Abstract
The detection of DNA damage activates DNA repair pathways and checkpoints to allow time for repair. Ultimately, these responses must be coordinated to ensure that cell cycle progression is halted until repair is completed. Several multiprotein complexes containing members of the structural maintenance of chromosomes family of proteins have been described, including the condensin and cohesin complexes, that are critical for chromosomal organization. Here we show that the Smc5/Smc6 (Smc5/6) complex is required for a coordinated response to DNA damage and normal chromosome integrity. Fission yeast cells lacking functional Smc6 initiate a normal checkpoint response to DNA damage, culminating in the phosphorylation and activation of the Chk1 protein kinase. Despite this, cells enter a lethal mitosis, presumably without completion of DNA repair. Another subunit of the complex, Nse1, is a conserved member of this complex and is also required for this response. We propose that the failure to maintain a checkpoint response stems from the lack of ongoing DNA repair or from defective chromosomal organization, which is the signal to maintain a checkpoint arrest. The Smc5/6 complex is fundamental to genome integrity and may function with the condensin and cohesin complexes in a coordinated manner.
Cell cycle checkpoints are surveillance mechanisms that monitor the order and fidelity of cell cycle events. Among these are the DNA integrity checkpoints, which monitor DNA replication and DNA damage. Ongoing or stalled replication activates a checkpoint that prevents initiation of mitosis, ensuring the interdependence of the S phase and mitosis to maintain ploidy. There is considerable overlap between this checkpoint and that which monitors DNA damage during the S and G2 phases of the cell cycle, at which time there is the added challenge of coordinating cell cycle progression with successful DNA repair (8).
The biology of the G2 DNA damage checkpoint in the fission yeast Schizosaccharomyces pombe has been extensively studied (40). This checkpoint arrests the cell cycle through maintenance of the inhibitory tyrosine-15 phosphorylation of Cdc2, which is achieved through Chk1-dependent signaling to the Cdc25 phosphatase and the Wee1 kinase that regulate Cdc2 (15, 39, 41, 42). The earliest molecular marker of the G2 DNA damage response is Rad3-dependent phosphorylation of its binding partner Rad26, the homolog of human ATR and ATRIP (9, 12). Phosphorylation of Chk1 following DNA damage is mediated by Rad3 and requires several other proteins encoded by the checkpoint rad genes, rad1, hus1, rad9, and rad17, together with the BRCT domain protein, Crb2 (29, 43, 56). These proteins, and the Rad3 and Rad26 homologs, have also been shown to independently localize to the sites of DNA damage in both Saccharomyces cerevisiae and human cells (23, 35, 58). Although Rad3/ATR activation is an early checkpoint response to DNA damage, the actual sensors of DNA lesions are unknown and may differ depending on the specific nature of the lesion (3, 59).
Experiments using fission yeast and a conditional allele of rad3 have shown that Rad3 is required only for initiation of the DNA damage checkpoint, providing evidence for the separation of checkpoint initiation and maintenance (29). This indicates that other proteins in addition to Rad3 must exist to coordinate checkpoint arrest with completion of DNA repair and entry into mitoses.
Chromosomal organization is a dynamic process to facilitate gene expression, DNA repair, sister chromatid cohesion, and chromosome condensation. One family of proteins involved in these processes is the structural maintenance of chromosomes (SMC) protein family (19). SMC proteins are highly conserved, existing as single homodimers in prokaryotes and as heterodimers in eukaryotes in complex with several non-SMC subunits. The SMC proteins have N- and C-terminal globular domains with Walker A and B ATP-binding motifs, respectively. These domains are separated by two coiled-coil domains that are interrupted by a flexible hinge and form intramolecular interactions, bringing the Walker A and B motifs into proximity to form a structure reminiscent of those of the ABC transporters (16, 17, 27, 34).
In eukaryotes, Smc1 and Smc3 are part of the cohesin complex, which is loaded onto sister chromatids during replication and (in human cells) remains on the kinetochore until the metaphase-anaphase transition (18, 26, 53). Smc1 and Smc3 form part of a ring structure that encircles sister chromatids and is stabilized by the Scc1 protein, the cleavage of which allows sisters to separate (16, 52). Mutation of Scc1 in fission yeast (rad21) results in a double-stranded DNA break repair defect, presumably as sister chromatids are required for recombinational repair, and also results in a partial loss of checkpoint arrest by an unknown mechanism (1, 6, 48). In human cells, Smc1 is phosphorylated by ATM in response to DNA damage and interacts with both Nbs1 and BRCA1 (22, 57).
Smc2 and Smc4 form a distinct complex with several non-SMC proteins termed the condensin, which is required for chromosome condensation together with type II topoisomerase (5, 20, 21, 50). Condensin is localized primarily to the cytoplasm during interphase and is actively recruited to chromosomes during mitosis, where it is localized to domains that are discrete from those occupied by topoisomerase II (28, 47). Mutations in a fission yeast non-SMC subunit, Cnd2, also result in defective responses to replication blocks and DNA damage (2).
Both the cohesin and condensin have been extensively studied in a number of systems and are universally required for chromosome integrity (19). A third SMC complex, containing Smc5 and Smc6, exists in all eukaryotes. This complex is less well defined and is referred to here as the Smc5/Smc6 (Smc5/6) complex. This complex was first defined by isolation of the fission and budding yeast Smc6 homologues, Rad18 and Rhc18, respectively, and more recently by characterization of Smc5 and Smc6 in human cells (13, 25, 49, 54). Genes encoding members of this complex are essential, and much of what is known about them comes from analysis of the hypomorphic rad18-X and rad18-74 mutant alleles in fission yeast. rad18-X and rad18-74 are hypersensitive to ionizing radiation and UV-C, do not repair double-strand breaks, and show impaired excision of UV-C lesions, most likely through defects in recombination (25, 54). Both rad18 alleles exhibit chromosome instability accompanied by an accumulation of mitotic defects and are synthetically lethal, with a temperature-sensitive allele of topoisomerase II, deletion of brc1, an allele-specific high-copy-number suppressor of rad18-74, and the DNA repair gene rad60 (37, 54). In fission yeast, moreover, Smc5 and Smc6 have been shown to interact with Rad60, albeit substoichiometrically (7). These genetic data are indicative of a requirement for Rad18 in chromosome organization, which may indirectly affect DNA repair processes.
