Abstract
In primary mammalian cells, DNA replication initiates in a small number of perinucleolar, lamin A/C-associated foci. During S-phase progression in proliferating cells, replication foci distribute to hundreds of sites throughout the nucleus. In contrast, we find that the limited perinucleolar replication sites persist throughout S phase as cells prepare to exit the cell cycle in response to contact inhibition, serum starvation, or replicative senescence. Proteins known to be involved in DNA synthesis, such as PCNA and DNA polymerase δ, are concentrated in perinucleolar foci throughout S phase under these conditions. Moreover, chromosomal loci are redirected toward the nucleolus and overlap with the perinucleolar replication foci in cells poised to undergo cell cycle exit. These same loci remain in the periphery of the nucleus during replication under highly proliferative conditions. These results suggest that mammalian cells undergo a large-scale reorganization of chromatin during the rounds of DNA replication that precede cell cycle exit.
DNA replication in the mammalian cell is organized into discrete nuclear foci that change in location as S phase proceeds (3, 17, 32). A fundamental question is whether the DNA synthesis occurring at these sites results from motion of the replication machinery along the DNA or from spooling of the DNA through fixed replication factories. Early work revealed that many eukaryotic origins spaced throughout a chromosomal region could fire synchronously, suggesting a nuclear coordination of replication initiation (13). It was later demonstrated, using autoradiography, that newly incorporated [3H]thymidine is attached to fixed positions along the nuclear matrix, suggesting the presence of replication complexes through which DNA is moved (33). Furthermore, visualization of these replication foci by electron microscopy showed spreading of label into adjacent chromatin with longer pulses of [3H]thymidine (12). While providing evidence for fixed sites of DNA synthesis, these findings do not directly discriminate between movement of replication machinery and that of replicating DNA.
A number of studies performed with immortalized cell lines using 5-bromo-2′-deoxyuridine (BrdU) labeling and immunofluorescence have shown that DNA synthesis initiates in hundreds of foci distributed throughout the nucleus (3, 32). Recent studies indicate that changes in replication patterns as S phase progresses reflect assembly and disassembly of sites of nucleotide incorporation, rather than motion, lending support to the fixed-replication-site model (6, 24, 38). Studies of the prokaryotic organism Bacillus subtilis, which utilizes a small number of replication foci, have provided the clearest evidence for the factory replication model (23, 28). Expression of a DNA polymerase (pol)-green fluorescent protein fusion protein showed the replication machinery to remain fixed in a few discrete positions throughout S phase. Chromosomal regions were observed to colocalize with the centrally located DNA pol and migrate away from it after replication, providing strong evidence for factory replication in B. subtilis (23).
We have observed that primary mammalian cells display a novel sequence of replication patterns when methods that conserve nuclear architecture are used to detect sites of BrdU incorporation (17). DNA replication initiates in a small number of perinucleolar foci that are associated with intranuclear lamin A/C structures. During S-phase progression in actively cycling cells, these foci disperse to hundreds of small sites throughout the nucleus, forming a distributed replication pattern. Towards the end of S phase, the replication foci cluster over regions containing heterochromatin. Here we report that this sequence of replication patterns is altered as cells prepare to exit the cell cycle. The perinucleolar replication pattern persists throughout the entire length of S phase prior to arrest via contact inhibition, serum starvation, or cellular senescence. Using the NPAT protein as a marker for the histone gene loci (29, 43), we provide an example of a chromosomal region being redistributed to these foci under conditions leading to growth arrest. These findings suggest that the nucleus undergoes a large-scale reorganization during the final S phase(s) prior to cell cycle exit, with much of the genome being recruited through a small number of perinucleolar replication foci.
MATERIALS AND METHODS
Cell culture and BrdU labeling.
WI-38 human diploid fibroblasts (Coriell) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). For asynchronous cell density experiments, actively cycling cells were split into different densities on 10-cm-diameter plates containing coverslips and allowed to grow for 2 days. BrdU labeling reagent (Amersham) was then added directly to the medium at a final concentration of 10 μM and left for 15 min. Coverslips were retrieved from the plates for immunofluorescence, while the remainder of the plate was harvested for fluorescence-activated cell sorter (FACS) analysis. For synchronous cell density experiments, cells cultured on large plates were arrested by contact inhibition, allowing cells to remain at confluence for several days. We find that WI-38 cells exist at approximately 2.0 × 106 cells/10-cm-diameter plate under these conditions. Cells were then released into the cell cycle by replating the same population of cells into low density on 10-cm-diameter plates or into high density on 6-cm-diameter plates. Parallel plates for immunofluorescence and FACS analysis were harvested at hourly time points after labeling with BrdU for 15 min. For the experiments examining longer BrdU pulses, asynchronous cells were split into high cell density on coverslips. After 2 days of growth at high density, cells were pulse-labeled for 15 min, 6 h, or 12 h prior to immunofluorescence.
For the senescence experiments, cells at 72 population doublings (PDLs) were stained for senescence-associated β-galactosidase activity (7). These cells, in addition to cells at 24 PDLs, were plated on coverslips at low density and pulse-labeled with BrdU for 15 min prior to immunofluorescence. To examine replication patterns in cells undergoing serum starvation, asynchronous cells were plated onto coverslips at low density. Cells were either shifted into DMEM containing 0.1% FCS or kept in DMEM containing 10% FCS. After 24 or 48 h, cells were pulse-labeled with BrdU for 15 min and prepared for immunofluorescence.
For double-labeling experiments, IMR-90 (American Type Culture Collection) human diploid fibroblasts were plated on coverslips at low density in DMEM with 10% fetal bovine serum (FBS). Two days later, the medium was replaced with DMEM with 10 or 0.1% FBS. Forty-eight hours later, the coverslips were treated with one of four protocols. Unless otherwise noted, 5-chloro-2′-deoxyuridine (CldU) was used at 20 μM and 5-iodo-2′-deoxyuridine (IdU) was used at 10 μM. In one protocol, cells were treated with CldU for 10 min and fixed immediately. In another, cells were labeled simultaneously with CldU and IdU for 6 h before fixation. For sequential labeling, cells were incubated in DMEM with CldU and either 10 or 0.1% FBS for 6 h. After 6 h, the CldU-containing medium was removed and replaced with medium containing IdU, and cells were fixed after 10 min. For the pulse-chase experiment, cells were labeled with IdU for 10 min, rinsed 3 times in phosphate-buffered saline (PBS) containing 100 μM CldU, and then chased in medium containing CldU for 6 h.
