression domain (as identified above) or a control peptide sequence derived from the RNA polymerase III-specific transcription factor hRRN3 (25). These peptide-treated extracts were then used in an in vitro transcription assay as in Fig. 1 (Fig. 2A). When we tested the HeLa nuclear extract that had been treated with the WT1 suppression domain peptide, GAL4-SD-SP1 was now able to activate transcription. This was not observed with a nuclear extract treated with the control peptide. Thus, consistent with previous observations, the isolated WT1 suppression domain is able to titrate a cosuppressor, thereby rendering the DNA-bound suppression domain inactive.
FIG. 2.
Evidence for a WT1 transcriptional cosuppressor. (A) One microgram of a synthetic peptide (SD, EQCLSAFTLHFSGQFTGT; residues 86 to 103 of WT1) or a control peptide (MAAPLLHTRLPGDAC; derived from hRRN3 [25]) was incubated for 30 min on ice with 250 μg of HeLa nuclear extract. Transcription assays were as described for Fig. 1. (B) Antibodies raised against the WT1 suppression domain peptide or a control antibody were incubated with the recombinant GAL4 fusion proteins (100 ng) for 30 min on ice prior to assembly of the transcription assay. (C) GST or a GST fusion of the WT1 suppression domain (residues 71 to 101; GST-SD) were purified from E. coli and examined by SDS-PAGE (5 μg of each protein). Molecular mass markers are shown at the left. (D) HeLa nuclear extract (10 mg) was fractionated over a column containing 5 mg of either GST or GST-SD linked to 1 ml of glutathione agarose. The flowthrough (FT) was used in transcription assays with the GAL4 fusion proteins as described for panel A. (E) HeLa nuclear extract that had been fractionated over a GST-SD column was used alone or supplemented with dialyzed eluates from the GST-SD column or a GST column. The extracts were then tested in an in vitro transcription assay as described for panel D.
We raised antibodies to the minimal WT1 suppression domain peptide used for Fig. 2A and purified them by using a column containing the same peptide. The anti-suppression domain antibody or a control antibody was then incubated with each GAL4 fusion protein before the remainder of the components of the transcription assay were added. The suppression domain antibody, but not the control antibody, was able to block the function of the WT1 suppression domain, thus allowing GAL4-SD-SP1 to activate transcription (Fig. 2B). These results are consistent with the requirement for a factor that interacts with the WT1 suppression domain to inhibit transcriptional activation.
The WT1 suppression domain acts by recruiting a transcriptional cosuppressor.
We next sought direct evidence that the WT1 suppression domain interacts with a cosuppressor. We purified either GST or a GST fusion of the WT1 suppression domain (GST-SD) linked to glutathione agarose and checked their purity by SDS-PAGE (Fig. 2C). These beads were then assembled into columns, over which HeLa cell nuclear extract was fractionated. The flowthrough fractions from the columns were then tested in transcription assays with GAL4, GAL4-SD-SP1, or GAL4-SP1 (Fig. 2D). The flowthrough from the GST column showed the same transcription effects with the GAL4 fusion proteins that we observed with the untreated HeLa cell nuclear extract. When we examined the flowthrough from the GST-SD column, the activities of GAL4 and GAL4-SP1 were the same as that observed with the flowthrough from the GST column. In contrast, however, GAL4-SD-SP1 was able to activate transcription in the flowthrough from the GST-SD column, indicating that the GST-SD column had depleted a cosuppressor from the HeLa nuclear extract.
After extensive washing, we eluted the proteins that were bound to the GST and GST-SD columns with buffer containing 1 M salt and dialyzed the eluates into buffer containing 100 mM salt. The eluates from the GST and GST-SD columns were then added to a HeLa cell nuclear extract that had been fractionated over a GST-SD column (Fig. 2E). Addition of the 1 M eluate from the GST column had no effect on the activities of the GAL4 fusion proteins in the GST-SD flowthrough. However, addition of the 1 M eluate from the GST-SD column rendered GAL4-SD-SP1 inactive but did not reduce the level of transcriptional activation by GAL4-SP1. Thus, cosuppressor activity is present in the 1 M salt eluate from the GST-SD column but not in the GST column eluate.
Identification of BASP1 as a component of the WT1 transcriptional cosuppressor.
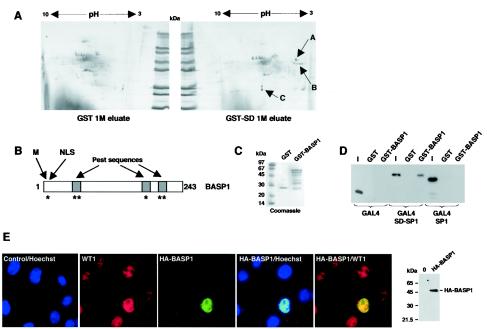
The 1 M eluates from the GST and GST-SD column were analyzed by two-dimensional gel electrophoresis followed by silver staining (Fig. 3A). Analysis of the profiles revealed three spots that were present in the GST-SD eluate but not the GST eluate. Spot A was positively identified by nanoelectrospray tandem mass spectrometry as containing only BASP1. The group of spots designated B could not be identified, but it is possible that these are related to spot A (see below). Spot C contained GST, and it is therefore likely that this is GST-SD that has shed from the column.
FIG. 3.
