Abstract
Background
A major outbreak of bloody diarrhea associated with Shiga toxin-producing Escherichia coli O104:H4 occurred early in 2011, to which an unusual number of hemolytic uremic syndrome cases were linked. Due to limited information regarding pathogenesis and/or virulence properties of this particular serotype, we investigated the contribution of the aerobactin iron transport system during in vitro and in vivo conditions.
Results
A bioluminescent reporter construct was used to perform real-time monitoring of E. coli O104:H4 in a mouse model of infection. We verified that our reporter strain maintained characteristics and growth kinetics that were similar to those of the wild-type E. coli strain. We found that the intestinal cecum of ICR (CD-1) mice was colonized by O104:H4, with bacteria persisting for up to 7 days after intragastric inoculation. MALDI-TOF analysis of heat-extracted proteins was performed to identify putative surface-exposed virulence determinants. A protein with a high similarity to the aerobactin iron receptor was identified and further demonstrated to be up-regulated in E. coli O104:H4 when grown on MacConkey agar or during iron-depleted conditions. Because the aerobactin iron acquisition system is a key virulence factor in Enterobacteriaceae, an isogenic aerobactin receptor (iutA) mutant was created and its intestinal fitness assessed in the murine model. We demonstrated that the aerobactin mutant was out-competed by the wild-type E. coli O104:H4 during in vivo competition experiments, and the mutant was unable to persist in the cecum.
Conclusion
Our findings demonstrate that bioluminescent imaging is a useful tool to monitor E. coli O104:H4 colonization properties, and the murine model can become a rapid way to evaluate bacterial factors associated with fitness and/or colonization during E. coli O104:H4 infections.
Background
Shiga toxin-producing Escherichia coli (STEC) are members of a category of pathogenic E. coli that can cause illness ranging from mild intestinal diarrheal disease to severe kidney complications, such as hemolytic uremic syndrome (HUS; reviewed in [1]). Cases and outbreaks of STEC have been associated with the consumption of contaminated food and water. Although more than 100 serogroups have been implicated, the major outbreaks are linked to a very small number of serotypes (reviewed in [2]). In 2011, an uncommon strain of pathogenic E. coli serotype O104:H4 caused an unusual number of gastroenteritis and HUS cases, occurring predominantly in adults. The strain originated in northern Germany and disseminated to other European countries [3-5]. The outbreak was originally thought to have been caused by a STEC strain, but was later shown to be produced as a result of an enteroaggregative E. coli (EAEC) strain that had acquired the genes for production of Shiga toxins [6-9].
The EAEC category is heterogeneous, and it is associated with cases of acute or persistent diarrhea in children and adults worldwide (reviewed in [10,11]). The virulence of EAEC is known to require a variety of virulence factors. The mechanism by which EAEC exerts pathogenesis; however, is thus far poorly characterized since EAEC strains are recovered from healthy as well as diseased subjects (reviewed in [10,11]). EAEC strains are recognized by their characteristic aggregative or "stacked-brick" adherence pattern and their ability to form biofilms. It has been proposed that host cellular changes during EAEC infection results in digestive-absorptive abnormalities, prolonging the diarrhea [12]. The ability of EAEC to obtain essential nutrients during this process and multiply successfully in this environment is crucial. EAEC, like most bacteria, must acquire iron to survive, since the inability to acquire this metal will disrupt biofilm formation properties and EAEC interaction with human epithelial cells [13]. Therefore, EAEC strains attempting to establish an infection must have the ability to scavenge iron and multiply within the host environment as fundamental requirements for the disease onset.
A wide variety of strategies for acquiring iron have been developed by pathogenic E. coli, the most common being the production of siderophores and the utilization of heme [14]. Okeke et al. showed that the most prevalent iron acquisition-encoding genes found in clinical EAEC isolates corresponded to those involved in the synthesis of siderophores, such as in the case of aerobactin [15]. Furthermore, this study found an association between geographical variation of the EAEC strains and their iron utilization genes with disease onset, indicating that most EAEC strains contain more than one iron transport system [15]. There is an urgent need to characterize additional virulence factors in E. coli O104:H4, besides the Shiga toxins, which might be associated with disease in the natural setting and not just in silico or in vitro. Therefore, we combined a murine model that mimics the enteropathogenicity of E. coli strains [16,17] with bioluminescent imaging (BLI) technology, a method recently optimized in our laboratory [18]. We hypothesized that the murine model of experimental infection using E. coli O104:H4 bacteria not only is an appropriate way to visualize the site of intestinal colonization, but will also aid in rapid screening of putative virulence factors in vivo. This BLI infection method provided us with the advantage of quantitatively assessing the E. coli O104:H4 burden and facilitated the development of new insights into tissue tropism during infection. Furthermore, BLI application reduced the number of animals required for competition experiments, aided in the localization of E. coli O104:H4 infection sites, and enabled us to quickly screen the role of the aerobactin iron transport system (iut/iuc system) as a virulence factor in this pathogen.
Results
In vivo bioluminescence imaging
The E. coli O104:H4 lux strain RJC001 was generated as described in Methods. We used the pCM17 plasmid containing the lux operon under the OmpC constitutive promoter. This plasmid was used for the following properties: to avoid the exogenous addition of luciferase substrate, it carries both a two-plasmid partitioning system and a post-segregational killing mechanism, and maintenance can be ensured for at least 7 days [19]. E. coli O104:H4 transformants were plated on the appropriate media, incubated at 37 °C, and monitored for bioluminescence. Colonies that did not display any apparent difference in the bioluminescent signal after patching on plates containing the appropriate antibiotic were further evaluated for their resistance to multiple antibiotics (E. coli O104:H4 displayed an extended-spectrum β-lactamase phenotype [20]), presence of multiple plasmids, and growth phenotype similar to that of the wild-type strain (data not shown). E. coli strain RCJ001 was selected because it displayed wild-type characteristics and showed a strong bioluminescence signal.