rad18-74 was isolated in a screen for checkpoint mutants, and, in addition to its common phenotypes with rad18-X, is unable to maintain a checkpoint arrest after DNA damage (54). Unlike rad18-X cells, which remain cell cycle arrested following irradiation, rad18-74 cells show a DNA damage checkpoint delay of essentially wild-type duration. However, these cells reenter the cell cycle with highly aberrant and lethal mitoses, suggesting that rad18-74 mutants are proficient in initiating a checkpoint but are unable to maintain checkpoint arrest. Similarly, spores lacking rad18 are unable to arrest cell cycle progression in response to DNA damage (54). However, the biochemical dissection of checkpoint regulation and the assessment of checkpoint kinetics in these types of experiments are not feasible due to the presence of wild-type spores and the nonsynchronous nature of spore germination. The Smc5/6 heterodimer appears to be in a complex with several additional proteins (13). SMC6 is localized to the nucleus in interphase but is excluded from condensed mitotic chromosomes (49). A non-SMC component of an Smc5/6 complex, Nse1, has been isolated in budding yeast. Nse1 is an essential nuclear protein, and mutations in Nse1 result in sensitivity to DNA damage, though the nature of this sensitivity has not been investigated (14).
We present here evidence that the Smc5/6 complex is required to ensure that cells remain checkpoint arrested, presumably until the completion of DNA repair, for the successful completion of mitosis. We assessed the DNA damage checkpoint of a dominant-negative rad18 allele and of a conditional null mutant. These mutants show cell cycle arrest of wild-type duration and wild-type kinetics for Chk1 phosphorylation and kinase activation. However, these mutants, like rad18-74, reenter the cell cycle and undergo lethal mitoses, indicating that Rad18 is required to maintain a checkpoint arrest subsequent to the Rad3-dependent activation of Chk1. We have also assayed the dependence of this complex on non-SMC components for checkpoint proficiency through the analysis of cells lacking Nse1. When fission yeast spores lacking Nse1 are germinated in the presence of DNA damaging agents, they are unable to undergo normal cell cycle arrest, producing a phenotype similar to cells lacking functional Rad18. We suggest that following the activation of Chk1, the Smc5/6 complex is required to coordinate the duration of the activated checkpoint with completion of DNA repair.
MATERIALS AND METHODS
Fission yeast methods.
All strains used were derivatives of 972 h− and 975 h+. Standard procedures and media were used for propagation and genetic manipulation (36). Methods for transformation, microscopy, and fluorescence-activated cell sorter (FACS) analysis have been described previously (38, 55). For nmt1 promoter induction experiments, exponentially growing cells were washed three times in medium lacking thiamine and then inoculated into defined medium lacking thiamine. For all Rad18 depletion experiments, Rad18 expression in the rad18-so strain was repressed by addition of 10 μg of thiamine/ml to exponentially growing cells. Cell numbers were determined using a Sysmex K-1000 cell counter.
Checkpoint analyses.
In all experiments, cells were grown to mid-logarithmic phase in defined medium. Cells were filtered onto polyvinylidene difluoride membranes (Millipore) and irradiated with 100 J of UV-C/m2 in a Stratalinker apparatus (Stratagene). Cells were subsequently reinoculated into prewarmed medium. Cell cycle progression was assessed by septation indices derived from the average of three counts of 100 cells. This was normalized to results for unirradiated cells (t = 0) for graphical representation. The number of abnormal mitoses was obtained by counting at least three samples of 100 ethanol-fixed cells, which had been stained with 4′,6′-diamidino-2-phenylindole (DAPI) and visualized by fluorescence microscopy.
Construction of rad18 mutants.
Construction of the rad18-dn allele has been previously described (54). To construct the rad18-so mutant, the rad18 cDNA was subcloned into a pREP81 vector carrying the lowest-strength nmt1 promoter (4, 31) and the sup3-5 tRNA suppressor. This plasmid was transformed into the rad18Δ heterozygous diploid (25). The resulting diploid transformants, which contained a single integrated copy of the plasmid, were sporulated, and ade6+ ura4+ haploid spores that were not viable in the presence of thiamine were selected. To assess the depletion of Rad18 protein from this strain, cell extracts were prepared in urea buffer (8 M urea, 100 mM Na2HPO4, 10 mM Tris-HCl, pH 8.0) and analyzed by Western blotting with a polyclonal antibody to Rad18 (1/500), a gift from Alan Lehmann. To assay rad18 mRNA levels, cells were homogenized and TRIzol (Gibco) was extracted according to the manufacturer's instructions. RNA (10 μg) was separated on a 1% agarose gel, transferred to a Zeta-probe (Bio-Rad) membrane, and probed with the rad18 cDNA.
For green fluorescent protein (GFP) localization, a NotI site was introduced into the 5′ region of the rad18+ and rad18-dn cDNAs by site-directed mutagenesis to enable subcloning into pSGP573 (a gift from Susan Forsburg). These plasmids were transformed into wild-type cells for fluorescence microscopy and the rad18-74 mutant to test functionality.
Construction and immunoprecipitation (IP) of endogenously HA-tagged Rad18.
A fragment encoding six consecutive histidines followed by three copies of the hemagglutinin (HA) epitope (39) was introduced in a BglII site that was introduced into the 5′ region of rad18 between codons 2 and 3 by site-directed mutagenesis (24). The construct was transformed into the rad18-so strain, and ura4− colonies were selected. This corresponded to replacement of the rad18::ura4, as confirmed by Southern blotting with a ura4 probe. The rad18-so construct was removed via backcrossing to the wild type.
Untagged or HA-tagged Rad18 cells were transformed with Myc-tagged Nse1 and expressed using the medium-strength nmt1 promoter under repressing conditions or empty vector (pRMH42; a gift from Antony Carr). Co-IP of HA-Rad18 and Myc-tagged Nse1 was carried out under the following buffer conditions: 50 mM Tris (pH 7.5), 80 mM β-glycerophosphate, 1% Triton X-100, 50 mM NaF, 1 mM dithiothreitol (DTT), 0.1 mM VO4, 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 2 μg of pepstatin/ml, and 1 mM phenylmethylsulfonyl fluoride. A total of 5 μg of anti-HA 12CA5 monoclonal antibody (Roche) or 5 μg of the anti-Myc 9E10 monoclonal antibody was added per milligram of protein, and buffer was used to achieve a reaction volume of 1 ml. Immunoprecipitates were incubated at 4°C with rotation ∼2 h prior to addition of 40 μl of protein-A Sepharose (50% slurry in buffer). Immunoprecipitates were incubated for a subsequent 90 min at 4°C with rotation. The Sepharose beads were washed three times with extraction buffer. The beads were boiled in 1× sodium dodecyl sulfate (SDS) sample buffer and separated on an SDS-10% polyacrylamide gel electrophoresis (PAGE) gel. Proteins were transferred to a nitrocellulose membrane, and the top half was probed with 12CA5 (1/200) to detect HA-Rad18. The bottom half was probed with 9E10 (1/200) to detect Myc-Nse1.