BASP1 associates with the WT1 transcriptional suppression domain and localizes with WT1 in the nuclei of M15 cells. (A) Proteins in the 1 M eluates from the GST and GST-SD columns were resolved by two-dimensional electrophoresis and examined by silver staining. Spots (A to C) that were unique to the GST-SD eluate are indicated. (B) Schematic of BASP1. M, myristoylation signal sequence; NLS, nuclear localization sequence. Regions of PEST sequences are shown by shading. Potential protein kinase C and casein kinase II phosphorylation sites are indicated by asterisks. (C) GST and GST-BASP1 were purified from E. coli and assessed by SDS-PAGE and Coomassie blue staining. Molecular mass markers are shown at left. (D) GST pull-down assay showing that GAL4-SD-SP1 interacts with GST-BASP1 but not with GST. GAL4-SP1 does not interact with GST-BASP1, suggesting that the interaction with GAL4-SD-SP1 is mediated via the suppression domain. Lanes I, 10% of the input into each assay. Detection of the GAL4 derivatives was by immunoblotting with anti-GAL4 antibodies. (E) A plasmid encoding HA-tagged BASP1 under the control of a CMV promoter was transfected into mouse M15 cells. Cells were fixed and probed with anti-WT1 antibody (and then secondary Cy3 antibody [red]) and anti-HA tag antibody (and then secondary FITC antibody [green]). The nuclei were counterstained with Hoechst stain (blue).
BASP1 was previously purified as an N-terminally myristoylated protein from the cytoplasm of brain cells (27). It belongs to a group of factors that includes neuronal tissue-enriched acidic protein (NAP22) (29) and neuronal growth-associated protein (GAP-43) (38). It was reported, however, that outside the brain, BASP1 is present in the kidney, spleen, and testis, which are also major sites of WT1 function. A schematic of BASP1 is shown in Fig. 3B. It contains an N-terminal myristoylation signal and also several potential protein kinase C and casein kinase II recognition sites. The phosphorylation sites are nested within PEST sequences, which are characteristic of high-turnover proteins (27). We also note that BASP1 contains a nuclear localization sequence close to the N terminus. The functional significance of any of these motifs in BASP1 is not known.
We obtained an EST that contained the entire BASP1-coding sequence. A GST-BASP1 fusion protein was produced and analyzed by SDS-PAGE and Coomassie blue staining (Fig. 3C). GST and GST-BASP1 linked to glutathione agarose beads were then used in binding assays with the recombinant GAL4 fusion proteins that were used in our in vitro transcription assays (Fig. 3D). Neither GAL4 or GAL4-SP1 interacted with GST-BASP1. However, we observed a robust interaction of GAL4-SD-SP1 with GST-BASP1, but not with GST, confirming that the WT1 suppression domain is indeed able to interact directly with BASP1.
We next transfected a plasmid containing HA-tagged BASP1-coding sequence under the control of a CMV promoter into mouse kidney M15 cells (Fig. 3E). M15 cells are derived from the mouse mesonephros, a progenitor of the kidney, and naturally express WT1 (17). Immunofluorescence was performed on the transfected cells to detect both the endogenous WT1 and HA-tagged BASP1. The cells were counterstained with Hoechst stain to show the nucleus. WT1 was present within the nuclei of all of the cells. In addition, HA-BASP1 was present entirely in the nuclei of all of the transfected cells (Fig. 3E and data not shown).
BASP1 expression and association with WT1.
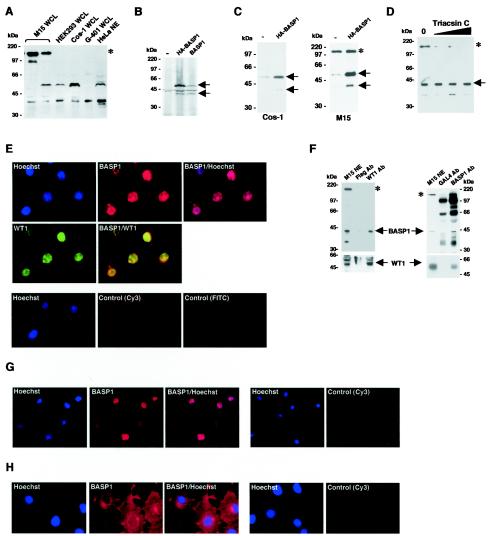
We next raised polyclonal antisera that recognize BASP1 and purified them by affinity chromatography with a column containing immobilized BASP1. The purified anti-BASP1 antibodies were then used to immunoblot a panel of cells lines (Fig. 4A). HeLa cell nuclear extract shows two major forms of BASP1, migrating at 52 and 40 kDa. Whole-cell extracts derived from HEK 293 cells and monkey kidney Cos-1 cells also contain the 52- and 40-kDa forms of BASP1. G-401 cells, derived from a human pediatric kidney tumor, contain predominantly the 40-kDa form of BASP1. Two different M15 whole-cell extracts are shown to represent a heterogeneity that we observed with extracts from these cells. M15 cells can contain both the 52- and 40-kDa forms, although in variable ratios. In addition, M15 cells contain a much more slowly migrating immunoreactive protein. The 52- and 40-kDa BASP1 products were produced by in vitro translation of either HA-BASP1 or BASP1 in a rabbit reticulocyte lysate, suggesting that they arise from natural processing of BASP1 (Fig. 4B). We also transfected the plasmid driving expression of HA-BASP1 into either Cos-1 cells or M15 cells and immunoblotted with the anti-BASP1 antibody (Fig. 4C). In both cell lines, transfection of the HA-BASP1 construct caused a significant increase in both the 52- and 40-kDa immunoreactive bands.
FIG. 4.