E. coli O104:H4 lux strain RJC001 was evaluated as a reporter strain in following intestinal infection of the ICR (CD-1) mouse model. A group of 10 ICR mice were infected intragastrically with 1 x 108 CFUs of E. coli strain RJC001 (Figure 1A). Every 24 hours, the animals were anesthetized and monitored for bioluminescent signal by using the IVIS spectrum, which collected and quantified the photons emitted by the E. coli O104:H4 lux infecting the animals. Three animals were sacrificed every 24 hours (except for 72 h and 7 d on which 2 animals were sacrificed), and intestines were harvested for ex vivo imaging. Over the course of the study, the bioluminescence signal increased in whole animals, peaking at 24 h and eventually decreasing with time (Figure 1A). The bioluminescent signal was significantly reduced when the intestines were imaged ex vivo; however, it was evident that bacteria colonize the murine cecum and persist there throughout the various time points (Figure 1B). A bioluminescent signal was undetectable at 168 h (7 days) post infection. Intestinal cecum sections from different time points were homogenized and plated on LB agar containing kanamycin to determine whether the reporter strain remained in the intestine or was eliminated with time. We recovered 4.8 x 106 ± 1.3 x 106 (at 24 h), 1.6 x 107 ± 4.7 x 106 (at 48 h), 3.2 x 107 ± 9.5 x 106 (at 72 h), and 2.3 x 103 ± 9.7 x 102 (at 168 h) CFUs of strain RJC001, confirming that colonization of the intestinal cecum occurred within 3 days of infection, and lower numbers of bacteria were recovered after 7 days. In our previous work, we reported that the threshold of bioluminescent detection is likely in the range of 1 x 103 - 1 x 104 bacteria [18]; therefore, the low numbers of the reporter strain recovered at 7 days explained the absence of the signal.
Figure 1.
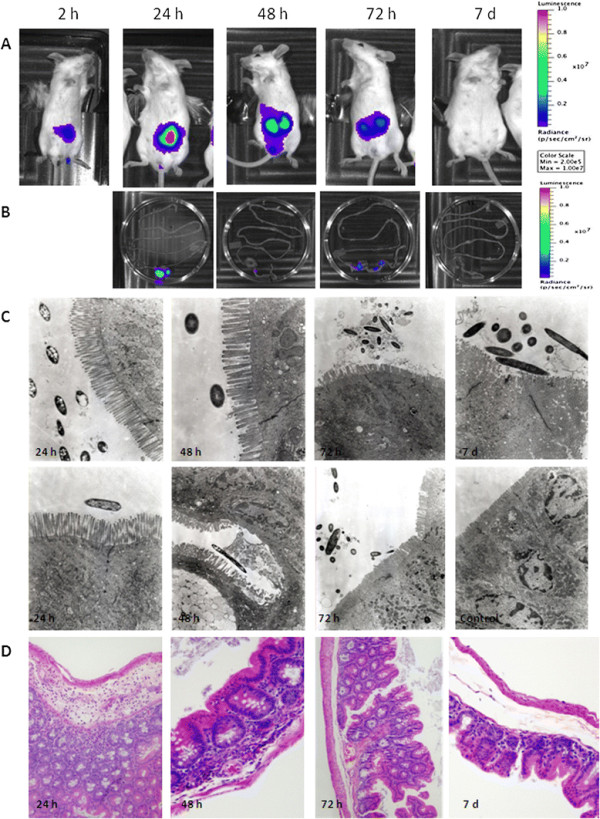
Bioluminescent imaging characterization and tissue analysis of mice infected withE. coliO104:H4luxstrain RJC001.A. RJC001 was inoculated via the intragastrical route into ICR (CD-1) mice. The in vivo bioluminescence (BLI) imaging was conducted at 2, 24, 48, 72 and 168 h (7 days; 7d) post-infection. The intensity of emission is represented as a pseudocolor image. B. At each time point, starting at 24 h, two animals were sacrificed, and intestines were harvested for ex vivo imaging and bacterial load determination, and fixed for electron microscopy and histological analysis. Images are representative of 4 replicate experiments. C. Ultrastructural studies of the cecum infected with E. coli O104:H4 lux strain. RJC001-infected cecum demonstrated a slight destruction of the cellular villi and some cell death at 24, 48 and 72 h post infection. Streptomycin-treated, non-infected tissue was used for comparison (control). Magnification corresponds to 31,000-47,000. D. Representative images from hematoxylin and eosin-stained mouse cecum at 24 h, 48 h, 72 h and 7 days post infection. Focal inflammatory (PMN) infiltrates in the submucosa were seen at 24 h and 48 h post infection. A couple of sections at 72 h and 7d showed very contained foci of residual necrosis surrounded by normal regenerated tissue, but the remainder of the tissue at the later time points was of normal appearance.
Histological characterization and electron microscopical analysis
We used the remainder of the cecum sections for histological and ultrastructural evaluation. Tissues were processed and examined by electron microscopy to determine whether infection with E. coli O104:H4 damaged intestinal epithelial cells. As shown in Figure 1C, bacteria were present in E. coli O104:H4-only infected tissues at all time points. Although, no close interaction with the epithelia was observed, destruction of the microvilli and cell death were detected in the sections analyzed at 48 h and 72 h post infection. Macroscopically, the pathological damage of the intestinal wall at these time points was depicted as bleeding upon contact. In contrast, no changes to tissue integrity were observed at 24 h post infection. At 7 days, integrity of the intestinal epithelial barrier recovered, despite an increase in the number of luminal bacteria. The bacteria appeared clustered and surrounded by extracellular matrices of unknown composition, an interesting feature observed at 72 h post infection (Figure 1C).
Histological examination of the H&E-stained infected tissues also revealed scattered inflammatory infiltrates in the submucosa at 24 and 48 h. Inflammatory infiltrates rarely extended to the mucosa and the muscularis. With the exception of rare foci showing residual necrosis and inflammation, the sections collected at 72 h and at 7 days appeared mostly unremarkable (Figure 1D).
Aerobactin receptor expression is induced on MacConkey agar
We have previously demonstrated that expression of novel putative virulence factors, such as the locus for diffuse adherence in atypical enteropathogenic E. coli[21] or the enterotoxigenic E. coli afimbrial adhesion locus (del Canto et al., manuscript in preparation), are induced when bacteria are grown on MacConkey agar at 37 °C. Furthermore, it is shown that if these factors are expressed on the bacterial surface, a simple extraction method using heat is sufficient in isolating the protein that can then be submitted for sequencing [21]. Therefore, we investigated proteins expressed differentially on MacConkey compared to LB agar in 3 E. coli O104:H4 strains: our prototype German E. coli O104:H4 isolate C3493 and 2 E. coli O104:H4 (strains 2050 and 2071) recovered from an outbreak in the Republic of Georgia. Coomassie-stained SDS-PAGE gel comparison of the heat-extracted protein profiles of the 3 E. coli O104:H4 grown in LB and MacConkey agar revealed one protein in all 3 strains with an apparent molecular weight of ~80 kDa when samples were grown on MacConkey agar (Figure 2, protein A). A second protein of ~55 kDa was also expressed in the E. coli O104:H4 strain 2071 (Figure 2, protein B). In contrast, these two proteins were absent from the crude heat-extracts of the 3 E. coli O104:H4 strains grown in LB agar alone. Both proteins were submitted for MALDI-TOF analysis and identified as the ferric aerobactin receptor (protein A, 731 aa, 80.9 kDa; 18% sequence coverage) and the E. coli chain A, dipeptide-binding protein (protein B, 507 aa, 57.4 kDa). The ferric aerobactin transport system is a well-known virulence factor in E. coli strains causing extraintestinal infections (reviewed in [22]), such as urinary tract infections [23]. Although its role as a virulence determinant in intestinal E. coli is not well understood, it has been proposed that it contributes to the strong colonizing capacity of those strains carrying the aerobactin genes [24]. For this reason, we evaluated the contribution of this iron transport system in the colonization capabilities of E. coli O104:H4.