Cloning of S. pombe nse1.
Nse1 was cloned from genomic DNA by PCR using the following oligonucleotide primers: 5′-GGAATTCCATATGGAGAAAGAGAGACAAGATGG-3′ (forward) and 5′-CGGGATCCTATAAACAAGTAAAGCGAACACCAG-3′ (reverse). To confirm the presence of the single intron in nse1, an additional reverse primer (5′-CGGGATCCTCAGTTAGTAAGTTTGCATTTGTGA-3′) was used in combination with the aforementioned forward primer. This does not produce a product when amplified from cDNA. These primers were used to amplify full-length nse1 containing NdeI and BamHI sites which were used to subclone nse1 into the pRMH42 and pREP1 vectors for co-IP and overexpression, respectively. The integrity of the nse1 clones was confirmed by sequencing and according to their ability to complement nse1::ura4 haploid spores.
Construction of nse1::ura4.
For construction of an nse1 knockout, PCR with primers 5′-ATGTTTATGTGCGTAAAC-3′ (forward) and 5′-GATTGCACTCAGATTTAA-3′ (reverse) was used to amplify a ∼2.6-kb product which contained the 1.8-kb ura4 fragment flanked on either side by approximately 400 nucleotides of sequence adjacent to the nse1 5′ and 3′ coding regions, respectively. The 2.6-kb PCR fragment was transformed into a diploid strain (ura4-D18/ura4-D18 leu1-32/leu1-32 ade6-M210/ade6-M216 h+/h−), and the resulting transformants were selected for uracil prototrophy as previously described (54). The ura4 gene replacement was confirmed by Southern blotting. Tetrads from 40 azygotic asci were dissected from an appropriate heterozygous diploid. In all cases, only two viable ura4− spores were observed.
Spore germination.
Spores were made from a wild-type strain with the genotype ura4-D18/ura4-D18 leu1-32/leu1-32 ade6-M210/ade6-M216 h+/h−, a previously described rad18Δ strain with the genotype rad18+/rad18::ura4 ura4-D18/ura4-D18 leu1-32/leu1-32 ade6-704/ade6-704 h+/h90, and the nse1Δ strain (25, 54). Approximately 6 × 106 spores were inoculated into defined medium lacking uracil (or medium including uracil for wild-type cultures) and incubated at 30°C until the appearance of polar growth (10 to 11 h). Cultures were then incubated for a further 8 h with 0.005% methyl methanesulfonate (MMS), with 0.31 μg of 4-nitroquinoline-N-oxide (4-NQO)/ml, or with no treatment. Cells were fixed in ethanol and DAPI stained for counting mitoses and cell images.
Chk1 phosphorylation and kinase assays.
For Chk1 IP, cells were lysed in IP buffer (10 mM NaPO4 [pH 7.0], 0.15 M NaCl, 1% NP-40, 10 mM EDTA, 50 mM NaF, 2 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 3 mg of tosylsulfonyl phenylalanyl chloromethyl ketone/ml, 10 mg of E64/ml, 100 mM benzamidine, 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 2 μg of pepstatin/ml) and 5 μg of anti-HA 12CA5 monoclonal antibody (Roche) was added per 1 mg of protein; IP buffer was used to achieve a reaction volume of 1 ml. Immunoprecipitates were incubated at 4°C on a rotator for 90 min prior to the addition of 20 μl of protein-A Sepharose (50% slurry in IP buffer). Immunoprecipitates were incubated for a subsequent 60 min at 4°C. The beads were washed five times with IP buffer and three times with 1× kinase buffer (50 mM Tris [pH 7.0], 1 mM DTT, 5 mM MgCl2, 0.4 mM MnCl2, 25% glycerol [Sigma G5516], 0.1% Triton X-100, 100 μM ATP). The washed Sepharose was suspended in 24 μl of kinase reaction buffer (2× kinase buffer, 12 μg of peptide [RIARAASMAALARK], [γ-32P]ATP [3,000 Ci/mmol]), incubated at 30°C for 15 min in an Eppendorf thermomixer, and immediately placed on ice. The Sepharose was pelleted, and 20 μl of the supernatant was spotted onto P81 phosphocellulose cation exchange paper (Whatman). The P81 paper was washed at least five times in 0.5% orthophosphoric acid until counts were no longer detectable in the washes, rinsed with ethanol, and air dried. The dried P81 papers were mixed with Ready Safe scintillation mix (Beckman) and counted in a Beckman liquid scintillation counter. For all Chk1 kinase assays, the activity of corresponding untagged strains was subtracted to remove background activity. These extracts were analyzed for Chk1 mobility shift by SDS-PAGE and Western blotting.
Cloning of human Nse1.
Human Nse1 was obtained by PCR from a human fetal cDNA library (Clontech) by using the following primers: 5′-GATCGGATCCATGACTGATGTCCACCGGCGC-3′ and reverse primer 5′-GATCGAATTCATGGCTAATGCTGCCTGGACC-3′. The resulting PCR product was digested with BamHI and EcoRI and subcloned into pBluescript KS+ (Stratagene). The M13 forward and reverse primers were used to confirm the sequence of human Nse1 with respect to information from the GenBank database (accession no. BC018938). BamHI- and EcoRI-digested Nse1 was then subcloned into the expression vector pKH3, producing an N-terminal, triple-HA-tagged Nse1 protein (30).
Generation of anti-SMC6 antibodies.
A portion of human SMC6 corresponding to nucleotides 2443 to 2832 (amino acids 725 to 847) was amplified by PCR and subcloned into pGEX-4T vector (Pharmacia). Large-scale production of glutathione S-transferase fusion proteins was performed, and the results were purified according to the manufacturer's instructions. Two rabbits were inoculated with 1 mg each of the purified glutathione S-transferase fusion proteins. Antibodies against SMC6 were affinity purified according to standard protocols as described previously (38, 39).
Cell lines and transfections.