Characterization of BASP1. (A) A panel of cell lines was analyzed by Western blotting with anti-BASP1 antibodies. Molecular mass markers are shown at the left. WCL, whole cell lysate; NE, nuclear extract. Immunoreactive bands common among the cell lines are indicated by arrows (52 and 40 kDa). The BASP1 form present only in M15 cells is indicated by an asterisk. (B) pcDNA3 (−), pcDNA3-HA-BASP1, or pcDNA3-BASP1 was added to rabbit reticulocyte lysate and [35S]methionine. The translated products were resolved by SDS-PAGE and visualized by autoradiography. The two products corresponding to BASP1 are indicated by arrows. (C) Cos-1 or M15 cells were transfected with either empty CMV expression vector (−) or CMV expression vector driving the production of HA-tagged BASP1 (3 μg each). Cells were harvested, and extracts were immunoblotted with affinity-purified anti-BASP1 antibodies. The two major forms of BASP1 (52 and 40 kDa) are indicated by arrows, and the slower-migrating form in M15 cells is indicated by an asterisk. (D) M15 cells were treated with Triacsin C (1.5, 3, and 6 μM) for 16 h, and whole-cell lysates immunoblotted with anti-BASP1 antibodies. 0, control cells. BASP1 is indicated by an arrow, and the slower-migrating form is indicated by an asterisk. (E) M15 cells were subjected to immunofluorescence with anti-WT1 and anti-BASP1 antibodies. WT1 is indicated by green, and BASP1 is indicated by red. Nuclei were counterstained with Hoechst stain. Control immunofluorescence (Cy3 and FITC) omitted the primary antibody. (F) Left panel, M15 nuclear extracts were subjected to immunoprecipitation with either anti-WT1 monoclonal antibodies or control anti-Flag monoclonal antibodies linked to agarose beads. The precipitates were resolved by SDS-PAGE and immunoblotted withpolyclonal anti-BASP1 antibodies (top panel) or anti-WT1 polyclonal antibodies (bottom panel). BASP1 products are indicated by an asterisk and arrow. WT1 forms are indicated by an arrow. M15, 4% of the input into the immunoprecipitation assay. Right panel, M15 nuclear extracts were subjected to immunoprecipitation with either purified anti-BASP1 polyclonal antibodies or a control anti-GAL4 purified polyclonal antibody linked to protein A-agarose beads. The precipitates were resolved by SDS-PAGE and immunoblotted with anti-BASP1 polyclonal antibodies (top panel) or anti-WT1 monoclonal antibodies (bottom panel). Specific BASP1 products are indicated by an asterisk and arrow. The nonspecific bands present in both the anti-BASP1 immunoprecipitation and the control (GAL4 immunoprecipitation) arise from the use of rabbit antibodies for both the immunoprecipitation and immunoblot. WT1 forms are indicated by an arrow. (G) G-401 cells derived from a human kidney tumor were subjected to immunofluorescence with anti-BASP1 antibodies (staining in red). The nuclei were counterstained with Hoechst stain. (H) Same as panel G, except that Cos-1 cells were used.
BASP1 has previously been shown to be N-terminally myristoylated. The N-terminal tag in HA-BASP1 would prevent this, suggesting that the 52- and 40-kDa BASP1 forms do not specifically arise from myristoylation. We considered the possibility, however, that the large immunoreactive product observed in M15 cells may result from myristoylation of BASP1. We therefore treated M15 cells with Triacsin C, an inhibitor of protein myristoylation (26). Figure 4D shows that after treatment of M15 cells with Triacsin C, there is a selective reduction of the slow-migrating immunoreactive product, suggesting that this form likely arises from myristoylation of BASP1.
We next performed immunofluorescence to localize the endogenous BASP1 in M15 cells (Fig. 4E). As we had observed with HA-BASP1, endogenous BASP1 in M15 cells is located largely within the nucleus along with WT1. Nuclear extracts were prepared from M15 cells and subjected to immunoprecipitation with either anti-WT1 antibodies (Fig. 4F, left panel) or anti-BASP1 antibodies (Fig. 4F, right panel). Precipitates were resolved by SDS-PAGE and immunoblotted to detect both WT1 and BASP1. The results show that an anti-WT1 monoclonal antibody, but not an anti-Flag monoclonal antibody, coprecipitates both WT1 and the 40-kDa form of BASP1. Neither the myristoylation-dependent form of BASP1 or a further-truncated BASP1 product were immunoprecipitated with anti-WT1 antibodies (left panel). In addition, purified anti-BASP1 polyclonal antibodies, but not GAL4 polyclonal antibodies, coimmunoprecipitated BASP1 and WT1. Taken together, the data suggest that endogenous WT1 and BASP1 can be associated within the same complexes in M15 cells.
We also performed immunofluorescence experiments to detect endogenous BASP1 in human pediatric kidney tumor G-401 cells (Fig. 4G), finding that BASP1 was again located entirely within the nucleus. When we examined Cos-1 cells, BASP1 was also present in the nucleus (Fig. 4H). However, the cytoplasm of Cos-1 cells showed a greater level of staining, which was particularly prominent around the nuclear periphery. Thus, the cellular localization of BASP1 is cell type specific; in the cell lines that we have analyzed, BASP1 is present in the nuclei of M15, G-401, and Cos-1 cells but showed a significant presence only in the cytoplasm in Cos-1 cells.
BASP1 is present in the developing mouse kidney.