Figure 2.
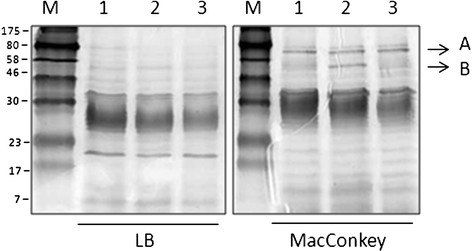
Detection of differentially expressed surface proteins inE. coliO104:H4 strains 15% SDS-PAGE of heat-extracted proteins fromE. coliO104:H4 strain 2050 (lanes 1), 2071 (lanes 2), and C3493 (lanes 3) grown on LB or MacConkey agar. The arrows indicate the location of the aerobactin transport receptor (Arrow A) and the chain A, dipeptide-binding protein (Arrow B).
Low iron concentration in MacConkey induces aerobactin receptor expression
MacConkey agar is considered a low iron-containing medium which has been used to identify high-affinity iron and zinc uptake systems [25]. Therefore, expression of the aerobactin receptor in the E. coli O104:H4 wild type and the iutA mutant was investigated by using heat-extracted preparations of bacteria grown on agar plates with and without the addition of the iron chelator 2,2’-dipyridyl (DP). Expression was monitored on MacConkey as well as LB agar supplemented with DP, because the addition of the iron chelator is known to induce expression of iron transport systems in E. coli[17]. No production of IutA (the 80.9 kDa aerobactin receptor) was observed on Coomassie-stained 12.5% SDS-PAGE gels containing LB agar-recovered bacterial extracts, while abundant IutA was evident in samples from MacConkey plates (Figure 3, panel A). In contrast, the iutA mutant lacked detectable expression of IutA on either media tested. To confirm that aerobactin receptor expression responded to iron depletion, the media was supplemented with 200 μM of DP. As shown in Figure 3, panel A, iron chelation resulted in the expression of IutA in bacteria grown on LB + DP as well as MacConkey + DP. As expected, the aerobactin receptor was absent in heat extracts obtained from the CSS001 strain (iutA::cat) grown on either of the iron-depleted media. However, for reasons that remain unclear, the expression of the IutA receptor does not appear to be further induced on MacConkey agar supplemented with DP.
Figure 3.
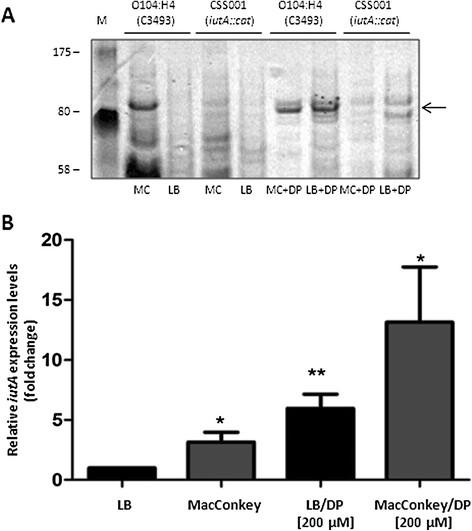
IutA protein induction and qRT-PCR analysis ofiutAexpression.A. Heat-extracted proteins of E. coli O104:H4 strains C3493 (German isolate) and CCSS001 (iutA::cat) grown on MacConkey (MC) or LB agar in the absence (MC or LB) or presence (MC + DP or LB + BS) of 2,2’-dipyridil (DP) were separated in 12.5% SDS-PAGE gels and stained with Coomassie brilliant blue. Molecular mass markers are indicated on the left and the heat-extracted IutA protein is depicted by an arrow on the right. B. E. coli O104:H4 strain C3493 was grown on LB, MacConkey, LB/DP and MacConkey/DP for approximately 18 h at 37 °C, RNA was extracted and cDNA synthesized. The fold variation of gene expression was obtained by the comparative cycle threshold (∆∆CT) method. The iutA expression expressed as a value of 1 represented bacteria grown in LB, and variations in expression in other media conditions are related to this value. The expression of iutA resulted in 2.15- (*, P = 0.01), 4.9- (*, P = 0.001) and 12.13-folds (*, P = 0.01), increase in bacteria grown on MacConkey, LB/DIP and MacConkey/DIP respectively. Student’s T-test was used for the statistical analysis.
Quantitative real-time PCR was performed to support the results obtained with the heat-extracted proteins and to quantify the expression of iutA in the E. coli O104:H4 wild-type strain, while grown in LB or MacConkey media with and without DP. Basal expression of iutA in the wild-type strain was set at a value of 1, and all other values of expression were related to this baseline. The expression of iutA was 2.1-fold higher in the wild-type strain grown in MacConkey as compared to LB (Figure 3B, P = 0.01). In the presence of DP, the iutA expression level in the wild-type strain increased (4.9-fold, P = 0.001) when grown in LB + DP and reached 12.1-fold when the wild-type strain was grown on MacConkey agar supplemented with DP (Figure 3B, P = 0.01). Overall, data confirmed that the aerobactin receptor is expressed on the surface of E. coli O104:H4 wild-type strain, while grown on MacConkey agar, and that expression increased in response to iron depletion.