Human HEK293T cells were maintained as a monolayer with Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal calf serum (CSL) and 100 U of penicillin-streptomycin/ml in 5% CO2. On the day before transfection, cells were plated at 106 cells per 10-cm-diameter plate. Cells were transfected (using Metafectene [Scientifix] according to the manufacturer's instructions) with 5 μg of indicated DNA. Cells were harvested 48 h posttransfection, washed with ice-cold phosphate-buffered saline, and snap frozen in liquid nitrogen.
SMC6/Nse1 cell extracts, IP, and Western blotting.
Cells were extracted in a lysis buffer containing 10 mM Tris-HCl (pH 8.0), 50 mM KCl, 5 mM MgCl2, and 0.5% NP-40 supplemented with protease inhibitors. To more effectively release endogenous SMC6, chromatin was sheared by passing the extracts through a 1-cm3 syringe fitted with a 27-gauge needle; extracts were clarified by centrifugation. 12CA5 antibody (Roche) (1 μg) was added to 500 μg of protein extracts and incubated with rotation at 4°C for 1 h. A total of 5 μl of a 50% solution of protein A Sepharose (Amersham) was added, and the mixture was incubated with rotation for another hour. Beads were washed three times with lysis buffer supplemented with 250 mM NaCl and three times with lysis buffer. The beads were boiled in the presence of Laemmli buffer and separated on an SDS-8% PAGE gel. Proteins were transferred to a nitrocellulose membrane (Schleicher and Schuell). Following transfer, the membrane was cut lengthwise just above the 85-kDa molecular-mass marker. The top half was probed with anti-SMC6 antibody at a 1:500 dilution followed by donkey anti-rabbit horseradish peroxidase-conjugated secondary antibody (Amersham) at a 1:2,500 dilution. The bottom half of the membrane was probed with anti-HA (12CA5) antibody at a 1:1,000 dilution and sheep anti-mouse horseradish peroxidase-conjugated secondary antibody (Amersham) at a 1:5,000 dilution. Proteins were visualized (using BioMax film [Kodak]) by ECL (Amersham) according to the manufacturer's instructions.
RESULTS
Overexpression of a dominant-negative mutant of rad18.
To date, the genetic analysis of the Smc5/6 complex has largely relied on rad18-X and rad18-74, two hypomorphic alleles in fission yeast, with the former being checkpoint proficient and the later being partially defective. Spore germination experiment results suggested that the absence of Rad18 signified the presence of a checkpoint defect, though these experiments were not amenable to biochemical or kinetic analyses (25, 54). We therefore sought to assess the checkpoint kinetics of additional alleles of rad18 to assess whether the checkpoint defects of rad18-74 are indicative of a function for the Smc5/6 complex or specifically represent a phenotype of this allele. We have previously reported the identification of another rad18 allele, rad18-dn. rad18-dn is a Walker A domain mutant that (when overexpressed from the wild-type nmt1 promoter) results in an inability to form colonies (54). With the aim of analyzing the checkpoint kinetics of rad18-dn, we wanted to further characterize this mutant allele.
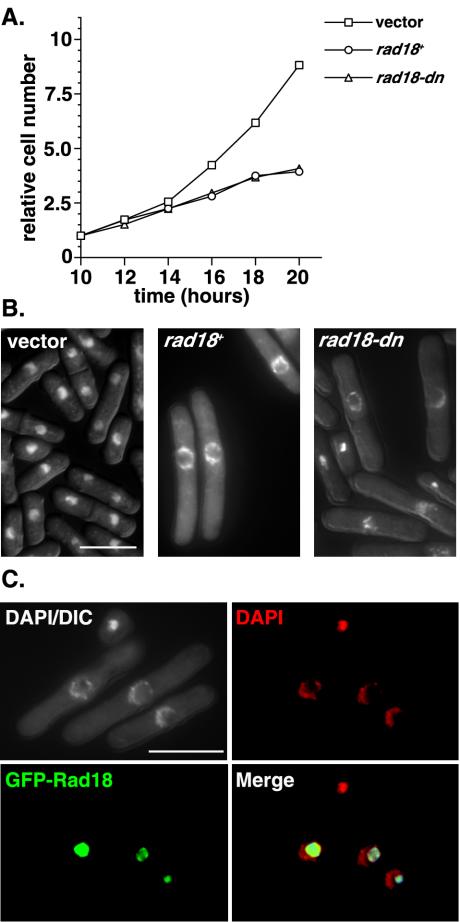
Wild-type cells were transformed with rad18-dn or wild-type rad18 (rad18+) expressed from the wild-type nmt1 promoter. Prolonged overexpression of rad18-dn in liquid culture results in cell cycle arrest (Fig. 1A). Unexpectedly, the same phenotype was also observed in cells overexpressing rad18+, despite the fact that these cells can form colonies on plates. Microscopic analysis of these cells showed significant cell elongation, suggesting a cell cycle arrest. In addition, a striking hollow-sphere-like nuclear staining pattern was observed, indicating gross structural changes to the nucleus (Fig. 1B). This nuclear phenotype was apparent in 35 to 40% of rad18-dn and rad18+ cells after 20 h of growth in the absence of thiamine, corresponding to ∼8 h of promoter derepression. A similar nuclear morphology was seen in cells expressing a GFP-H2A histone to allow visualization of chromatin without the use of DAPI (data not shown). FACS analysis of these cells was also consistent with G2/M cell cycle arrest, with low levels of >2C and <1C DNA. By microscopy, this was accounted for by ∼5% of cells with a cut phenotype, in which a mitotic defect results in bisection of the nucleus by the medial septum at cytokinesis.
FIG. 1.
Overexpression of wild-type or dominant negative Rad18 leads to cell cycle arrest and a hollow-sphere-like nuclear phenotype. (A) Wild-type cells expressing vector, wild-type Rad18 (rad18+), or a dominant-negative form of Rad18 (rad18-dn) from the full-strength nmt1 promoter were grown in the absence of thiamine for 10 h to induce promoter derepression. Cell number was measured every 2 h for a subsequent 10 h. (B) DAPI-stained ethanol-fixed cells after 20 h of growth in the absence of thiamine were visualized by fluorescence microscopy. Note the cell elongation and large hollow-sphere-shaped nuclear staining visible after prolonged overexpression of either rad18+ or rad18-dn. (C) To determine Rad18 localization, GFP-tagged Rad18 was expressed from the wild-type nmt1 promoter. After approximately 8 h of promoter derepression (∼20 h of growth in medium lacking thiamine), the majority of GFP-Rad18 (green) does not colocalize with DNA (DAPI; red) in methanol-acetone-fixed cells. The same localization pattern was also observed for the GFP-tagged dominant-negative Rad18 (unpublished data). DIC, differential interference contrast; Merge, merged image of DAPI and GFP-Rad18 results.