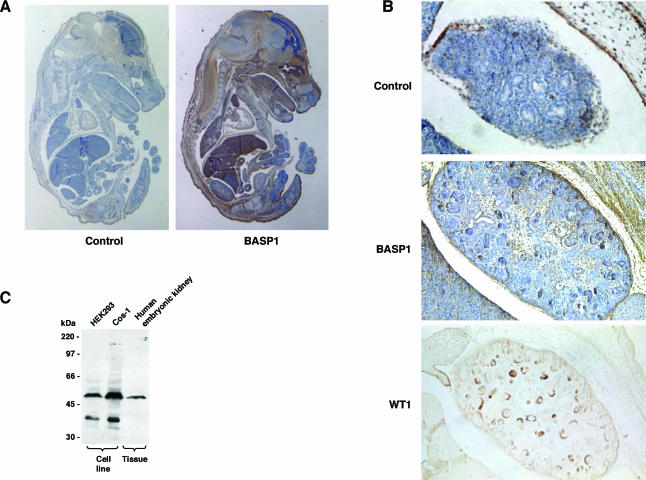
WT1 is a transcription factor that plays a central role in development. In light of the limited information regarding the biology of BASP1, we sought to determine the expression pattern of BASP1 in the mouse embryo. Sections from the mouse embryo recovered at 15.5 days postcoitum were probed with either anti-BASP1 antiserum or preimmune serum (control) (Fig. 5A). BASP1 was present in many regions of the mouse embryo. It was highly expressed in the central nervous system and also in the lungs, heart, thymus, liver, and tongue. WT1 is also expressed in several regions of the developing embryo, and its expression pattern in the developing kidney is well characterized (1). Figure 5B shows the kidney region of the mouse embryos from Fig. 5A. Significantly, BASP1 was present within the forming nephron structures of the developing kidney, which are also regions of high WT1 expression. Consistent with this, immunoblotting of an HEK tissue sample confirmed the presence of the 52-kDa form of BASP1 (Fig. 5C).
FIG. 5.
Expression of BASP1 in the developing mouse embryo. (A) Sections from 15.5-day-postcoitum mouse embryos were subjected to immunohistochemistry with either preimmune serum (control) or anti-BASP1 antibodies. Staining was visualized with diaminobenzidine, and counterstaining was with hematoxylin. Magnification, ×12.5. (B) Immunohistochemistry of mouse embryonic kidney sections with preimmune serum (control) or anti-BASP1 antibodies (magnification, ×100; sections are the same as those shown in panel A). An adjacent section stained with an anti-WT1 antibody, with counterstain omitted to emphasize the staining pattern, is also shown (magnification, ×100). Enlarged images of the BASP1 and WT1 staining are included to highlight specific renal vesicles. (C) The indicated cell line extracts and a whole-cell extract derived from HEK tissue were subjected to immunoblotting with affinity-purified anti-BASP1 antibodies. Molecular mass markers are shown at the left.
BASP1 expression in the adult kidney.
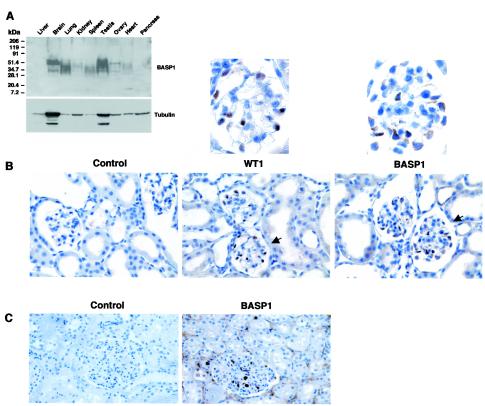
We next used a human adult multitissue blot to determine the regions of BASP1 expression (Fig. 6A). The tissue samples had been resolved by gradient electrophoresis, which likely causes the pleiotropic migration of the different forms of BASP1 and is consistent with the observations of others (27). High levels of BASP1 were detected in the adult brain, lungs, and testis (Fig. 6A). BASP1 was also present in lower levels in the kidney, spleen, ovary, and heart. Where comparable, these data are consistent with the embryo staining pattern, except for the liver expression, which was high in the mouse embryo but is not detected in the adult multitissue blot.
FIG. 6.
Expression of BASP1 in adult tissues and the adult kidney. (A) A human multitissue blot was probed with affinity-purified anti-BASP1 antibodies. Molecular mass markers are shown at the left. The blot was then stripped and reprobed with antitubulin antibodies. (B) Adult mouse kidney sections were probed with either anti-WT1 antibodies or affinity-purified anti-BASP1 antibodies. The control panel omitted the primary antibody. Detection was via diaminobenzidine. The sections were counterstained with hemalum. Magnification, ×200. The glomeruli indicated by arrows are shown at the top at a higher magnification (×600). (C) Same as panel B, except that the kidney section was from a human adult and only the affinity-purified BASP1 antibody was used.
In the adult kidney, WT1 expression is restricted to the highly specialized podocyte cells that line the glomerulus. By immunohistochemistry, this produces a very distinctive pattern of WT1 staining (Fig. 6B). Significantly, we found that the expression of BASP1 in the adult mouse kidney shows a restriction very similar to that in the glomerulus, including the podocytes. The higher-magnification image in Fig. 6B shows that BASP1 is present in both the nuclei and cytoplasm of these cells. Analysis of human adult kidney sections produced comparable results (Fig. 6C). Taken together with the results in Fig. 5, our data show that BASP1 is expressed temporally and spatially in concert with WT1 in the embryonic and adult kidney.
BASP1 exhibits WT1 transcriptional cosuppressor activity.