Contribution of aerobactin to intestinal colonization
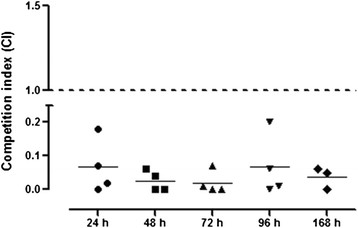
Given that the aerobactin transport system has been proposed as a contributor to the strong intestinal colonizing capability of some strains [24], the influence of the mutation of this iron transport system in E. coli O104:H4 intestinal colonization in mice was assessed. In a wild-type background, deletion of iutA aerobactin receptor gene had a significant effect upon colonization of the cecum (Figure 4). Starting at 24 h post-infection, the wild-type strain outcompeted the iutA mutant [geometric mean (95% confidence interval)]; [0.042 (0.01-0.178)]), suggesting that aerobactin production makes a contribution to colonization early during infection. Consistent with the results at 24 h, the CIs of the iutA mutant at 48 h [0.047 (0.01-0.183)], 72 h [0.01 (0.01-0.137)], 96 h [0.030 (0.01-0.177)], and 168 h [0.005 (0.01-0.140)], were drastically diminished as compared to the wild-type strain. Data suggested that the in vivo intestinal colonization of the E. coli O104:H4 strain required the aerobactin transport system, and the defects observed were due to the inability of the strain to acquire iron.
Figure 4.
TheiutAmutant is outcompeted byE. coliO104:H4 strain C3493 in the murine intestine. Female ICR mice were intragastrically inoculated with 1:1 mixtures of (A) E. coli O104:H4 strain C3493 and its isogenic iutA mutant CSS001. Animals were euthanized at 24, 48, 72, 96 and 168 h, and the number of colonies recovered from the cecum and counted on antibiotic-containing media was used to calculate the competition index (CI). The CI is the ratio of mutant to wild-type CFU in output samples/mutant to wild type CFU in the inoculum. A CI value of 1 (shown by the black line) indicates that the mutant competes equally with the wild-type strain. Bars represent the geometric mean with the 95% confidence interval. The CIs of samples from the same intestinal site were compared by the Mann Whitney non-parametric test.
Discussion
Shiga toxin-producing E. coli O104:H4 is a recently identified emerging pathogen that caused an outbreak resulting in a large number of HUS cases and fatalities in adults. Although the serotype O104:H4 was previously isolated in 2001 from a child presenting HUS [9] and in 2006 from a woman who contracted HUS in Korea [26], the unprecedented number of cases, lethality, and complications resulting from the infection identifies this strain as a public threat to human health. The intestinal disease that arises from the E. coli O104:H4 causing the outbreak seems to be the result of a hybrid infection that developed from recombination of the Shiga toxin genes from STEC O157:H7 into an EAEC strain, which became evident after sequencing the genome of this isolate [3-5]. Despite the extensive body of literature available regarding STEC and EAEC infections and the study of the pathogenic mechanisms, no data are available on the virulence mechanisms of hybrid strains, as in the case of E. coli O104:H4. Data collected by our group and others demonstrated that in vivo bioluminescence imaging is a valuable tool for providing insights into mechanisms of pathogenesis, with the goal of identifying new virulence or colonization properties [18,19]. In the current study, it was demonstrated that E. coli O104:H4 infection in the streptomycin-treated mouse colonization model can be monitored by using RJC001, a bioluminescent strain of E. coli O104:H4.
BLI has been used to study the mechanisms of pathogenesis and treatment efficacies for a number of infectious enteric bacteria. One of the first investigations using BLI was conducted to monitor the virulence differences among strains of Salmonella enterica serovar Typhimurium [27]. In that study, the authors showed the utility of the bioluminescence system by visualizing the efficacy of antibiotic treatment in infected animals. BLI in E. coli has also been used to track EAEC colonization in the streptomycin-treated mouse intestine [28], and the study proposed that the BLI system offers a simple and direct method to study in vitro and in vivo competition between mutants and parental strain. Furthermore, the streptomycin-treated mouse colonization model was previously used to investigate the role of other iron uptake systems (e.g. ferrous iron uptake [Feo] system) in E. coli K12 [29], and it was demonstrated that iron is an essential source for E. coli growth in the murine intestine. It is well known that the maintenance of intestinal colonization requires many properties, among which metabolic competence is of the utmost importance. Therefore, when two strains are in competition for a limited nutrient, like iron, the one that is able to use it more efficiently should outcompete the other [30]. For this purpose, we combined the power of BLI with in vivo murine competition experiments to demonstrate that the aerobactin transport system is required for colonization of E. coli O104:H4.
The aerobactin transport system is a well-established virulence factor in extra-intestinal E. coli infections, but the role of this siderophore system during intestinal infection by pathogenic E. coli strains has never been fully established. However, several lines of evidence suggest that this iron transport system might be an important virulence factor for some intestinal pathogenic E. coli. A previous epidemiological study performed by our group to identify the distribution of iron utilization genes in collections of EAEC strains isolated during case control studies in Nigeria and Brazil, indicated that the aerobactin transport system is present in >75% of the strains analyzed [15]. Interestingly, a significant association was found between the aerobactin transport and the heme transport systems with more strains from cases than from controls in the Nigerian collection [15]. A recent study has also investigated whether virulence determinants, commonly present in extraintestinal pathogenic E. coli, are associated with the fitness of E. coli strains in the infant bowel microbiota [31]. The authors found that accumulation of specific sets of virulence markers, including aerobactin and fimbrial adhesin genes in each individual strain [24], correlated positively with its time of persistence in the colon of infant patients. Therefore, they proposed that some bacterial traits contributing to extra-intestinal infections have evolved to increase the fitness of E. coli in the intestine [31]. Interestingly, E. coli strains that persist and are considered members of the commensal flora can become pathogenic under the appropriate inflammatory conditions in the intestine [32]. For example, members of a newly classified group known as adherent and invasive E. coli (AIEC) are commonly found in ileal lesions of Crohn's Disease patients, and they represent isolates that do not have the classical virulence factors found in other E. coli pathotypes. Recent studies trying to identify those virulence determinants in AIEC that might contribute to the initiation or persistence of CD indicated that the genome of AIEC strains is closely related to those E. coli strains causing extraintestinal infections [17]. Interestingly, all of the AIEC genomes sequenced today possess the aerobactin genes, and it was also demonstrated that iron uptake mediated by the aerobactin system is important for AIEC intracellular survival and mouse intestinal colonization [17]. Overall, the data point to the possibility that the aerobactin transport system participates in the maintenance of the bacteria within the anaerobic environment of the gut. Therefore, this iron transport system in E. coli O104:H4 becomes an important “fitness” determinant, as in the utilization of ferric iron, it confers a competitive advantage to this and other pathogenic bacteria over those organisms that do not possess this transport system.