Rad18 has previously been shown to localize to the nucleus, colocalizing with DAPI in interphase cells (54). To address whether Rad18 still localized with the DNA in these cells, GFP-Rad18 was expressed from the nmt1 promoter under repressing conditions (i.e., in the presence of thiamine). This plasmid was able to complement the MMS sensitivity of rad18-74 and was localized to the nucleus in the presence of thiamine in wild-type cells, indicting that the GFP-Rad18 fusion is functional in the presence of thiamine. After 20 h of growth in the absence of thiamine, the majority of GFP-Rad18 did not colocalize with DNA but, rather, localized to the inner nucleoplasm that failed to stain with DAPI (Fig. 1C). The same localization pattern was observed after 20 h of GFP-rad18-dn overexpression. These data suggest that Rad18 overexpression is lethal, resulting in gross structural changes to the nucleus, and that these phenotypes are independent of ATPase activity.
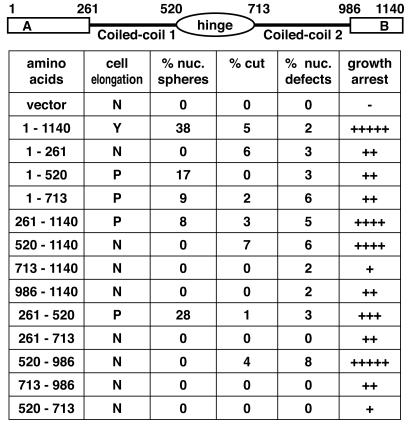
We performed a structure-function analysis to determine which domains of Rad18 were required for the overexpression phenotypes (Fig. 2). The nuclear-sphere phenotype was separable from the growth arrest effects. Of the constructs in which expressed proteins could be detected by Western blotting, the first coiled-coil domain appeared to be both necessary and sufficient for the nuclear spheres. An exception to this was a construct that included both the coiled-coil domain and the central hinge. The specificity of this effect suggests that the spheres are a result of a specific interaction, possibly through a non-SMC component of this complex. Conversely, the overexpression of multiple domains resulted in growth arrest, primarily in interphase with some defective mitoses, suggesting a generalized dominant-negative effect on the Smc5/6 complex.
FIG. 2.
The growth arrest and nuclear phenotypes of Rad18 overexpression are separable. To determine which domain of Rad18 produced the distinct hollow-sphere nuclear phenotype, various structural domains of Rad18 were overexpressed from the nmt1 promoter for 20 h in the absence of thiamine (corresponding to approximately 8 h of promoter derepression). Overexpression of full-length Rad18 produces a cell cycle delay, as evidenced by cell elongation; however, cell elongation was either not observed (N) or reduced (P) in the other constructs. Growth arrest was observed with overexpression of several different domains (−, no arrest; + to +++++, 20, 40, 60, 80, or 100% arrest compared to overexpression of full-length Rad18). The nuclear phenotypes are represented as the averages of at least three counts of 100 ethanol-fixed cells that had been stained with DAPI. Note that overexpression of amino acids 261 to 520 (the first coiled-coil domain) alone was sufficient to produce the nuclear-sphere phenotype.
rad18-dn cells fail to maintain a DNA damage checkpoint.
To investigate whether the checkpoint maintenance defects of rad18-74 were characteristics of impaired Smc5/6 complex function, we wanted to address whether the rad18-dn mutant displayed a similar checkpoint response. Three means of checkpoint analysis were undertaken for these and the subsequent experiments described below. We attempted to assess checkpoint kinetics synchronously, first by centrifugal elutriation. However, although we were able to obtain a homogeneous population of cells (as determined on the basis of cell size), the rad18-dn strain (and the rad18-so strain; see below) exhibited heterogeneous cell sizes; thus, the selected population was not synchronous with respect to cell cycle phase. cdc25-22 block-and-release synchronization was undertaken but was found to be an inappropriate method. This protocol was found to potentiate a strong checkpoint defect with all rad18 mutant alleles (except rad18-X) which was not evident by other assay methods (see below). Moreover, this checkpoint defect was also observed in wild-type cells after a longer incubation (4.5 h) at a nonpermissive temperature. Therefore, we assessed the DNA damage checkpoint kinetics in asynchronous cultures.
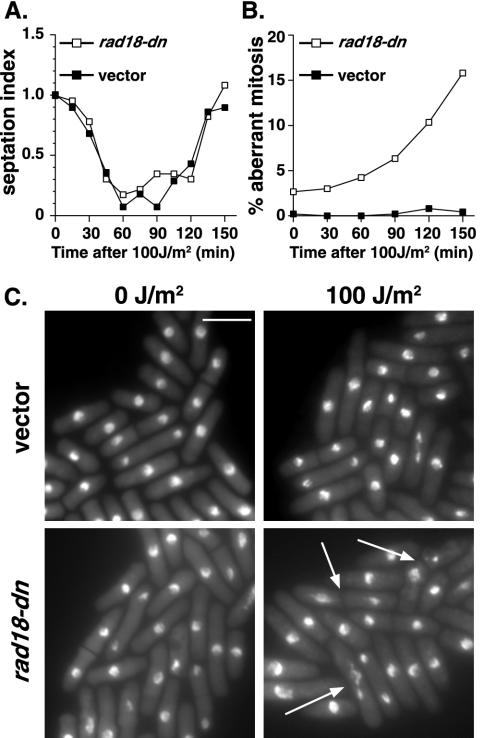
Asynchronous checkpoint analysis of rad18-dn was performed after 14 h of promoter derepression. The rate of cell cycle progression following irradiation was assessed by measuring the septation index, a discrete marker of cytokinesis, which occurs ∼20 min following mitosis. At this time, rad18-dn cells are growing normally and have yet to manifest defective nuclear phenotypes. Cells expressing rad18-dn and vector controls were irradiated with 100 J of UV-C/m2. Vector control cells showed a wild-type cell cycle delay in response to irradiation, as evidenced by a transient loss of septation, with minimal effects on cell viability or mitotic fidelity (Fig. 3A and B). rad18-dn cells also showed a wild-type cell cycle delay in response to DNA damage but on reentry into the cell cycle showed highly aberrant, lethal mitoses. As with the rad18-74 allele, the predominant mitotic defects were those of stretched DNA along the divisional plane and of nuclei bisected with a septum (Fig. 3C).