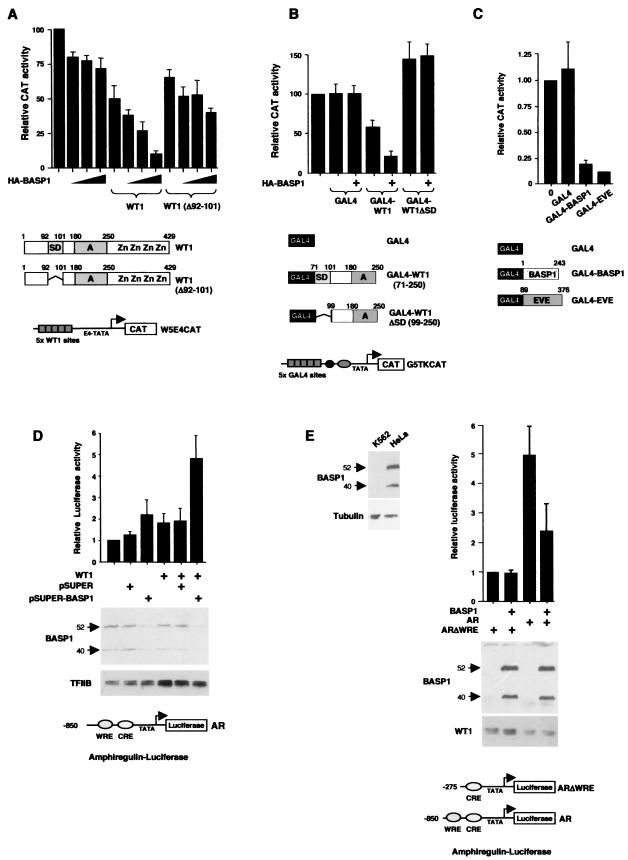
WT1 can repress transcription at highly active promoters. We therefore considered that if BASP1 is indeed part of a WT1 transcriptional cosuppressor, it might augment the transcriptional repression of an active promoter in the presence of WT1. A reporter containing five WT1 DNA-binding sites was transfected into 293 cells either alone or with a plasmid driving expression of intact WT1 (−/− isoform) or a WT1 deletion mutant that lacks the minimal suppression domain (Δ92-101) (Fig. 7A). The results show that increased coexpression of HA-BASP1 significantly augmented transcriptional repression by intact full-length WT1. In comparison, BASP1 showed a small effect in the absence of WT1 and also in the presence of the WT1 derivative lacking a suppression domain (Δ92-101). We also analyzed a GAL4 fusion protein containing WT1 residues 71 to 250, which has previously been shown to repress the “high basal” thymidine kinase (tk) promoter linked to GAL4 DNA-binding sites (G5tkCAT) (21, 24, 37). G5tkCAT was transfected into HEK 293 cells along with a plasmid that expresses either the GAL4 DNA-binding domain, GAL4-WT1 (residues 71 to 250), or a GAL4-WT1 (residues 99 to 250) which lacks the suppression domain. In the absence of HA-BASP1, the GAL4-WT1(71-250) construct caused a modest level of transcriptional repression, consistent with that previously reported by us (24) (Fig. 7B), but the GAL4-WT1 derivative lacking an intact suppression domain (residues 99 to 250) did not. Cotransfection of HA-BASP1 augmented transcriptional repression by GAL4-WT1 (residues 71 to 250) but had no effect when cotransfected with the plasmid expressing either GAL4 or GAL4-WT1 (residues 99 to 250). Taken together, the results in Fig. 7A and B suggest that BASP1 can cooperate with the suppression domain of WT1 to augment transcriptional repression at an active promoter. Consistent with such a cosuppressor function, a GAL4-BASP1 fusion protein was able to repress transcription to a level similar to that observed with the well-characterized repression domain from Even Skipped (GAL4-EVE) (Fig. 7C) (24).
FIG.7.
BASP1 exhibits transcriptional cosuppressor activities. (A) The reporter W5E4CAT (2 μg) was transfected into 293 cells along with 2 μg of empty CMV vector or a CMV vector driving expression of either intact WT1 (−/− isoform) or a deletion mutant of WT1 (Δ92-101). The HA-BASP1 plasmid was added in increasing amounts (1, 2, and 4 μg). After 72 h, the cells were harvested and lysed and CAT activity was measured. The activity of the W5E4CAT reporter alone is set at a value of 100% activity, and other measurements are presented relative to this. The results are the means from three experiments with standard deviations. (B) The reporter G5tkCAT (1 μg) was transfected into HEK 293 cells along with a plasmid (1 μg) driving the expression of either GAL4(1-93), GAL4(1-93)-WT1 (residues 71 to 250) or GAL4(1-93)-WT1 (residues 99 to 250). Where indicated, the plasmid driving expression of HA-BASP1 was also included (2 μg). CAT assays and data presentation are as in panel A. (C). The G5tkCAT reporter (1 μg) was transfected into 293 cells as for panel A, except that 2 μg of a plasmid driving expression of either GAL4(1-93), GAL4(1-93)-BASP1, or GAL4(1-93)-EVE was also included. Data presentation is as in panel A. (D) The amphiregulin promoter-luciferase reporter (AR) (shown at bottom) was transfected into HeLa cells with CMV-WT1 where indicated. Either pSUPER or pSUPER driving BASP1 RNAi expression was also included where indicated (see Materials and Methods for full details of the transfection protocol). Relative luciferase activity is presented in arbitrary units. Bottom panel, cell lysates were immunoblotted with either anti-BASP1 antibodies or anti-TFIIB antibodies. (E) At left, K562 and HeLa whole-cell extracts were immunoblotted with anti-BASP1 (top) and antitubulin (bottom) antibodies. The 52- and 40-kDa BASP1 forms are indicated. K562 cells were transfected with an amphiregulin promoter reporter that either contains (AR) or lacks (ARΔWRE) the WT1 DNA-binding site (1 μg). Where indicated, a plasmid driving expression of BASP1 was included (1 μg). Luciferase activity was measured and presented as in panel D. The immunoblots below the graph show the expression of the transfected BASP1 and the endogenous WT1.