Although the mouse model does not accurately reflect the intestinal infection or complications seen in humans infected with EAEC, STEC or E. coli O104:H4, it still remains a relatively practical way to investigate the pathogenesis of E. coli strains, especially when compared to more resource-consuming animal models of EAEC/STEC infection, such as the gnotobiotic piglet [33,34] and the rabbit [35,36]. Previous studies have shown that an EAEC O104:H4 strain 55989Str can colonize the streptomycin-treated mouse gut extensively for at least 3 weeks [37]. Even though no sign of disease was evident in the infected animals, the same model was recently used to study the replication of three bacteriophages specific for an EAEC O104:H4 strain, and the mouse intestinal samples enabled the investigators to examine the long-term dynamic interactions between bacteriophages and bacteria within a mammalian host [38]. In the case of STEC, the mouse model has been developed and used to monitor STEC disease and pathology, as well as the impact of Stx in the promotion of intestinal colonization [39]. In our case, the incorporation of BLI analysis proved a useful tool in facilitating the development of an E. coli O104:H4 pathogenesis model, as it significantly reduced the number of animals required to identify the intestinal site of E. coli O104:H4 persistence and colonization. Although the lux-encoded plasmid system that we utilized failed to monitor the infection beyond 7 days and the signal decreased significantly with ex vivo intestines, as previously reported [19], it proved to be a useful way of quantifying colonization of this strain while lacking experimental information about putative pathogenic genes. Currently, we are improving our reporter E. coli O104:H4 strain by mobilizing a constitutively expressed lux operon into its chromosome, providing a stable system that can be used to monitor intestinal colonization and persistence properties for an extended period of time.
Conclusions
Our findings demonstrate that bioluminescent imaging is a useful tool to monitor E. coli O104:H4 colonization properties and present the murine model as a rapid means of evaluating the bacterial factors associated with fitness and/or colonization during E. coli O104:H4 infections.
Methods
Bacterial strains and mutant construction
All strains used in this study are derivatives of the E. coli O104:H4 strain C3493, isolated from a stool sample of a patient with HUS during the 2011 E. coli O104:H4 outbreak in Europe. The sample was obtained from the Enteric Diseases Laboratory Branch, Center of Disease Control and Prevention (CDC, Atlanta, GA). Furthermore, 2 E. coli O104:H4 strains 2050 and 2071, recovered from an outbreak in the Republic of Georgia, were also obtained from the CDC. Unless indicated, strains were grown overnight in Luria-Bertani (LB) medium at 37 °C, shaking at 225 rpm. The aerobactin transport iutA mutant CSS001 was constructed by PCR amplification and cloning of a fragment containing the iutA gene, disrupted with the cam cassette and cloned into the pCVD442 suicide vector. The mutagenesis approach was previously described [23]. The iutA mutant was confirmed by PCR by using the oligonucleotides listed in Table 1, under the following conditions: 1 cycle at 94 °C for 3 min, and then 30 cycles at 94 °C for 1 min, 60 °C for 1 min, and 72 °C for 1 min. For the spatial-temporal location of E. coli O104:H4 in mice, the transformed RJC001 was constructed by electroporation with 3 μg of pCM17 plasmid, containing the luxCDABE operon driven by the OmpC promoter (constitutive expression), which was previously used to visualize pathogenic E. coli[19]. The plasmid was generously donated by J.B. Kaper. Transformants were selected on LB agar plates supplemented with kanamycin (50 mg/ml), and BLI was confirmed by using the IVIS Spectrum (Caliper Corp., Alameda, CA).
Table 1.
qRT-PCR primers used in this study
| Primer name | Sequence | Characteristics | References |
|---|---|---|---|
| 5RTRRSB |
5’-TGCAAGTCGAACGGTAACAG-3’ |
qRT-PCR rrsb gene |
[40] |
| 3RTRRSB |
5’-AGTTATCCCCCTCCATCAGG-3’ |
||
| rpoS Fw |
5’-AGTCAGAATACGCTGAAAGTTCATG-3’ |
qRT-PCR rpoS gene |
[41] |
| rpoS Rv |
5’-AAGGTAAAGCTGAGTCGCGTC-3’ |
||
| iutAFw |
5’- GATCATAGTGTCTGCCAGCC-3’ |
qRT-PCR iutA gene |
This study |
| iutARv |
5’- GCTCTTTACCGCCCTGAATC-3’ |
||
| iutAO104_F |
5’-ATGGAGTTTGAGGCTGGCAC-3’ |
iutA mutant confirmation |
This study |
| iutAO104_R | 5’-GCTTACTGTCGCTGACGTTC-3’ |
Growth curves
Cultures containing no antibiotics were grown overnight at 37 °C, 225 rpm. On the next day, 1:500 dilutions of overnight were inoculated into 30 mL of pre-warmed, sterile LB media. The growth of CSS001 was compared to the growth of wild-type E. coli O104:H4 strain C3493. Sampling was performed at approximately 1-h intervals during the first 9 h of the assay, and a final sample was analyzed 24 h from the start of the experiment. The growth of the E. coli wild- type and CSS001 strains was monitored by plating serial dilutions (log10 CFU/ml) from the time points on LB media with and without 2,2’-dipyridyl as well as by OD600 readings (Additional file 1: Figure S1).
Mice
Female ICR (CD-1) mice of 20 to 25 g were obtained from Charles River Laboratories and housed in the pathogen-free animal facility at UTMB upon arrival for 72 h prior to experiments. Animal studies were performed in accordance with the Animal Care and Use Committee’s guidelines at UTMB as recommended by the National Institute of Health.
Bacterial bioluminescence and competition experiments
For the BLI experiments, animals were inoculated with a suspension of RJC001 (E. coli O104:H4 lux; 1 × 108 CFUs) and, for the competition experiments, with a mixture of E. coli O104:H4 wild-type strain and CSS001 (E. coli O104:H4 iutA::cat; 5 × 107 CFUs per strain) in a final volume of 0.4 ml delivered by gavage (20-gauge needle), thereby using the mouse intestinal model to study enteropathogenicity of E. coli strains previously described by our group [16,17]. Briefly, animals received streptomycin (5 g/L in drinking water) for 48 h prior to oral inoculation with the E. coli strains and were food restricted for 12 h before oral inoculation. The concentration of the initial inoculum was determined by plating on selective antibiotic LB media by using the dot plate method [42]. Groups of mice (n = 10) were maintained for 7 days, and at different time points (24 h, 48 h, 96 h, and 169 h post-inoculation), groups of two or four animals were euthanized, and the cecum of each animal was collected, weighed, and homogenized for bacterial load enumeration. After homogenization, centrifugation at 3,000 xg for 30 seconds was done in order to sediment the cell debris, allowing for collection of accurate volumes needed to make serial dilutions. Samples were plated on LB agar, LB + streptomycin (100 mg/mL), and LB + streptomycin + kanamycin (50 mg/mL) to determine total bacterial cell counts from those of E. coli O104:H4 or the iutA mutant strain. The vast majority of bacteria recovered from the cecum corresponded to the O104:H4 isogenic strains (data not shown). The replicates plated for each mouse were averaged, and competitive indices were calculated as previously described [43]. Groups were compared by using the Mann Whitney non-parametric test.