Immortalized cell lines, including NIH 3T3, U20S, and SAOS-2 cells, as well as pRB−/−/p107−/−/p130−/− mouse embryo fibroblasts (MEFs) (a gift of J. Sage and T. Jacks), were plated onto coverslips at low and high cell densities, allowed to grow for 2 days, and then pulse-labeled with BrdU for 15 min prior to immunofluorescence.
FACS analysis.
Cells were harvested for FACS analysis after being labeled with BrdU for 15 min. In brief, trypsinized cells were fixed in 80% ethanol prior to denaturation by treatment with 2 M HCl-0.5% Triton X-100 at room temperature for 30 min. Following a neutralization step in 0.1 M Na2B4O7 (pH 8.5), cells were incubated with an anti-BrdU primary antibody (Becton Dickinson) for 30 min and then with a fluorescein-conjugated secondary antibody (Vector or Molecular Probes) for another 30 min. Cells were treated with RNase, and DNA was stained with propidium iodide prior to two-dimensional FACS analysis.
Immunofluorescence and antibodies.
After being pulse-labeled with BrdU, cells were fixed for 10 min in 1× PBS-4% paraformaldehyde and then permeabilized in 1× PBS-0.5% Triton X-100. Standard indirect immunofluorescence was performed with the indicated primary antibodies and fluorescein- or Texas red-conjugated secondary antibodies (Vector). All quantitative data were collected by using standard fluorescence microscopy. Images were collected by using charge-coupled device (CCD) and deconvolution microscopy (Scanalytics, Deltavision, or Zeiss Axiovision), except for those in Fig. 6, 7, and 9, which are CCD images taken with no deconvolution. All images except that in Fig. 6D were recompiled from images on several sections throughout the nucleus and thus reflect the staining pattern of the entire nucleus compressed into a two-dimensional image. The image in Fig. 7D displays contiguous 0. 5-μm axial slices through the nucleus.
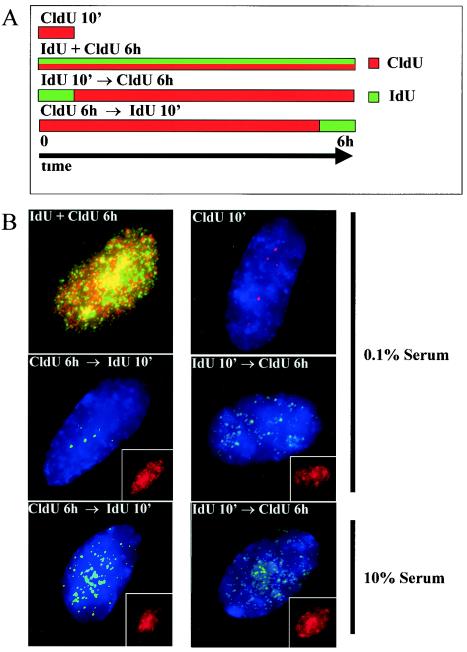
FIG. 6.
Double labeling of serum-starved cells indicates that focal replication patterns persist late into S phase. (A) Schematic representation of double-labeling experiments. (B) Pulse-labeling experiments were performed with IMR-90 fibroblasts kept for 48 h in 10 or 0.1% FBS. CldU-labeled DNA is shown in red, and IdU-labeled DNA is shown in green. The top four panels are representative images of serum-starved cells, and the bottom two panels are representative images of cells maintained with high serum concentrations. The insets in the bottom four panels show CldU signal from the same cell. Longer exposure times were necessary to detect IdU signal in pulse-chase experiments. Images were collected by use of a CCD camera without deconvolution and are presented as a compressed two-dimensional view of the entire nucleus.
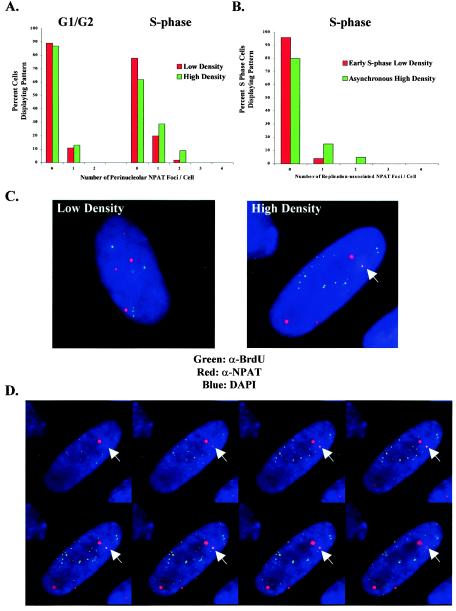
FIG. 7.
Redistribution of histone gene loci to perinucleolar replication foci during S phase in high-density cells. (A) Localization of histone gene loci relative to the nucleolus in asynchronous low- and high-density WI-38 cells. An antibody to NPAT (α-NPAT) is used to mark the histone gene clusters. At high cell density there is a significant redistribution of NPAT foci to perinucleolar regions in S-phase cells. NPAT foci associated with chromosome 6 are more intense than those associated with chromosome 1, and therefore they are distinguishable. (B) Comparison of histone gene loci to perinucleolar replication foci in early-S-phase low-density cells and asynchronous high-density cells. Whereas overlap between NPAT and the perinucleolar foci is minimal at low cell density, there is a significant increase in overlap at high cell density, with up to two NPAT foci coinciding with perinucleolar sites of replication. (C) Representative volume view images displaying the relative lack of colocalization between NPAT and perinucleolar replication foci at low cell density (left) and an increase in overlap at high cell density (right). Colocalization of NPAT (red) and BrdU (green) appears yellow. DAPI, 4′,6′-diamidino-2-phenylindole. (D) Slices of 0.25 μm through the nucleus of the high-density cell shown in panel C, demonstrating that the overlap between NPAT and BrdU occurs in the same plane and is not a result of volume averaging.
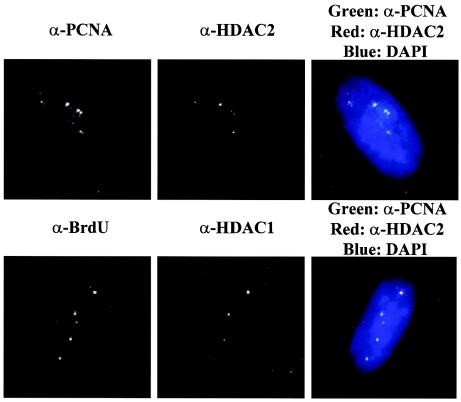
FIG. 9.