We next sought to determine if BASP1 can regulate the transcriptional activation function of WT1 at a bona fide WT1-responsive target promoter. The amphiregulin promoter has previously been shown to be a target of WT1 during kidney development (18). A luciferase reporter under the control of the amphiregulin promoter was transfected into HeLa cells in the absence or presence of WT1 (Fig. 7D). WT1 elicited a low level of activation of the amphiregulin promoter. However, when we cotransfected a construct that produces a small interfering RNA to target the BASP1 message, we observed a robust activation of the amphiregulin reporter that was dependent upon the presence of WT1. Immunoblotting confirmed that BASP1 expression was specifically eliminated by the expression of the BASP1 small interfering RNA. These data confirm that endogenous BASP1 silences the WT1 transcriptional activation function.
During our analysis of different cell lines, we found that K562 cells lack detectable levels of BASP1 compared to others (Fig. 7E, left panel). K562 cells normally express WT1 (30), and we therefore speculated that, due to the absence of significant levels of BASP1, the endogenous WT1 would exhibit transcriptional activator function. The results in Fig. 7E show that the high activity of the amphiregulin-luciferase reporter in K562 cells is dependent upon the region of the promoter containing the WT1 response element (WRE). Significantly, transfection of a plasmid driving expression of BASP1 causes a downregulation of the amphiregulin reporter that is also dependent upon the presence of the WRE. Thus, BASP1 can regulate the activity of endogenous WT1 at the amphiregulin promoter.
DISCUSSION
It has previously been proposed that a cosuppressor modulates the function of the WT1 transcriptional activation domain (24, 37). Consistent with this notion, analysis of both cell lines and tissues demonstrated that WT1 exists within large protein complexes (13, 16, 23). Factors that have been shown to interact with the −KTS isoform of WT1 and modulate its transcription activity include p53, adenovirus E1b, Par4, CBP, and Ciao 1 (reviewed in reference 33). None of these factors, however, was found to interact with the WT1 suppression domain.
We identified BASP1 as a factor present in a functional WT1 transcriptional cosuppressor fraction. WT1 and BASP1 associate in cells that naturally express both proteins and show temporal and spatial coexpression during kidney development. Downregulation of endogenous BASP1 by RNA interference elicited transcriptional activation of the amphiregulin promoter by WT1. Moreover, ectopic expression of BASP1 in a cell line with naturally low levels of BASP1 suppresses transcriptional activation of the amphiregulin promoter by endogenous WT1. Taken together, our results strongly suggest that BASP1 is a natural regulator of WT1 transcription function. BASP1 shows a more diverse expression pattern in the embryo than that observed for WT1, signifying an additional (but as-yet-unknown) function(s) for this protein. Consistent with this, our immunoprecipitation data suggest that only a portion of BASP1 is associated with WT1.
BASP1 was previously isolated from neuronal cells and was found to be a myristoylated protein (27). This does not make a role in transcription unlikely. Indeed, the Oct1/2-specific transcriptional coactivator, Bob1, exists as both myristoylated and nonmyristoylated forms (40). Only the nonmyristoylated isoform acts as a coactivator for Oct1. Our results suggest that BASP1 also exists in both myristoylated and nonmyristoylated forms and that myristoylation produces a form of BASP1 that does not interact with WT1. We found that BASP1 was present in the nuclei of mouse M15 and human G-401 cells. In Cos-1 cells we observed significant staining of BASP1 in the cytoplasm, particularly around the nuclear membrane. Transfection of WT1 into Cos-1 cells does not cause BASP1 to localize to the nucleus (data not shown). In addition, the presence of myristoylated BASP1 in M15 cells suggests that the cell line-dependent cellular localization of BASP1 cannot be explained simply by myristoylation. Thus, further studies will be required to understand how BASP1 distributes between the nucleus and the cytoplasm.
WT1 plays a critical role in kidney development and is required for the condensation of the metanephric mesenchyme onto the ureteric bud, its proliferation, and then differentiation into the epithelia of the nephron. BASP1 expression overlaps with WT1 within these same intermediate structures in the developing kidney. WT1 expression levels decrease in the latter stages of kidney development and are very low in the adult kidney, where WT1 expression has been confined to a very specialized cell type, the podocytes. Expression of WT1 is required for the maintenance of podocyte function (8). BASP1 expression is also confined to the podocytes of the adult kidney. These data strongly suggest that WT1 and BASP1 are involved in the same pathway of kidney development and perhaps in the maintenance of podocyte function. The −KTS isoform of WT1 plays a greater role in podocyte development and maintenance than the +KTS isoform (9, 10). This suggests that the podocytes are a critical site for the function of WT1 as a transcriptional regulator. Perhaps the presence of BASP1 in the podocytes provides temporal specificity to the transcriptional activity of WT1 in these cells. The WT1 target amphiregulin gene shows the same expression profile within the developing and adult kidney (18). It is therefore possible that BASP1 modulates transactivation of the amphiregulin promoter by WT1 during kidney development. This mechanism perhaps affords a tighter control of WT1 target gene expression and may be critical in regulating proliferation and differentiation. Further insights into such a mechanism will require a determination of WT1, BASP1, and amphiregulin expression at different stages of kidney development.
As mentioned above, an attractive role for the WT1 suppression domain is that it provides specificity to the function of the WT1 transcriptional activation domain. In a specific cell context, the WT1-BASP1 interaction may be disrupted or BASP1 may be downregulated, hence releasing the function of the WT1 transcriptional activation domain. It is tempting to speculate that the PEST sequences within BASP1 may be part of this regulatory process. Our data also suggest that BASP1 cellular localization could be a potential point of regulation. For example, exclusion of BASP1 from the nucleus could relieve suppression of the WT1 transcriptional activation domain. Finally, a role for BASP1 as a modulator of transcriptional activity may help to explain the variability of WT1 in the regulation of proposed target genes in different cell lines (33). Further studies of BASP1 will shed light on the function of the WT1 cosuppressor and the specific transcriptional activator function of WT1.