Bioluminescent quantification
For in-vivo imaging, mice were anesthetized with 2-3% isofluorane in an oxygen-filled induction chamber and then transferred to an isolation chamber placed inside the imaging chamber. Bioluminescent images were acquired by using an IVIS Spectrum (Caliper Corp., Alameda, CA) as we previously described [18]. The ex vivo images of the intestine were acquired at each time point immediately after euthanasia. Bioluminescent signal is represented in the images with a pseudocolor scale ranging from red (most intense) to violet (least intense) indicating the intensity of the signal. Scales were manually set to the same values for every comparable image (in-vivo and ex-vivo) to facilitate comparison of intensity of the bioluminescence at each time point.
Electron microscopy analysis and histopathology
Segments of the mouse cecum infected with the wild-type E. coli O104:H4 strain were collected, washed gently with PBS, and fixed in a mixture of 2.5% formaldehyde, 0.1% glutaraldehyde, 0.03% trinitrophenol, and 0.03% CaCl2 in 0.05 M cacodylate buffer (pH 7.2) as previously described [16]. Samples were processed further by postfixing in 1% OsO4, stained en bloc in 2% aqueous uranyl acetate (in 0.1 M maleate buffer, pH 5.2), dehydrated in ethanol, and embedded in Poly/Bed 812 (Polysciences, Warrington, PA). Ultrathin sections were examined in a Philips 201 electron microscope. One observer, masked to the origin of the samples, examined the sections and took random photomicrographs of each sample. For histological analysis, segments of intestinal cecum were instilled with formalin, processed, and paraffin-embedded. Hematoxylin and eosin-stained slides, containing 1–3 sections of cecum, were examined by a pathologist without knowledge of the origin of the specimens.
Heat-extracted proteins
The strains were grown overnight on LB, MacConkey (Oxoid) agar with or without addition of 200 μM of 2,2’-dipyridyl (DP). The bacterial colonies were suspended in 1x PBS (pH 7.4) and concentrations adjusted spectrophotometrically (600 nm) to 4 x 109 CFU. Bacterial suspensions were incubated at 60 °C for 30 minutes, and then samples were pelleted by centrifugation at 3000 xg for 10 minutes. The supernatant was transferred to a new tube, SDS-sample buffer was added, and samples were boiled at 100 °C for 10 minutes [21]. The samples were separated in 12.5% or 15% SDS-PAGE gels [44]. Expression of distinctively different protein bands were excised from the gel and their identity determined by MALDI-TOF analysis (UTMB Protein core facility) The sequence coverage and the location of the matched peptides are displayed in Additional file 2: Figure S2.
RNA isolation and cDNA synthesis
Total RNA was obtained from bacteria grown on LB and MacConkey agar with or without 200 μM of 2,2’-dipyridyl; after the bacterial colonies were recovered from the plates and suspended in 4 ml of PBS. The samples were stabilized with RNAProtect reagent (QIAGEN, Valencia, CA) and harvested by centrifugation at 3,500 rpm for 20 minutes. The samples were re-suspended in 10 mM Tris–HCl (in 0.1% DEPC-H2O). RNA was purified by using the High Pure RNA Isolation Kit treated with DNaseI (Roche, Mannheim, Germany), quantified, and qualitatively analyzed on 2% agarose gels. Five μg of total RNA was used for cDNA synthesis by the SuperScript First-Strand Synthesis System (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions. A negative control with no reverse transcriptase was also included. The resulting cDNA was utilized for qRT-PCR.
Quantitative real-time RT-PCR (qRT-PCR)
The qRT-PCR was performed by using the iQ™ SYBR supermix and the CFX96 System Test (Bio-Rad, Hercules, CA). We used rrsB and rpoS genes to normalize our data and a value of 1 to standardize iutA gene expression in the wild-type strain grown in LB (primers used are listed in Table 1). For each reaction, 1 μl of reverse-transcribed cDNA was subjected to PCR amplification in a 12.5-μl final volume, containing 500 nM of each primer, and 6.5 μl of 2x SYBR supermix. The following conditions were used for amplification: 1 cycle at 95 °C for 30 s, then 40 cycles at 95 °C for 5 sec, and 60 °C for 30 sec. To ensure the specificity of the PCR products, we performed melting curve analysis by heating products from 65 °C to 95 °C in increments of 0.5 °C every 5 s while monitoring the fluorescence. These assays were performed in triplicate for each strain. Student’s t test was used for statistical analysis.
Authors’ contributions
AGT designed experiments and drafted the manuscript. RJC, MRL, CAB, CSS, and RKJ contributed to the conduct of experiments and reviewing the manuscript. ES conducted and provided histological analysis. VLP conducted and provided electron microscopy analysis. NS and JBK contributed with strains and reagents. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Growth curves ofE. coliO104:H4 isogenic strains. Growth curve of wild-type E. coli O104:H4 strain C3493 and its isogenic mutant CSS001 (ΔiutA) in LB or LB supplemented with 2,2’-dipyridyl (LB + DP) at 37 °C and represented as A. CFU/mL and B. OD600.
Figure S2. MALDI-TOF identified peptides matching the aerobactin receptor. Peptides were identified by MALDI-TOF and subjected to BLAST search analysis which resulted in identification of the Ferric aerobactin receptor precursor from Escherichia coli (gi|218692454) with a score 0f 158 and an expected value of 1.5e-11. The sequence coverage was 18% and the matched peptides are depicted as bold letters.
Contributor Information
Alfredo G Torres, Email: altorres@utmb.edu.
Roberto J Cieza, Email: rjcieza@utmb.edu.
Maricarmen Rojas-Lopez, Email: marojasl@utmb.edu.
Carla A Blumentritt, Email: cablumen@utmb.edu.