HDAC1 and HDAC2 colocalize with perinucleolar replication foci in asynchronous high-density cells. A comparison of BrdU and PCNA localization with that of HDAC1 and HDAC2, respectively, in asynchronous high-density WI-38 cells pulse-labeled with BrdU for 15 min is shown. The merged images appear on the right, with only CCD imaging and no deconvolution. The nucleus is stained by DAPI (4′,6′-diamidino-2-phenylindole). Colocalization between BrdU (green) and HDAC1 (red) and between PCNA (green) and HDAC2 (red) appears yellow. α-, anti-.
BrdU was detected with mouse anti-BrdU antibody (Amersham) containing DNase I. CldU- and IdU-labeled DNAs were detected with rat anti-BrdU clone BU1/75(ICR1) (Novus) and mouse anti-BrdU clone B44 (Becton Dickinson), respectively (6, 25), with DNase I treatment as described by Kennedy et al. (17). Antibodies to PCNA (Zymed), ANA-N (Sigma), pRB (PharMingen), DNA pol δ (BD Biosciences), and p130 (Santa Cruz) were used to detect the locations of the respective antigens. A rabbit polyclonal antibody was used to detect NPAT (43). Antibodies to HDAC1 and HDAC2 were kindly provided by Ed Seto.
RESULTS
Perinucleolar replication foci remain fixed throughout S phase prior to cell cycle exit.
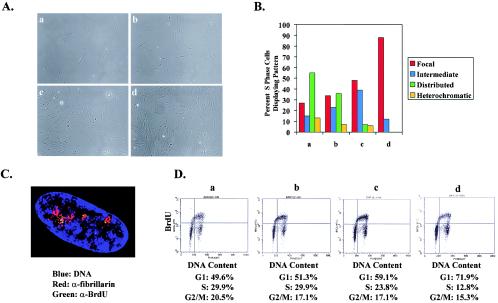
During the course of experiments examining replication patterns in primary human diploid fibroblasts, we noted that cells plated at higher cell densities display enrichment for the perinucleolar replication pattern. To explore this observation further, asynchronous WI-38 cells were split into four different cell densities and then pulse-labeled with BrdU for 15 min prior to fixation (Fig. 1A). At the lowest cell density, the focal, intermediate, distributed, and heterochromatic replication patterns were all represented, with only a fraction of cells displaying the early-S-phase perinucleolar foci (Fig. 1B) (17). In contrast, nearly every BrdU-positive cell at the highest cell density exhibited the focal replication pattern (Fig. 1C). By using two-dimensional FACS analysis to compare BrdU incorporation with DNA content, it was apparent that the proportion of S-phase cells declined with increasing cell density, indicating that contact-inhibited cells were indeed exiting from the cell cycle (Fig. 1D). However, the distribution of BrdU-positive cells across S phase was equivalent at each density, indicating that the high-density cells were not simply delayed in early S phase. Rather, as cells approach contact inhibition, the perinucleolar sites of DNA synthesis persist throughout the entire length of S phase.
FIG. 1.
DNA replication is limited to perinucleolar foci throughout S phase as cells approach contact inhibition. (A) Distribution of replication patterns in asynchronous WI-38 cells plated at different cell densities. Panels a to d show phase-contrast images of cells from which the data were collected. (B) Replication patterns of cells grown at different densities. Lowercase letters correspond to the phase-contrast images in panel A. The data shown are characteristic of those from multiple experiments. Representative images of focal, intermediate, distributed, and heterochromatic patterns have been reported previously (17). (C) A representative cell undergoing DNA replication at high cell density. The image was acquired by deconvolution microscopy and represents a compressed two-dimensional (volume) view of the entire nucleus. The nucleolus is marked with antibodies to fibrillarin (α-fibrillarin). Note that green BrdU foci appear in regions near the nucleolus. Some BrdU foci appear yellow but in fact were above or below the nucleolus when examined in individual images that were used to compose the volume view depicted. (D) Cell cycle distributions of cells analyzed above, ranging from the lowest cell density on the left to the highest cell density on the right. Two-dimensional FACS analysis was used to plot BrdU incorporation versus DNA content and thus to assess the position of BrdU-labeled cells in S phase.
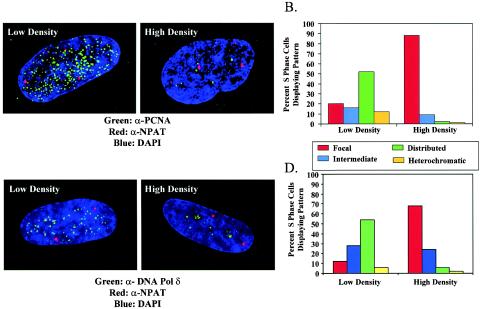
To exclude the possibility that this phenomenon might be due to an artifact of BrdU detection that occurs as cells become dense, we examined the localization of the replication proteins PCNA and DNA pol δ at different cell densities. PCNA has been shown to coincide with sites of DNA replication and, in contrast to BrdU, does not require denaturation of DNA for its detection (17, 18, 37). In order to identify cells in S phase, we utilized the observation that the protein NPAT is consistently detected as exactly two nuclear spots in G1- and G2-phase cells but is detected as four spots throughout S phase (29, 43). Consistent with this observation, at both low and high cell densities, BrdU incorporation is restricted to cells containing four NPAT spots. When PCNA localization was examined in the low-density cells with four NPAT spots, the typical distribution of replication patterns was observed (Fig. 2A and B). In contrast, when cells were grown to a very high cell density, nearly every cell containing four NPAT spots exhibited focal PCNA staining. These foci were demonstrated to be largely in perinucleolar regions (not shown). Similar findings were obtained by using antibodies to DNA pol δ. The fraction of S-phase cells, defined by four NPAT spots, exhibiting focal replication was dramatically increased in cells grown to a very high density (Fig. 2C and D). These results support the notion that the replication machinery remains fixed at the perinucleolar foci during the DNA synthesis that precedes contact inhibition.
FIG. 2.
PCNA and DNA pol δ localization in asynchronous WI-38 cells plated at low or high cell density. (A and B) PCNA localization in S-phase cells propagated at low or high density was determined. S-phase cells were identified because they displayed four nuclear spots upon staining with antibodies directed against NPAT (α-NPAT). DAPI, 4′,6′-diamidino-2-phenylindole. (C and D) DNA pol δ localization was determined in an identical manner under similar growth conditions. All images were acquired by deconvolution microscopy and represent a compressed two-dimensional view of the entire nucleus.