Acknowledgments
We thank Carol Lyon and Angus Lamond for help with two-dimensional electrophoresis, Craig Smith for providing kidney sections, and Joost Zomerdijk for providing a control peptide. We thank Daniel Haber for providing us with the amphiregulin promoter. We thank Neil Perkins, Andy Sharrocks, Paul Shore, and members of the lab for comments on the manuscript.
This work was funded by the AICR, BBSRC, Cancer Research UK (SP2410/0101), MRC, and Wellcome Trust. S.G.E.R. is a Wellcome Trust Senior Research Fellow.
REFERENCES
- 1.Armstrong, J. F., K. Pritchard-Jones, W. A. Bickmore, N. D. Hastie, and J. B. L. Bard. 1992. The expression of the Wilms' tumour gene, WT1, in the developing mammalian embryo. Mech. Dev. 40:85-97. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2002. Short protocols in molecular biology, 5th ed., vol. 1, p. 16. John Wiley and Sons, Inc., New York, N.Y.
- 3.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 4.Davies, R., A. Moore, A. Schedl, E. Bratt, K. Miyagawa, M. Ladomery, C. Miles, A. Menke, V. van Heyningen, and N. Hastie. 1999. Multiple roles for the Wilms' tumor suppressor, WT1. Cancer Res. 59:1747S-1750S. [PubMed] [Google Scholar]
- 5.Drummond, I. A., S. L. Madden, P. Rohwernutter, G. I. Bell, V. P. Sukhatme, and F. J. Rauscher. 1992. Repression of the insulin-like growth factor-II gene by the Wilms-tumor suppressor WT1. Science 257:674-678. [DOI] [PubMed] [Google Scholar]
- 6.Englert, C., X. Hou, S. Maheswaran, C. Ngwu, G. G. Re, A. J. Garvin, M. R. Rosner, and D. A. Haber. 1995. WT1 suppresses synthesis of the epidermal growth-factor receptor and induces apoptosis. EMBO J. 14:4662-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Englert, C. 1998. WT1—more than a transcription factor? Trends Biochem. Sci. 23:389-393. [DOI] [PubMed] [Google Scholar]
- 8.Guo, J.-K., A. L. Menke, M.-C. Gubler, A. R. Clarke, D. Harrison, A. Hammes, N. D. Hastie, and A. Schedl. 2002. WT1 is a key regulator of podocyte function: reduced expression levels cause crescentic glomerulonephritis and mesangial sclerosis. Hum. Mol. Genet. 11:651-659. [DOI] [PubMed] [Google Scholar]
- 9.Hammes, A., J.-K. Guo, G. Lutsch, J.-R. Leheste, D. Landrock, U. Ziegler, M-C. Gubler, and A. Schedl. 2001. Two splice variants of the Wilms' tumour 1 gene have distinct functions during sex determination and nephron formation. Cell 106:319-329. [DOI] [PubMed] [Google Scholar]
- 10.Hastie, N. D. 2001. Life, sex, and WT1 isoforms—three amino acids can make all the difference. Cell 106:391-394. [DOI] [PubMed] [Google Scholar]
- 11.Hawkes, N. A., and S. G. E. Roberts. 1999. The role of human TFIIB in transcription start site selection in vitro and in vivo. J. Biol. Chem. 274:14337-14343. [DOI] [PubMed] [Google Scholar]
- 12.Herzer, U., A. Crocoll, D. Barton, N. Howells, and C. Englert. 1999. The Wilms tumour suppressor gene wt1 is required for development of the spleen. Curr. Biol. 9:837-840. [DOI] [PubMed] [Google Scholar]
- 13.Iben, S., and B. Royer-Pokora. 1999. Analysis of native WT1 protein from frozen human kidney and Wilms' tumors. Oncogene 18:2533-2536. [DOI] [PubMed] [Google Scholar]
- 14.Kreidberg, J. A., H. Sariola, J. M. Loring, M. Maeda, J. Pelletier, D. Housman, and R. Jaenisch. 1993. WT-1 is required for early kidney development. Cell 74:679-691. [DOI] [PubMed] [Google Scholar]
- 15.Kudoh, T., T. Ishidate, M. Moriyama, K. Toyoshima, and T. Akiyama. 1996. Constitutive expression of the Wilms tumor suppressor gene WT1 in F9 embryonal carcinoma cells induces apoptotic cell death in response to retinoic acid. Proc. Natl. Acad. Sci. USA 92:4517-4521. [PubMed] [Google Scholar]
- 16.Ladomery, M. R., J. Slight, S. McGhee, and N. D. Hastie. 1999. Presence of WT1, the Wilms' tumor suppressor gene product, in nuclear poly(A)(+) ribonucleoprotein. J. Biol. Chem. 274:36520-36526. [DOI] [PubMed] [Google Scholar]
- 17.Larsson, S. H., J. P. Charlieu, K. Miyagawa, K. Engelkamp, M. Rassoulzadegan, A. Ross, F. Cuzin, V. van Heyningen, and N. D. Hastie. 1995. Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell 81:391-401. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. B., K. Huang, R. Palmer, V. B. Truong, D. Herzlinger, K. A. Kolquist, J. Wong, C. Paulding, S. K. Yoon, W. Gerald, J. D. Oliner, and D. A. Haber. 1999. The Wilms' tumor suppressor WT1 encodes a transcriptional activator of amphiregulin. Cell 98:663-673. [DOI] [PubMed] [Google Scholar]
- 19.Lee, T. H., P. Moffett, and J. Pelletier. 1999. The Wilms' tumor suppressor gene (wt1) product represses different functional classes of transcriptional activation domains. Nucleic Acids Res. 27:2889-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Little, M., G. Holmes, and P. Walsh. 1999. WT1: what has the last decade told us? Bioessays 21:191-202. [DOI] [PubMed] [Google Scholar]