Cristiane S Souza, Email: crisssouza@butantan.gov.br.
R Katie Johnston, Email: rkjohnst@utmb.edu.
Nancy Strockbine, Email: Nancy.Strockbine@cdc.hhs.gov.
James B Kaper, Email: Jkaper@medicine.umaryland.edu.
Elena Sbrana, Email: elsbrana@utmb.edu.
Vsevolod L Popov, Email: vpopov@utmb.edu.
Acknowledgements
The work in the AGT laboratory was supported by UTMB discretionary funds and partially by NIH/NIAID grant 5U01AI082103. The authors would like to thank Dr. Douglas Botkin for technical advice and support. We are grateful to Mardelle Susman for many helpful editorial suggestions on this manuscript. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID or NIH.
References
- Farfan MJ, Torres AG. Molecular mechanisms mediating colonization of Shiga toxin-producing Escherichia coli strains. Infect Immun. 2011;80:903–913. doi: 10.1128/IAI.05907-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro JP, Kaper JB. Diarrheogenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–210. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank C, Werber D, Cramer JP, Askar M, Faber M, ander Heiden M, Bernard H, Fruth A, Prager R, Spode A. et al. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med. 2011;365:1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- Askar M, Faber MS, Frank C, Bernard H, Gilsdorf A, Fruth A, Prager R, Hohle M, Suess T, Wadl M. et al. Update on the ongoing outbreak of haemolytic uraemic syndrome due to Shiga toxin-producing Escherichia coli (STEC) serotype O104, Germany, May 2011. Euro Surveill. 2011;16(pii):19883. doi: 10.2807/ese.16.22.19883-en. [DOI] [PubMed] [Google Scholar]
- Scheutz F, Nielsen EM, Frimodt-Møller J, Boisen N, Morabito S, Tozzoli R, Nataro JP, Caprioli A. Characteristics of the enteroaggregative Shiga toxin/verotoxin-producing Escherichia coli O104:H4 strain causing the outbreak of haemolytic uraemic syndrome in Germany, May to June 2011. Euro Surveill. 2011;16(pii):19889. doi: 10.2807/ese.16.24.19889-en. [DOI] [PubMed] [Google Scholar]
- Brzuszkiewicz E, Thürmer A, Schuldes J, Leimbach A, Liesegang H, Meyer FD, Boelter J, Petersen H, Gottschalk G, Daniel R. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC) Arch Microbiol. 2011;193:883–891. doi: 10.1007/s00203-011-0725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde H, Qin J, Cui Y, Li D, Loman NJ, Hentschke M, Chen W, Pu F, Peng Y, Li J. et al. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N Engl J Med. 2011;365:718–724. doi: 10.1056/NEJMoa1107643. [DOI] [PubMed] [Google Scholar]
- Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin CS, Iliopoulos D. et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365:709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W. et al. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One. 2011;6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores J, Okhuysen PC. Enteroaggregative Escherichia coli infection. Curr Opin Gastroenterol. 2009;25:8–11. doi: 10.1097/MOG.0b013e32831dac5e. [DOI] [PubMed] [Google Scholar]
- Harrington SM, Dudley EG, Nataro JP. Pathogenesis of enteroaggregative Escherichia coli infection. FEMS Microbiol Lett. 2006;254:12–18. doi: 10.1111/j.1574-6968.2005.00005.x. [DOI] [PubMed] [Google Scholar]
- Andrade JA, Freymüller E, Fagundes-Neto U. Adherence of enteroaggregative Escherichia coli to the ileal and colonic mucosa: an in vitro study utilizing the scanning electron microscopy. Arq Gastroenterol. 2011;48:199–204. doi: 10.1590/S0004-28032011000300009. [DOI] [PubMed] [Google Scholar]
- Alves JR, Pereira AC, Souza MC, Costa SB, Pinto AS, Mattos-Guaraldi AL, Hirata-Júnior R, Rosa AC, Asad LM. Iron-limited condition modulates biofilm formation and interaction with human epithelial cells of enteroaggregative Escherichia coli (EAEC) J Appl Microbiol. 2010;108:246–255. doi: 10.1111/j.1365-2672.2009.04417.x. [DOI] [PubMed] [Google Scholar]
- Grass G. Iron transport in Escherichia coli: all has not been said and done. Biometals. 2006;19:159–172. doi: 10.1007/s10534-005-4341-2. [DOI] [PubMed] [Google Scholar]
- Okeke IN, Scaletsky IC, Soars EH, Macfarlane LR, Torres AG. Molecular epidemiology of the iron utilization genes of enteroaggregative Escherichia coli. J Clin Microbiol. 2004;42:36–44. doi: 10.1128/JCM.42.1.36-44.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen ST, Blumentritt CA, Slater TM, Patel SD, Tutt CB, Estrella-Jimenez ME, Pawlik J, Sower L, Popov VL, Schein CH. et al. Testing the efficacy and toxicity of adenylyl cyclase inhibitors against enteric pathogens using in vitro and in vivo models of infection. Infect Immun. 2010;78:1740–1749. doi: 10.1128/IAI.01114-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash JH, Villegas A, Kropinski AM, Aguilar-Valenzuela R, Konczy P, Mascarenhas M, Ziebell K, Torres AG, Karmali MA, Coombes BK. Genome sequence of adherent-invasive Escherichia coli and comparative genomic analysis with other E. coli pathotypes. BMC Genomics. 2010;11:667. doi: 10.1186/1471-2164-11-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey S, Johnston K, Mott TM, Judy BM, Kvitko BH, Schweizer HP, Estes DM, Torres AG. in vivo Bioluminescence Imaging of Burkholderia mallei Respiratory Infection and Treatment in the Mouse Model. Front Microbiol. 2011;2:174. doi: 10.3389/fmicb.2011.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee KJ, Cheng H, Harris A, Morin C, Kaper JB, Hecht G. Determination of spatial and temporal colonization of enteropathogenic E. coli and enterohemorrhagic E. coli in mice using bioluminescent in vivo imaging. Gut Microbes. 2011;2:34–41. doi: 10.4161/gmic.2.1.14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielaszewska M, Mellmann A, Zhang W, Köck R, Fruth A, Bauwens A, Peters G, Karch H. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis. 2011;11:671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- Torres AG, Tutt CB, Duval L, Popov V, Nasr AB, Michalski J, Scaletsky IC. Bile salts induce expression of the afimbrial LDA adhesin of atypical enteropathogenic Escherichia coli. Cell Microbiol. 2007;9:1039–1049. doi: 10.1111/j.1462-5822.2006.00850.x. [DOI] [PubMed] [Google Scholar]