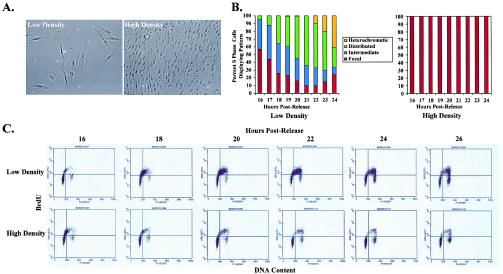
Given that we observed a restriction of DNA replication to perinucleolar foci prior to growth arrest at confluence, we wondered whether this might be associated with a change in the kinetics of S-phase transit. We therefore synchronized WI-38 cells via contact inhibition and released them into S phase by replating them into low or high cell densities (Fig. 3A). Whereas release of cells into low density revealed the previously observed sequence of replication patterns (17), cells released into high density exhibited focal replication patterns at every time point after release (Fig. 3B). Two-dimensional FACS analysis demonstrated that only 10% of cells traversed S phase at high density, compared to approximately 60% of cells at low density, but the rates of movement through S phase were similar in both instances (Fig. 3C). Thus, contact inhibition, while altering the nuclear organization of DNA replication, does not affect the kinetics of S-phase passage.
FIG. 3.
Nuclear reorganization of DNA replication at high cell density is not associated with a change in S-phase kinetics. (A) Phase-contrast images of the low- and high-density cells used. (B) Replication patterns observed in WI-38 cells at hourly time points after synchronous release from contact inhibition into low versus high cell density. (C) FACS analysis demonstrating progression of these cells through S phase at low and high cell densities. Cells begin to enter S phase at 16 h after release in both cases. Approximately 60% of cells have progressed through S phase into G2/M by 26 h at low density, compared to only 10% of cells at high density; however, the rates of movement through S phase are similar in both instances.
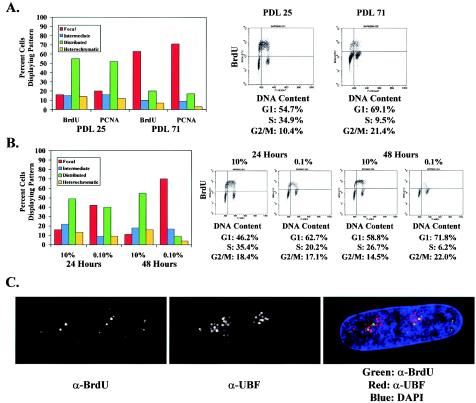
To determine whether the persistence of the perinucleolar foci throughout S phase is confined to conditions leading to density arrest or whether it is more generally associated with cell cycle exit, we examined replication patterns in WI-38 cells undergoing replicative senescence or serum starvation. Cells were continuously passaged to PDL 72, at which point their doubling time had significantly increased, they began to display the typical morphology of senescent cells, and they exhibited senescence-associated β-galactosidase activity (7) (not shown). After pulse-labeling cells with BrdU for 15 min, we compared the replication patterns in old and young cells (PDL 25), both plated at low cell density. Similar to cells undergoing arrest by contact inhibition, most of the BrdU-positive senescing cells displayed the focal replication pattern, whereas young control cells exhibited the typical distribution of replication patterns (Fig. 4A). PCNA localization in cells containing four NPAT foci yielded similar results. Two-dimensional FACS analysis comparing BrdU incorporation with DNA content demonstrated that significantly fewer senescent cells were undergoing DNA replication but that the BrdU-positive cells were spread evenly across S phase. Thus, replicative senescence, similar to contact inhibition, is preceded by replication in perinucleolar foci throughout S phase.
FIG. 4.
The shift towards perinucleolar replication foci is more generally associated with cell cycle exit. (A) Comparison of replication patterns in low-density asynchronous WI-38 cells at PDL 25 and in cells undergoing senescence at PDL 71. Replication patterns are shown by both BrdU incorporation and PCNA localization in cells containing four NPAT foci. FACS analysis demonstrates that fewer cells are in S phase as cells enter senescence but that these cells are evenly spread throughout S phase. (B) Analysis of replication patterns in asynchronous WI-38 cells shifted from 10% serum into 0.1% serum for 24 or 48 h. Replication patterns of serum-starved cells are compared to those of control plates incubated in 10% serum for similar lengths of time. FACS analysis reveals that serum-starved cells exit the cell cycle by 48 h. (C) Representative cell undergoing DNA replication in low-serum medium. The image was acquired by deconvolution microscopy. The nucleolus is marked with antibodies to UBF (α-UBF), a nucleolar transcription factor. Green BrdU foci appear largely in regions around the nucleolus. DAPI, 4′,6′-diamidino-2-phenylindole.
To examine replication patterns in cells undergoing arrest by serum starvation, asynchronous WI-38 fibroblasts plated at low cell density were shifted into medium containing 0.1% serum, and cells were pulse-labeled with BrdU for 15 min after 24 or 48 h of starvation (Fig. 4B). As a control, parallel plates were incubated in medium containing 10% serum and examined at the same time points. By 48 h there was a significant decline in the fraction of BrdU-positive serum-starved cells. This was associated with a shift towards the focal replication pattern at 24 h, with a majority of cells displaying the perinucleolar replication foci by 48 h (Fig. 4B and C). In contrast, control cells continued to display the full range of replication patterns at 24 and 48 h. Taken together, these results indicate that impending cell cycle exit, whether it is due to contact inhibition, replicative senescence, or serum starvation, is associated with persistence of perinucleolar replication foci during S-phase progression.
Changes in replication patterns reflect nuclear reorganization prior to cell cycle exit.
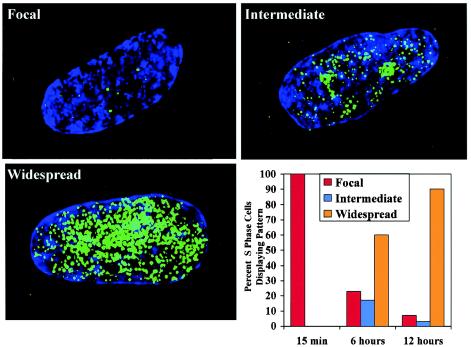
The findings described above imply that most or all of the genome is replicated through a small number of perinucleolar foci prior to arrest by contact inhibition. If this is the case, then longer BrdU pulses should label progressively more of the nucleus. We therefore labeled asynchronous high-density WI-38 cells with BrdU for 15 min, 6 h, or 12 h prior to fixation. As described above, labeling cells with BrdU for 15 min revealed a predominance of the focal replication pattern in S-phase cells (Fig. 5). In contrast, cells labeled for 6 h demonstrated spreading of BrdU throughout nucleus, and by 12 h nearly every BrdU-positive nucleus exhibited widely distributed label. The presence of a few cells with focal replication patterns at longer pulse-labeling times is expected, given that a fraction of cells enter S phase towards the end of the pulse. In sum, long BrdU incorporation times result in widespread labeling of high-density cells, while short pulses of BrdU label only a small number of perinucleolar foci. These findings are consistent with the idea that the perinucleolar foci represent the principal replication sites prior to cell cycle exit.