- 21.Madden, S. L., D. M. Cook, J. F. Morris, A. Gashler, V. H. Sukhatme, and F. J. Rauscher. 1991. Transcriptional repression mediated by the WT1 Wilms-tumor gene-product. Science 253:1550-1553. [DOI] [PubMed] [Google Scholar]
- 22.Madden, S. L., D. M. Cook, and F. J. Rauscher. 1993. A structure-function analysis of transcriptional repression mediated by the WT1, Wilms' tumour suppressor protein. Oncogene 8:1713-1720. [PubMed] [Google Scholar]
- 23.Maheswaran, S., C. Englert, S. B. Lee, R. M. Ezzel, J. Settleman, and D. A. Haber. 1998. E1B 55K sequesters WT1 along with p53 within a cytoplasmic body in adenovirus-transformed kidney cells. Oncogene 16:2041-2050. [DOI] [PubMed] [Google Scholar]
- 24.McKay, L. M., B. Carpenter, and S. G. E. Roberts. 1999. Regulation of the Wilms' tumour suppressor protein transcriptional activation domain. Oncogene 18:6546-6554. [DOI] [PubMed] [Google Scholar]
- 25.Miller, G., K. I. Panov, J. K. Friedrich, L. Trinkle-Mulcahy, A. I. Lamond, and J. C. B. M. Zomerdijk. 2001. hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoters. EMBO J. 20:1373-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morikawa, Y., S. Hinata, H. Tomoda, T. Goto, M. Nakai, C. Aizawa, H. Tanaka, and S. Omura. 1996. Complete inhibition of human immunodeficiency virus gag myristoylation is necessary for inhibition of particle budding. J. Biol. Chem. 271:2868-2873. [DOI] [PubMed] [Google Scholar]
- 27.Mosevitsky, M. I., J. P. Capony, G. Y. Skladchikova, V. A. Novitskaya, V. A. Plekhanov, and V. V. Zakharov. 1997. The BASP1 family of myristoylated proteins abundant in axonal termini. Primary structure analysis and physico-chemical properties. Biochimie 79:373-384. [DOI] [PubMed] [Google Scholar]
- 28.Natoli, T. A., A. McDonald, J. A. Alberta, M. E. Taglienti, D. E. Housman, and J. A. Kreidberg. 2002. A mammal-specific exon of WT1 is not required for development or fertility. Mol. Cell. Biol. 22:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, S., Y. Kim, B. Kim, C. Seong, Y. Oh, K. Baek, and J. Yoon. 1998. Characterization of bovine and human cDNAs encoding NAP-22 (22kDa neuronal tissue-enriched acidic protein) homologs. Mol. Cells 8:471-477. [PubMed] [Google Scholar]
- 30.Reddy, J. C., and J. D. Licht. 1996. The WT1 Wilms' tumor suppressor gene: how much do we really know? Biochim. Biophys. Acta Rev. Cancer 1287:1-28. [DOI] [PubMed] [Google Scholar]
- 31.Renshaw, J., L. King-Underwood, and K. Pritchard-Jones. 1997. Differential splicing of exon 5 of the Wilms tumour (WTI) gene. Genes Chrom. Cancer 19:256-266. [DOI] [PubMed] [Google Scholar]
- 32.Richard, D. J., V. Schumacher, B. Royer-Pokora, and S. G. E. Roberts. 2001. Par 4 is a coactivator for a splice-isoform specific transcriptional activation domain in WT1. Genes Dev. 15:328-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scharnhorst, V., A. J. van der Eb, and A. G. Jochemsen. 2001. WT1 proteins: functions in growth and differentiation. Gene 273:141-161. [DOI] [PubMed] [Google Scholar]
- 34.Shevchenko, A., M. Wilm, O. Vorm, O. N. Jensen, A. V. Podtelejnikov, G. Neubauer, P. Mortensen, and M. Mann. 1996. A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem. Soc. Trans. 24:893-896. [DOI] [PubMed] [Google Scholar]
- 35.Wagner, K. D., N. Wagner, V. P. I. Vidal, G. Schley, D. Wilhelm, A. Schedl, C. Englert, and H. Scholz. 2002. The Wilms' tumour gene Wt1 is required for normal development of the retina. EMBO J. 21:1398-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, Z. Y., Q. Q. Qiu, and T. F. Deuel. 1993. The Wilms-tumor gene-product WT1 activates or suppresses transcription through separate functional domains. J. Biol. Chem. 268:9172-9175. [PubMed] [Google Scholar]
- 37.Wang, Z. Y., Q. Q. Qiu, M. Gurrieri, J. Huang, and T. F. Deuel. 1995. WT1, the Wilms' tumour suppressor gene product represses transcription through an interactive nuclear protein. Oncogene 10:1243-1247. [PubMed] [Google Scholar]
- 38.Wiederkehr, A., J. Staple, and P. Caroni. 1997. The motility-associated proteins GAP-43, MARCKS, and CAP-23 share unique targeting and surface activity-inducing properties. Exp. Cell Res. 236:103-116. [DOI] [PubMed] [Google Scholar]
- 39.Wilm, M., A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, and M. Mann. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379:466-469. [DOI] [PubMed] [Google Scholar]
- 40.Yu, X., L. Wang, Y. Luo R. G. Roeder. 2001. Identification and characterization of a novel OCA-B isoform: implications for a role in B cell signaling pathway. Immunity 14:157-167. [PubMed] [Google Scholar]