FIG. 3.
A dominant-negative allele of rad18 is unable to maintain a DNA damage checkpoint arrest. Asynchronously growing cells expressing a vector control or rad18-dn from the nmt1 promoter were grown for 14 h in the absence of thiamine and irradiated with 100 J of UV-C/m2. (A) The kinetics of cell cycle arrest was measured by septation index analysis every 15 min for 2.5 h. (B) Abnormal mitoses were scored from ethanol-fixed cells stained with DAPI at the indicated times. Following UV-C irradiation, rad18-dn cells exhibited increased numbers of aberrant mitoses compared to the vector control cells. The data represent the means of three independent experiments. (C) DAPI-stained cells of the indicated strains exposed to 0 J of UV-C/m2 or at 150 min postexposure to 100 J of UV-C/m2. Arrows indicate cells undergoing abnormal mitoses. Bar, 10 μm.
The checkpoint kinetics of rad18-dn are consistent with previous analysis of rad18-74 and support the idea of a checkpoint defect in which the time period during which cell cycle arrest is maintained is not sufficient to repair DNA or organize chromosomes appropriately for mitosis and hence is not sufficient to ensure mitotic fidelity. That is, as the physiological role of this checkpoint is to prevent mitosis in the presence of DNA damage and not merely to control the duration of a dose-dependent cell cycle delay, these data are consistent with a defective cell cycle response to DNA damage.
Conditional depletion of Rad18.
Confirmation of a requirement for the Smc5/6 complex in checkpoint maintenance ideally required conditional inactivation of the complex in cycling cells. We initially attempted to construct temperature-sensitive rad18 alleles; however, no conditional alleles were recovered from over 100,000 mutagenized plasmids, and so we designed an alternative approach to conditionally deplete Rad18 from cells. Strains were constructed in which the only copy of rad18 present was expressed from a single integrated copy of rad18 expressed from the weakest nmt1 promoter. These cells were inviable when nmt1w::rad18 expression was repressed by the presence of thiamine. We refer to this shutoff strain as the rad18-so strain.
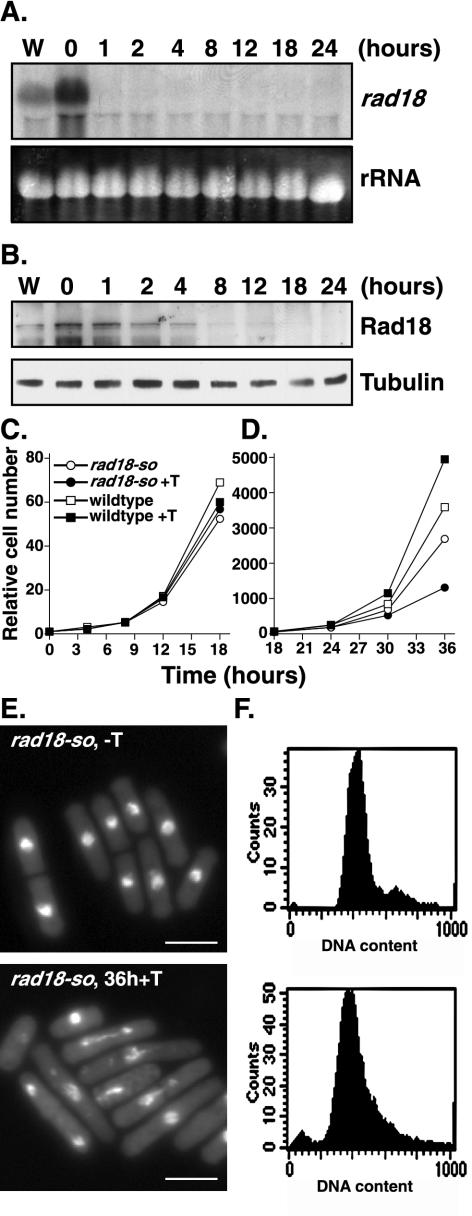
Northern blot analysis of rad18-so showed the rad18 mRNA fell to below detectable levels in ∼1 h (Fig. 4A). Western blot analysis showed that the protein persisted to ∼12 h post-addition of thiamine and was not detectable by 12 to 18 h (Fig. 4B). The rad18-so cells grew at a rate indistinguishable from that of controls for the first 24 h post-addition of thiamine (Fig. 4C) and then slowed over the next 12 h (Fig. 4D), with cells eventually arresting growth and hence unable to form colonies. Microscopic analysis of rad18-so cells 36 h post-addition of thiamine showed a phenotype similar to that of rad18Δ cells derived from sporulated heterozygous diploids and to those of several of the nonnuclear spheres forming overexpression alleles (Fig. 2). Cells showed variability through significant elongation, suggesting an interphase cell cycle delay, and many cells arrested in aberrant mitotic states, with incompletely segregated chromosomes stretched along the division plane and frequently accompanied by a medial septum (Fig. 4E). FACS analysis of DNA content confirmed the cells were arrested in the G2 and/or M phase (Fig. 4F). Therefore, these data confirm that rad18-so is conditionally null for Rad18 function.
FIG. 4.
Characterization of a conditional allele of rad18. A depletion allele of rad18 (designated rad18-so) was constructed by integrating a rad18 cDNA expressed from the weakest nmt promoter by sup3-5 integration into a rad18::ura4 background. (A) Northern blot of RNA extracted from wild-type (W) or rad18-so cells in the absence of thiamine (0 h) or after the indicated time in the presence of thiamine (1 to24 h) and probed with rad18. The results showed undetectable levels of rad18 mRNA after an hour of promoter repression (1 h). (B) Western blot analysis of the indicated strains (assayed as described for panel A) probed with anti-Rad18 antibodies. Note that the Rad18 protein remains detectable for several cell cycles after promoter repression (+T). (C and D) Relative cell numbers of wild-type and rad18-so cells grown in the presence and absence of thiamine over the indicated time period. Note that over the first 18 h of promoter repression the rad18-so strain exhibits relatively normal growth rates (C), while in the subsequent 18 h its growth rate is significantly reduced (D). (E) Ethanol-fixed rad18-so cells were stained with DAPI after 0 or 36 h in thiamine. After 36 h in thiamine, rad18-so cells displayed various nuclear abnormalities, including stretched and fragmented DNA and cells in which the nuclear material has been bisected by the septum, indicating mitotic failure. (F) FACS analysis of rad18-so cells at 0 or 36 h in thiamine shows the cells were arrested in G2 phase and mitosis.
rad18-so cells fail to maintain a DNA damage checkpoint.