FIG. 5.
Prolonged BrdU labeling of high-density cells results in a widespread nuclear signal. S-phase cells were labeled for 15 min, 6 h, or 12 h with BrdU, followed by indirect immunofluorescence to detect replicated DNA. Images characterized as focal, intermediate, or widespread are shown. The fraction of S-phase cells with focal, intermediate, or widespread BrdU incorporation was determined for each period of labeling. The data shown are characteristic of those from multiple experiments.
To further test the hypothesis that a large portion of the genome is replicated through a few perinucleolar replication factories prior to cell cycle exit, we performed double-labeling experiments to monitor the location of pulse-labeled DNA over a 6-h window. IMR-90 primary fibroblasts, which behave identically to WI-38 cells in replication assays, were seeded on coverslips at low density and then shifted to 0.1% serum for 48 h. Control cells were maintained in 10% serum. The cells were then labeled according to four protocols (Fig. 6A). Consistent with the results obtained with BrdU, simultaneous labeling of cells with IdU and CldU for 6 h revealed widespread labeling throughout the nucleus in both control and serum-starved cells (Fig. 6B, top left panel). Similarly, pulse-labeling the two populations of cells with CldU for 10 min revealed replication patterns consistent with BrdU labeling, including largely focal patterns for serum-starved cells (Fig. 6B, top right panel). To demonstrate that focal patterns of nucleotide incorporation persist into late S phase in cells preparing to exit the cell cycle, we labeled serum-starved and control cells for 6 h with CldU, followed by a 10-min pulse of IdU. In serum-starved cells, CldU incorporation was detected throughout the nucleus, whereas IdU-substituted DNA was detected only in discrete foci in the nuclear interior (Fig. 6B, middle left panel). In contrast, cells kept with a high serum concentration displayed a distributed pattern of IdU incorporation, characteristic of mid- to late S phase in proliferating cells (17) (Fig. 6B, bottom left panel). Thus, as cells prepare to exit the cell cycle, DNA replication is focal, even late in S phase.
The finding that serum-starved cells replicate large portions of the genome in perinucleolar foci throughout S phase implies that peripheral chromosomal elements must relocate to the nuclear interior to undergo DNA replication. To test this prediction, we labeled serum-starved cells for 10 min with IdU and chased for 6 h with CldU. After the chase, the IdU signal, although weak, was detectable in spots throughout the nucleus, including the nuclear periphery (Fig. 6B, middle right panel). The CldU signal from the same cell indicates that after 6 h, DNA throughout nucleus had incorporated labeled nucleotide (Fig. 6B, middle right panel, inset). Similar results were obtained for control cells (Fig. 6B, bottom right panel). These data suggest that although labeled nucleotide from pulses is detected in foci, after 6 h, the DNA replicated in these foci has spread through the nucleus.
To further address this hypothesis, we took advantage of the observation that NPAT specifically localizes to the histone gene clusters on chromosomes 1 and 6 (29, 43). Thus, we could use antibodies to NPAT to monitor the locations of two specific chromosomal loci during S phase. The two NPAT spots in non-S-phase cells mark the cluster on chromosome 6, while the four NPAT spots in S-phase cells mark the clusters on both chromosomes 1 and 6. It should be noted that although these loci replicate during S phase, the sister chromatids apparently do not segregate, and thus the two loci are detected as four spots throughout S phase and as two spots in G2 phase, which again associate specifically with the histone loci on chromosome 6 (43). It is not known why the histone loci on chromosome 1 can be visualized with antibodies to NPAT only in S phase, although this may be related to increased levels of NPAT during S phase, as NPAT transcription is regulated in an E2F-dependent manner (9).
Previous analysis of the locations of histone loci indicated that they often adopt peripheral locations in the nucleus (43). Consistent with this observation, we found that the histone loci rarely localized to perinucleolar regions in low-density, asynchronously dividing WI-38 cells (Fig. 7A). At high cell density, we observed no change in the fraction of histone loci that exhibited perinucleolar localization in G1/G2-phase cells. In contrast, a greater proportion of histone loci were associated with perinucleolar regions in S-phase cells. It should also be noted that we observed increased localization of histone loci to the nuclear interior during S phase at low density, although they less consistently aligned with nucleoli. Together, these results are in accord with the prediction that peripheral chromosomal loci are redirected to perinucleolar regions for replication at high density.
As a more direct test of this hypothesis, we compared the localization of the histone gene loci to that of perinucleolar BrdU foci in low- and high-density S-phase cells. At low cell density there was minimal overlap, indicating that histone gene loci are not replicated in early S phase in these cells (Fig. 7B and C). In contrast, at high cell density, NPAT spots were found in association with the perinucleolar replication foci in a significant fraction of cells. Axial slices through the nucleus demonstrated that colocalization between NPAT and BrdU occurs in the same plane (Fig. 7D). Interestingly, we detected a higher-than-expected fraction of cells with two histone gene loci localized to BrdU foci, suggesting that perhaps the two loci on chromosome 1 or 6 are replicating concurrently.
We conclude from these experiments that histone loci relocalize from the nuclear periphery to perinucleolar regions to undergo replication at high cell density. Hence, the shift towards perinucleolar replication prior to contact inhibition is associated with the recruitment of distant regions of the genome through this limited number of foci.
Redistribution of replication to perinucleolar foci in high-density immortalized cells.
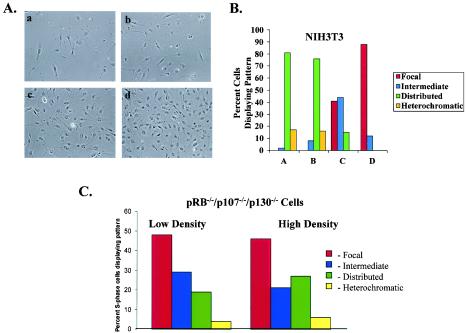
We previously reported that cellular immortalization alters the spatial organization of DNA replication relative to that in primary mammalian cells (17). A number of immortalized cell lines initiate DNA synthesis in a distributed fashion throughout the nucleus. Many of these cell lines, however, retain the ability to arrest via contact inhibition. We therefore sought to determine whether we might observe a similar shift in replication towards the perinucleolar foci in immortalized cells at high cell density. Replication patterns in asynchronous NIH 3T3 cells plated at various cell densities were analyzed (Fig. 8A). As previously reported, at low cell density a vast majority of BrdU-positive NIH 3T3 cells exhibited distributed replication patterns, with virtually no evidence of focal replication (17) (Fig. 8B). At higher cell densities, cells containing perinucleolar replication foci appeared, and at the highest cell density nearly every BrdU-positive cell displayed focal replication. NIH 3T3 cells arrested at confluence over several days displayed dramatically reduced levels of BrdU incorporation, consistent with exit from the cell cycle (not shown). A number of other immortalized cell lines, including other 3T3 derivatives and human cell lines (U20S and SAOS-2), respond to contact inhibition and exhibit perinucleolar replication foci throughout S phase at high cell density (data not shown). Thus, even in cells that have escaped senescence and typically replicate their DNA in a distributed fashion, there is a dramatic shift towards replication in the perinucleolar foci associated with contact inhibition.