- Braun V. Iron uptake by Escherichia coli. Front Biosci. 2003;8:s1409–1421. doi: 10.2741/1232. [DOI] [PubMed] [Google Scholar]
- Torres AG, Redford P, Welch RA, Payne SM. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect Immun. 2001;69:6179–6185. doi: 10.1128/IAI.69.10.6179-6185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrouzian FL, Adlerberth I, Wold AE. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect. 2006;8:834–840. doi: 10.1016/j.micinf.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Patzer SI, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Oh K, Jeon S, Cho S, Lee D, Hong S, Cho S, Park M, Jeon D, Kim S. Escherichia coli O104:H4 from 2011 European outbreak and strain from South Korea. Emerg Infect Dis. 2011;17:1755–1756. doi: 10.3201/eid1708.110879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contag CH, Contag PR, Mullins JI, Spilman SD, Stevenson DK, Benaron DA. Photonic detection of bacterial pathogens in living hosts. Mol Microbiol. 1995;18:593–603. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x. [DOI] [PubMed] [Google Scholar]
- Foucault ML, Thomas L, Goussard S, Branchini BR, Grillot-Courvalin C. In vivo bioluminescence imaging for the study of intestinal colonization by Escherichia coli in mice. Appl Environ Microbiol. 2010;76:264–274. doi: 10.1128/AEM.01686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojiljkovic I, Cobeljic M, Hantke K. Escherichia coli K-12 ferrous iron uptake mutants are impaired in their ability to colonize the mouse intestine. FEMS Microbiol Lett. 1993;108:111–115. doi: 10.1111/j.1574-6968.1993.tb06082.x. [DOI] [PubMed] [Google Scholar]
- Freter R, Brickner H, Fekete J, Vickerman MM, Carey KE. Survival and implantation of Escherichia coli in the intestinal tract. Infect Immun. 1983;39:686–703. doi: 10.1128/iai.39.2.686-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostblom A, Adlerberth I, Wold AE, Nowrouzian FL. Pathogenicity island markers, virulence determinants malX and usp, and the capacity of Escherichia coli to persist in infants' commensal microbiotas. Appl Environ Microbiol. 2011;77:2303–2308. doi: 10.1128/AEM.02405-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieza RJ, Cao A, Cong Y, Torres AG. Immunomodulation for GI infections. Expert Rev Anti Infect Ther. 2012;10:391–400. doi: 10.1586/eri.11.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzipori S, Montanaro J, Robins-Browne RM, Vial P, Gibson R, Levine MM. Studies with enteroaggregative Escherichia coli in the gnotobiotic piglet gastroenteritis model. Infect Immun. 1992;60:5302–5306. doi: 10.1128/iai.60.12.5302-5306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean-Nystrom EA, Pohlenz JF, Moon HW, O'Brien AD. Escherichia coli O157:H7 causes more-severe systemic disease in suckling piglets than in colostrum-deprived neonatal piglets. Infect Immun. 2000;68:2356–2358. doi: 10.1128/IAI.68.4.2356-2358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang G, Pulimood AB, Mathan MM, Mathan VI. Enteroaggregative Escherichia coli infection in a rabbit model. Pathology. 2001;33:341–346. [PubMed] [Google Scholar]
- Ritchie JM, Thorpe CM, Rogers AB, Waldor MK. Critical roles for stx2, eae, and tir in enterohemorrhagic Escherichia coli-induced diarrhea and intestinal inflammation in infant rabbits. Infect Immun. 2003;71:7129–7139. doi: 10.1128/IAI.71.12.7129-7139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Jéhanne V, du Merle L, Bernier-Fébreau C, Usein C, Gassama-Sow A, Wane A-A. et al. Role of deoxyribose catabolism in colonization of the murine intestine by pathogenic Escherichia coli strains. Infect Immun. 2009;77:1442–1450. doi: 10.1128/IAI.01039-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maura D, Morello E, du Merle L, Bomme P, Le Bouguénec C, Debarbieux L. Intestinal colonization by enteroaggregative Escherichia coli supports long-term bacteriophage replication in mice. Environ Microbiol. 2011. Nov 28 [Epub ahead of print] [DOI] [PubMed]
- Mohawk KL, O'Brien AD. Mouse models of Escherichia coli O157:H7 infection and shiga toxin injection. J Biomed Biotechnol. 2011;2011:258185. doi: 10.1155/2011/258185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverton LQ, Kaper JB. Temporal expression of enteropathogenic Escherichia coli virulence genes in an in vitro model of infection. Infect Immun. 2005;73:1034–1043. doi: 10.1128/IAI.73.2.1034-1043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir ER, Warthan M, Brown SP, Nataro JP, Guerrant RL, Hoffman PS. Nitazoxanide inhibits biofilm production and hemagglutination by enteroaggregative Escherichia coli strains by blocking assembly of AafA fimbriae. Antimicrob Agents Chemother. 2010;54:1526–1533. doi: 10.1128/AAC.01279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Nace GW, Irwin PL. A 6 x 6 drop plate method for simultaneous colony counting and MPN enumeration of Campylobacter jejuni, Listeria monocytogenes, and Escherichia coli. J Microbiol Methods. 2003;55:475–479. doi: 10.1016/S0167-7012(03)00194-5. [DOI] [PubMed] [Google Scholar]
- Lloyd SJ, Ritchie JM, Rojas-Lopez M, Blumentritt CA, Popov VL, Greenwich JL, Waldor MK, Torres AG. A double long polar fimbria mutant of Escherichia coli O157:H7 expresses curli and exhibits reduced in vivo colonization. Infect Immun. 2012;80:914–920. doi: 10.1128/IAI.05945-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Growth curves ofE. coliO104:H4 isogenic strains. Growth curve of wild-type E. coli O104:H4 strain C3493 and its isogenic mutant CSS001 (ΔiutA) in LB or LB supplemented with 2,2’-dipyridyl (LB + DP) at 37 °C and represented as A. CFU/mL and B. OD600.
Figure S2. MALDI-TOF identified peptides matching the aerobactin receptor. Peptides were identified by MALDI-TOF and subjected to BLAST search analysis which resulted in identification of the Ferric aerobactin receptor precursor from Escherichia coli (gi|218692454) with a score 0f 158 and an expected value of 1.5e-11. The sequence coverage was 18% and the matched peptides are depicted as bold letters.