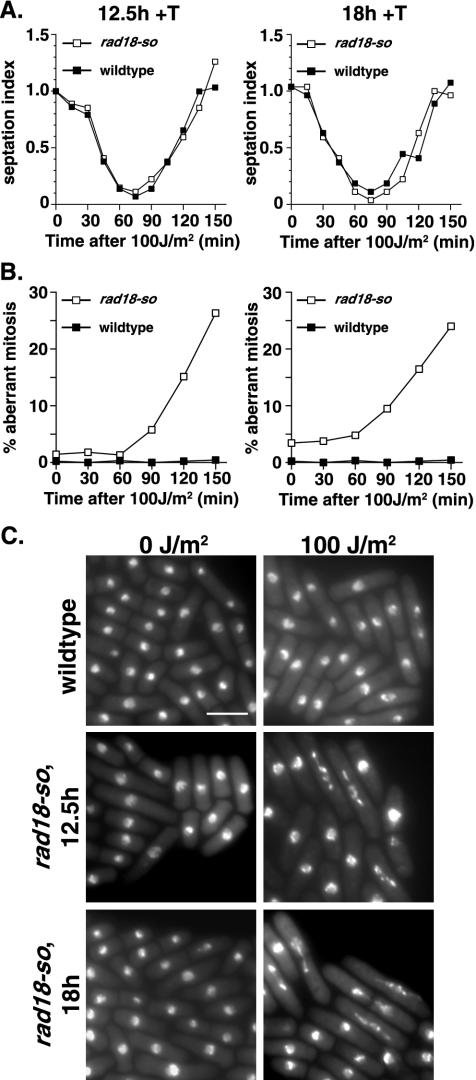
As the rad18-so strain enabled us to generate a pure population of cells lacking detectable Rad18, we next asked the effect of this on DNA damage checkpoint kinetics. In all assays, the wild-type cells contained an integrated empty vector to control for any effects of the sup3-5 integration marker. Control or rad18-so cells were grown in the presence of thiamine to repress Rad18 expression for either 12.5 or 18 h before irradiation. The duration of promoter repression was chosen to target a point at which rad18-so cells were growing exponentially, but little or no Rad18 was detectable by Western blotting. Moreover, significant growth changes or nuclear abnormalities were not evident for several subsequent cell cycles (Fig. 4C and D). Under conditions of exposure to 100 J of UV-C/m2, control cells produce a wild-type checkpoint delay (as evidenced by the transient loss of septa) with minimal effects on cell survival. Similarly, the rad18-so mutant displayed a delay in cell cycle progression of the same duration, indicating a checkpoint arrest in response to UV-C (Fig. 5A). However, rad18-so cells differed from controls in that the subsequent mitoses were highly aberrant (Fig. 5B). Microscopic analysis of these cells showed DNA stretched along the divisional plane, which in many cases was bisected by a septum, suggesting that these cells have progressed into mitosis with incompletely repaired DNA or disorganized chromosomes (Fig. 5C). The cell cycle delay and phenotypic abnormalities in rad18-so did not differ significantly between 12.5 and 18 h of Rad18 depletion. In the absence of thiamine, the rad18-so allele is checkpoint proficient in response to DNA damage (data not shown) despite approximately twofold overexpression.
FIG. 5.
rad18-so is unable to maintain an arrest in response to DNA damage. To deplete Rad18, thiamine was added to wild-type cells containing vector only or to nmtW-rad18-so cells. Mid-logarithmic cells were irradiated with 100 J of UV-C/m2 after 12.5 or 18 h of growth in thiamine (+T), which is prior to the appearance of cell cycle delay. (A) The kinetics of cell cycle arrest was measured using a septation index assay. (B) Abnormal mitoses were scored from DAPI-stained ethanol-fixed cells that showed high rates of abnormal mitoses in the rad18-so cells compared to those seen with control cells. The data represent the means of three independent experiments. (C) Ethanol-fixed cells were DAPI stained to assess the percentage of abnormal mitoses by fluorescence microscopy. The indicated strains were photographed in the absence of irradiation (0 J/m2) or at 150 min postir-radiation with 100 J of UV-C/m2 (100 J/m2). Note the predominance of stretched nuclear material and bisected cells, which comprise more than 80% of the total mitoses (∼25% of total cells) in rad18-so cells at 150 min post-UV irradiation. Bar, 10 μm.
These data, along with those determined for the rad18-74 and rad18-dn mutants, show a consistent checkpoint response marked by an initial cell cycle arrest persisting for approximately the same duration as that seen with controls, though followed by highly aberrant, lethal mitoses. These data support the notion that in the absence of fully functional Rad18, cells are unable to maintain a checkpoint arrest and enter mitosis either with unrepaired or partially repaired DNA or with inappropriately organized chromosomes.
rad18 checkpoint mutants show proficient DNA damage checkpoint initiation.
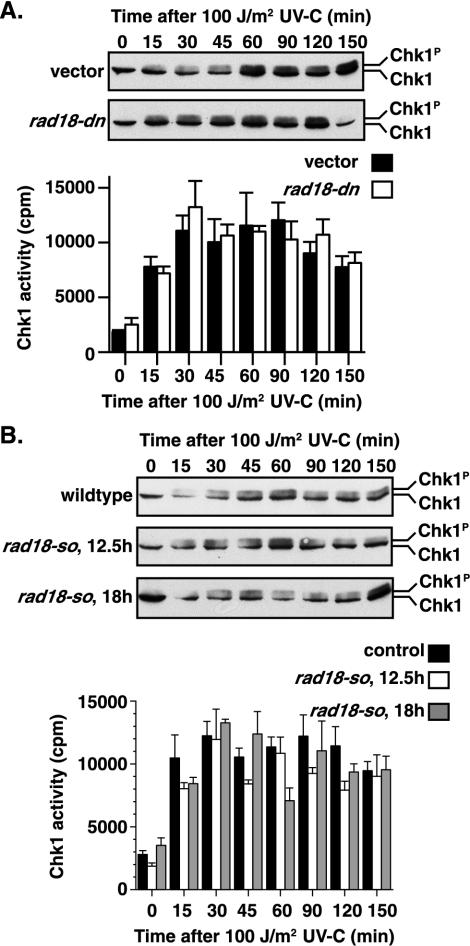
Initiation of the G2 DNA damage-signaling cascade culminates in activation of the Chk1 protein kinase. In response to DNA damage, Chk1 is phosphorylated, resulting in a slower-migrating form of Chk1 on SDS-PAGE, which is commonly used as a surrogate marker of checkpoint activation (56). The rad18-74 and rad18-X hypomorphic alleles show wild-type levels of Chk1 phosphorylation in response to DNA damage, suggesting that they are proficient in Rad3-mediated checkpoint initiation (54). To date, however, markers of checkpoint induction have not been analyzed in cells lacking Rad18. We therefore assayed both Chk1 phosphorylation and kinase activity in rad18-dn and rad18-so alleles during the checkpoint delay.