FIG. 8.
Immortalized cells shift to perinucleolar replication prior to cell cycle exit if they arrest in response to contact inhibition. (A) Panels a to d are phase-contrast images of cells in which replication patterns were determined by indirect immunofluorescence. (B) Distribution of replication patterns in asynchronous NIH 3T3 cells plated at different cell densities. The data shown are characteristic of those from multiple experiments. (C) Replication patterns observed in pRB−/−/p107−/−/p130−/− cells plated at low and high cell densities.
We next examined the behavior of cells that fail to respond to cell growth arrest signals at high density. MEFs generated from mice lacking wild-type pRB, p107, and p130 fail to exit the cell cycle under a variety of growth-inhibitory conditions (35). A significant fraction of S-phase cells exhibited perinucleolar replication foci during proliferation at low cell density (Fig. 8C). However, in contrast to all other cell types examined, these cells did not display enrichment for the perinucleolar replication pattern at high density. Instead, a large proportion of cells continued to exhibit distributed replication patterns. The failure of pRB−/−/p107−/−/p130−/− MEFs to display increased perinucleolar replication could be due to the inability of these cells to respond to cell cycle exit signals, or it might reflect a role for the retinoblastoma family of proteins in regulating replication patterns. In either case, this finding underscores the association between replication in perinucleolar foci throughout S phase and cell cycle exit.
Chromatin-modifying factors target perinucleolar foci prior to cell cycle exit.
The perinucleolar and distributed replication foci are associated with different sets of proteins during S-phase progression at low cell density (17). Both pRB and HDAC family members target the perinucleolar foci in G1 and early S phases but fail to colocalize with distributed foci later in S phase in proliferating low-density cells (17, 21). To address whether these proteins might continue to remain associated with the replication machinery prior to contact inhibition, we compared the localizations of HDAC and pRB family members relative to the perinucleolar replication foci in asynchronous high-density WI-38 cells pulse-labeled with BrdU. HDAC1 and -2 demonstrated significant colocalization with the perinucleolar replication foci, marked by BrdU and PCNA, respectively, in all cells examined (Fig. 9). In addition, pRB family members and other SIN3A components target a majority of the perinucleolar replication foci in high-density cells (not shown). Therefore, DNA replicated through the perinucleolar foci at high cell density is continuously exposed to proteins that have the potential to modify chromatin structure.
DISCUSSION
We recently reported that primary mammalian cells exhibit a novel replication pattern in early S phase, consisting of a small number of foci that are perinucleolar and associated with intranuclear lamin A/C structures (17). Moreover, whereas it had previously been assumed that the spatial and temporal organizations of DNA replication during S-phase progression are similar among most cells, we demonstrated that differences in replication patterning occur in primary and immortalized cells, presumably due to mutations arising during the immortalization process. We now find that changes in cell growth conditions can alter the nuclear organization of DNA replication. In cells preparing to exit the cell cycle via contact inhibition, serum starvation, or replicative senescence, perinucleolar replication foci persist throughout S phase. Longer periods of BrdU incorporation reveal spreading of label throughout the nucleus, suggesting that most or all of the genome is replicated through these foci. In addition, the histone gene clusters provide an example of a chromosomal locus that is redistributed to the perinucleolar foci under conditions leading to arrest. Taken together, these findings support the idea that mammalian cells undergo factory replication through a small number of perinucleolar foci prior to cell cycle exit.
Our results contrast with those from Dimitrova and Berezney, who argue that replication patterns are invariant across cell types and changes in physiologic conditions (5). Those authors agree that DNase I-based detection of BrdU is the method that best maintains nuclear architecture, and they corroborate our previous findings that the lamin framework is destroyed by HCl treatment (5, 17). It is thus difficult to imagine that the BrdU patterns generated by HCl treatment are accurate, and it is easier to see why treatment with HCl does not allow fine changes in the nuclear organization of DNA replication to be observed.
It is unclear why Dimitrova and Berezney were unable to observe the perinucleolar replication foci in primary fibroblasts after DNase I treatment; however, one possible explanation for the differences arises from the use of an ethanol solution at pH 2.0 as a fixative prior to DNase I exposure (5). These conditions differ dramatically from our reported methods and may provide an acidic environment sufficient to mimic the destructive effects of HCl treatment (17). As shown above, these foci are less apparent in low-density cycling cells, but they become strikingly visible as cells approach confluence and as they reenter S phase following release from arrest by contact inhibition. Furthermore, the localization of replication proteins to perinucleolar foci at high density and in early S phase provides strong evidence that these structures are not simply an artifact of incomplete DNase I digestion, since the detection of these proteins does not rely upon denaturation of DNA (Fig. 1C).
The observation that the perinucleolar replication foci persist throughout S phase prior to exit from the cell cycle suggests that large-scale chromosomal movements must occur in the process of replicating the genome through these structures. Previous studies have yielded conflicting results regarding motion of chromosomes during interphase (22). One intriguing study observed a heterochromatic homogenously staining region to move from the nuclear periphery into the nuclear interior and then back to the periphery between 4 and 6 h into S phase (26). Ultrastructural analysis by electron microscopy revealed that the site within the nuclear interior is closely apposed to the nucleolus. A more recent study of another chromosome site found it to shuttle between the nuclear periphery and the interior at 1 to 1.5 h into S phase (39). Our results suggest that such movement may be dependent upon cell growth conditions, and they could help to explain the different observations reported in the literature. Also, we noted that histone loci appear to be redirected to the nuclear interior during S phase at low cell density, although they do not target perinucleolar regions. This observation leads us to believe that replication may occur in specific internal locations in late S phase during rapid proliferation. However, these locations are more numerous and qualitatively different than the perinucleolar sites that are associated with cell cycle exit.
Asynchronous cells plated at progressively higher densities exhibit a greater percentage of cells with perinucleolar replication foci the denser cells become (Fig. 1A). In addition, cells released synchronously into moderately high density distribute the perinucleolar replication foci later in S phase. These results indicate that the persistence of replication in the perinucleolar foci is a progressive transition that occurs over the last few S phases that precede cell cycle exit. We also find that the restriction of PCNA to perinucleolar foci throughout S phase occurs at higher cell densities than does the restriction of BrdU to the perinucleolar foci (data not shown). This may simply reflect a higher sensitivity of the PCNA antibody for its epitope than the BrdU antibody, or it could mean that PCNA remains associated with the replicated DNA at these densities. Regardless, at the highest cell densities, both PCNA and BrdU are limited to the perinucleolar foci throughout S phase. Interestingly, the factory replication described for B. subtilis was most clearly observed in cells growing at reduced rates. When succinate was used as a carbon source to slow cell growth, a majority of cells exhibited only one focus of pol C-green fluorescent protein at mid-cell, whereas the use of glucose resulted in cells with up to four or five replication foci (23). This might suggest a conserved mechanism in prokaryotes and eukaryotes leading to the restriction of replication foci under conditions promoting growth inhibition.
Both PCNA and CAF-1 have been implicated in the epigenetic inheritance of chromosome states. Mutation of either PCNA or CAF-1 subunits in yeast results in partial loss of silencing at telomeres and the silent-mating-type loci (8, 16, 42). Mutations in the Drosophila PCNA homologue, mus209, result in suppression of position effect variegation (11). In mammalian cells, PCNA has been shown to mark newly replicated DNA for CAF-1-mediated chromatin assembly, suggesting a mechanism for how chromatin structure may be epigenetically inherited (37). PCNA also targets the DNA methyltransferase DNMT-1 to replication foci (4), and it may coordinate the inheritance of methylation patterns with chromatin assembly (37). Consistent with these reports, we find that PCNA and CAF-1 localize to both the perinucleolar and distributed foci, poised to maintain the epigenetic inheritance of chromatin structure in cycling cells (17).
Recently, it has also been shown that heterochromatin may be propagated by the ability of HP1 to recognize methylated H3 Lys 9 and to interact with SUV39H1, which methylates this residue (2, 20, 34). Similarly, euchromatin may be perpetuated by the ability of bromodomain-containing histone acetyltransferases to bind acetylated lysines (10, 40). It will be interesting to examine the localization of these proteins relative to the focal, distributed, and heterochromatic replication foci.
It is less clear how reprogramming of chromatin structure might occur during S-phase progression. It has long been recognized that DNA replication results in the disruption of nucleosomes and may provide a window of opportunity to effect changes in chromatin structure (41). Transcriptional activators and repressors, if present at sites of DNA replication, would have access to their DNA binding sites and could function to modify nucleosomal structure or acetylation status during chromatin assembly. Numerous examples of S-phase-dependent gene activation or silencing have been reported (1, 15, 30), although replication-independent transcriptional modification clearly exists as well (19, 27).
We suggest that replication of the genome through the perinucleolar factories may serve as a means of globally restructuring chromatin and modifying the transcriptional status within the nucleus. Cell cycle exit requires the expression of a different set of genes than for actively cycling cells (14, 36) and largely involves silencing of genes promoting proliferation. We find that pRB and HDAC complexes localize to the perinucleolar replication sites throughout S phase, poised to establish repressive chromatin structures enveloping E2F-responsive genes and perhaps others. Moreover, the nuclear dynamics involved in replicating the genome through these foci would provide the opportunity to reorganize chromatin within the nucleus in a way that might favor the repression and expression of different sets of genes. Interestingly, Narita et al. have recently shown that primary cells approaching senescence undergo pRB-dependent, large-scale changes in chromatin structure (31). It is also intriguing to speculate that this mode of replication could couple cell cycle exit to cellular differentiation. Transcriptional regulators of differentiation could potentially target these foci and modify chromatin assembly towards the expression of cell-type-specific genes. It will be interesting to examine the localization of transcription factors relative to replication foci in cells undergoing differentiation.
Finally, these observations reveal that the nuclear organization of DNA replication is regulated and is altered both by cell cycle exit and cellular immortalization. Elucidating the mechanisms governing this regulation should provide insight into the control of these events, which are fundamental to the process of oncogenesis.
Acknowledgments
We thank J. Sage and T. Jacks for helpful discussions, for editorial comments on the manuscript, and for providing pRB−/−/p107−/−/p130−/− MEFs. We also express our sincere gratitude to M. Classon for helpful discussions and scientific advice. Deconvolution microscopy was performed at the Whitehead Institute Microscopy Facility and at the WM Keck Center for Cellular Imaging at the University of Washington.
This work was funded by an NIH grant to E.H. D.A.B. was supported by a Howard Hughes Research Training Fellowship for Medical Students. B.A.K. was supported in part by PHS NRSA T32 GM07270 from NIGMS. J.Z. was supported by an American Cancer Society Fellowship. E.H. is an American Cancer Society Research Professor. B.K.K. was supported by a Leukemia Society of America Fellowship and is presently a Searle Scholar.
REFERENCES
- 1.Almouzni, G., and A. P. Wolffe. 1993. Replication-coupled chromatin assembly is required for the repression of basal transcription in vivo. Genes Dev. 7:2033-2047. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 3.Berezney, R., D. D. Dubey, and J. A. Huberman. 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108:471-484. [DOI] [PubMed] [Google Scholar]
- 4.Chuang, L. S.-H., H. I. Ian, T. W. Koh, H. H. Ng, G. Xu, and B. F. Li. 1997. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277:1996-2000. [DOI] [PubMed] [Google Scholar]
- 5.Dimitrova, D. S., and R. Berezney. 2002. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized, and transformed mammalian cells. J. Cell Sci. 115:4037-4051. [DOI] [PubMed] [Google Scholar]
- 6.Dimitrova, D. S., and D. M. Gilbert. 2000. Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nat. Cell Biol. 2:686-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubeli, O. Pereira-Smith, M. Peacocke, and J. Campisi. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA. 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enomoto, S., and J. Berman. 1998. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 12:219-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao, G., A. P. Bracken, K. Burkard, D. Pasini, M. Classon, C. Attwooll, M. Sagara, T. Imai, K. Helin, and J. Zhao. 2003. NPAT expression is regulated by E2F and is essential for cell cycle progression. Mol. Cell. Biol. 23:2821-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassan, A. H., p. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 11.Henderson, D. S., S. S. Banga, T. A. Grigliatti, and J. B. Boyd. 1994. Mutagen sensitivity and suppression of position-effect variegation result from mutations in mus209, the Drosophila gene encoding PCNA. EMBO J. 13:1450-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hozak, P., A. B. Hassan, D. A. Jackson, and P. R. Cook. 1993. Visualization of replication factories attached to the nucleoskeleton. Cell 73:361-373. [DOI] [PubMed] [Google Scholar]
- 13.Huberman, J. A., and A. D. Riggs. 1968. On the mechanism of DNA replication in mammalian chromosomes. J. Mol. Biol. 32:327-341. [DOI] [PubMed] [Google Scholar]
- 14.Iyer, V. R., M. B. Eisen, D. T. Ross, G. Schuler, T. Moore, J. C. F. Lee, J. M. Trent, L. M. Staudt, J. J. Hudson, M. S. Boguski, D. Lashkari, D. Shalon, D. Botstein, and P. O. Brown. 1999. The transcriptional program in the response of human fibroblasts to serum. Science 283:83-87. [DOI] [PubMed] [Google Scholar]
- 15.Kamakaka, R. T., M. Bulger, and J. T. Kadonaga. 1993. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 7:1779-1795. [DOI] [PubMed] [Google Scholar]
- 16.Kaufman, P. D., R. Kobayashi, and B. Stillman. 1997. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 11:345-357. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy, B. K., D. A. Barbie, M. Classon, N. Dyson, and E. Harlow. 2000. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 14:2855-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kill, J. R., J. M. Bridger, K. H. Campbell, G. Maldonado-Codina, and C. J. Hutchison. 1991. The timing of the formation and usage of replicase clusters in S-phase nuclei of human diploid fibroblasts. J. Cell Sci. 100:869-876. [DOI] [PubMed] [Google Scholar]
- 19.Kirchmaier, A. L., and J. Rine. 2001. DNA replication-independent silencing in S. cerevisiae. Science 291:646-650. [DOI] [PubMed] [Google Scholar]
- 20.Lachner, M., D. O' Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 21.Lai, A., B. K. Kennedy, D. A. Barbie, N. R. Bertos, X. J. Yang, M. C. Theberge, S. C. Tsai, E. Seto, Y. Zhang, A. Kuzmichev, W. S. Lane, D. Reinberg, E. Harlow, and P. E. Branton. 2001. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol. Cell. Biol. 21:2918-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamond, A. I., and W. C. Earnshaw. 1998. Structure and function in the nucleus. Science 280:547-553. [DOI] [PubMed] [Google Scholar]
- 23.Lemon, K. P., and A. D. Grossman. 1998. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science 282:1516-1519. [DOI] [PubMed] [Google Scholar]
- 24.Leonhardt, H., H. P. Rahn, P. Weinzierl, A. Sporbert, T. Cremer, D. Zink, and M. C. Cardoso. 2000. Dynamics of DNA replication factories in living cells. J. Cell Biol. 149:271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, F., J. Chen, E. Solessio, and D. M. Gilbert. 2003. Spatial distribution and specification of mammalian replication origins during G1 phase. J. Cell Biol. 161:257-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, G., G. Sudlow, and A. S. Belmont. 1998. Interphase cell cycle dynamics of a late-replicating, heterochromatic homogenously staining region: precise choreography of condensation/decondensation and nuclear positioning. J. Cell Biol. 140:975-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Y. C., T. H. Cheng, and M. R. Gartenberg. 2001. Establishment of transcriptional silencing in the absence of DNA replication. Science 291:650-653. [DOI] [PubMed] [Google Scholar]
- 28.Losick, R., and L. Shapiro. 1998. DNA replication. Bringing the mountain to Mohammed. Science 282:1430-1431. [DOI] [PubMed] [Google Scholar]
- 29.Ma, T., B. A. Van Tine, Y. Wei, M. D. Garrett, D. Nelson, P. D. Adams, J. Wang, J. Qin, L. T. Chow, and J. W. Harper. 2000. Cell cycle-regulated phosphorylation of p220NPAT by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 14:2298-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, A. M., and K. Nasmyth. 1984. Role of DNA replication in the repression of silent mating type loci in yeast. Nature 312:247-251. [DOI] [PubMed] [Google Scholar]
- 31.Narita, M., S. Nunez, E. Heard, M. Narita, A. W. Lin, S. A. Hearn, D. L. Spector, G. J. Hannon, and S. W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113:703-716. [DOI] [PubMed] [Google Scholar]
- 32.Newport, J., and H. Yan. 1996. Organization of DNA into foci during replication. Curr. Opin. Cell Biol. 8:365-368. [DOI] [PubMed] [Google Scholar]
- 33.Pardoll, D. M., B. Vogelstein, and D. S. Coffey. 1980. A fixed site of DNA replication in eucaryotic cells. Cell 19:527-536. [DOI] [PubMed] [Google Scholar]
- 34.Richards, E. J., and S. C. R. Elgin. 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 106:489-500. [DOI] [PubMed] [Google Scholar]
- 35.Sage, J., G. J. Mulligan, L. D. Attardi, A. Miller, S. Chen, B. Williams, E. Theodorou, and T. Jacks. 2000. Targeted disruption of the three Rb-related genes leads to loss of G1 control and immortalization. Genes Dev. 14:3037-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shelton, D. N., E. Chang, P. S. Whittier, D. Choi, and W. D. Funk. 1999. Microarray analysis of replicative senescence. Curr. Biol. 9:939-945. [DOI] [PubMed] [Google Scholar]
- 37.Shibahara, K., and B. Stillman. 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96:575-585. [DOI] [PubMed] [Google Scholar]
- 38.Sporbert, A., A. Gahl, R. Ankerhold, H. Leonhardt, and M. C. Cardoso. 2002. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell 10:1355-1365. [DOI] [PubMed] [Google Scholar]
- 39.Tumbar, T., and A. S. Belmont. 2001. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat. Cell Biol. 3:134-139. [DOI] [PubMed] [Google Scholar]
- 40.Turner, B. M. 2002. Cellular memory and the histone code. Cell 111:285-291. [DOI] [PubMed] [Google Scholar]
- 41.Wolffe, A. P. 1991. Implications of DNA replication for eukaryotic gene expression. J. Cell Sci. 99:201-206. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, Z., K. Shibahara, and B. Stillman. 2000. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408:221-225. [DOI] [PubMed] [Google Scholar]
- 43.Zhao, J., B. K. Kennedy, B. D. Lawrence, D. A. Barbie, A. G. Matera, J. A. Fletcher, and E. Harlow. 2000. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14:2283-2297. [PMC free article] [PubMed] [Google Scholar]