We assayed the ability of the rad18-dn mutant to produce damage-induced Chk1 phosphorylation. Following exposure to 100 J of UV-C/m2, rad18-dn showed a Chk1 mobility shift similar to vector controls (Fig. 6A, top panels). We also assayed Chk1 phosphorylation in rad18-so after 12.5 or 18 h of promoter repression to deplete Rad18 in response to 100 J of UV-C/m2. rad18-so cells, in similarity to control cells, showed a Chk1 mobility shift following DNA damage (Fig. 6B, top panels). Therefore, these data support the idea of proficient Chk1 phosphorylation in response to DNA damage in all rad18 alleles.
FIG. 6.
Cells lacking functional Rad18 are able to initiate a DNA damage checkpoint arrest. HA-Chk1 (Chk1) is phosphorylated (Chk1P) in response to irradiation in rad18-dn and rad18-so cells in a manner similar to that seen with the controls (Chk1). Western blot analysis with the 12CA5 antibody was used to detect the forms of HA-Chk1, including a slower-migrating form that is phosphorylated in response to DNA damage. Chk1 activity is also induced in response to DNA damage. Chk1-HA was immunoprecipitated from vector or rad18-dn cell extracts and assayed for protein kinase activity (A [bottom panel]). Chk1-HA increased in response to DNA damage in rad18-so cells in a manner similar to wild-type cells (B [bottom panel]). The data shown represent the means of three independent experiments, with error bars showing the standard errors. In all cases, time zero corresponds to unirradiated cells. Wild-type cells grown in thiamine for 12.5 or 18 h did not differ significantly in their responses to UV-C treatment.
We next assayed Chk1 activity in rad18-dn, rad18-so, and control strains following irradiation. HA-tagged Chk1 was immunoprecipitated and assayed for Chk1 kinase activity following exposure to 100 J of UV-C/m2. In several independent experiments, Chk1 kinase activity was consistently induced following DNA damage in both rad18-so and rad18-dn strains in a manner similar to that employed with control cells (Fig. 6). It should be noted that in asynchronous time courses, Chk1 phosphorylation and activity persist beyond the point of mitotic entry. A transient drop in Chk1 activity, lasting ∼5 to 10 min, is only assayable in highly synchronized cultures as cells trasverse mitosis, whereupon the activity returns in the next cell cycle by an unknown mechanism (C. Latif, N. R. den Elzen, and M. J. O'Connell, submitted for publication). Such synchrony is unobtainable in the rad18-so and rad18-dn strains. Nevertheless, these data show that following DNA damage, the rad18-so and rad18-dn mutants exhibit wild-type Chk1 activation in asynchronous cultures. This result, together with those of previous analyses of rad18-74, strongly supports the idea of proficient DNA damage checkpoint initiation in cells lacking functional Rad18 and suggests that Rad18 is required for the maintenance of a checkpoint arrest following DNA damage.
nse1, a non-SMC component of the S. pombe Smc5/6 complex.
Are the checkpoint maintenance defects of the rad18 mutants characteristic of Rad18 per se, or do they reflect a function of the Smc5/6 complex? Rad18 has been reported to exist in complex with several other proteins, including its fission yeast SMC partner, Spr18 (Smc5) (13). On the basis of data from investigations of the cohesin and condensin complexes, it has been inferred that Rad18 acts in complex with several other non-SMC proteins. To date, the only reported non-SMC component of the Smc5/6 complex is budding yeast Nse1 (14). We sought to isolate the fission yeast homologue of Nse1.
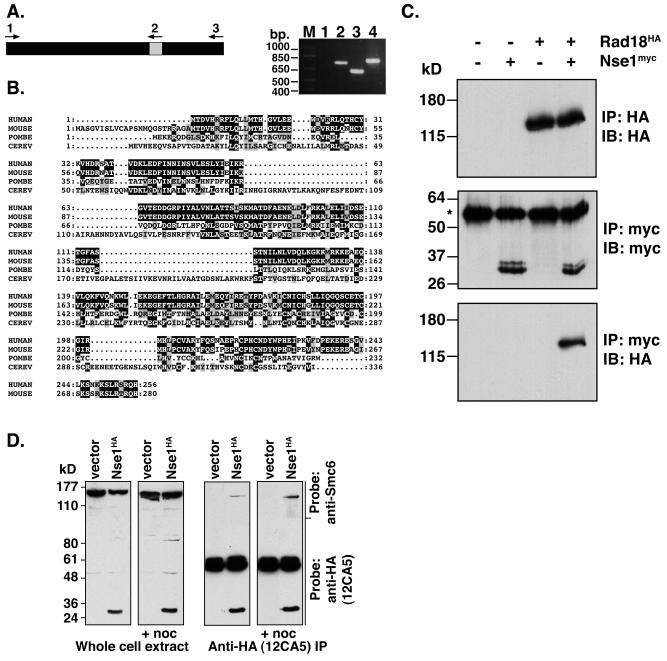
nse1 was cloned by PCR on the basis of its sequence homology to budding yeast Nse1. The nse1 gene contains a single intron, which was confirmed by PCR from both cDNA and genomic templates (Fig. 7A). This was necessary because the nse1 sequence differs from that listed in the S. pombe genome database, for which the 3′ end of the gene was reported to finish prematurely. nse1 is conserved across species, and the mouse, human, and fission yeast homologs contain several regions with high levels of homology (Fig. 7B). To assess whether fission yeast Nse1 interacted with Rad18, HA-tagged rad18 cells expressed using the endogenous rad18 promoter were transformed with Myc-tagged Nse1 expressed using the medium-strength nmt1 promoter in the presence of thiamine (repressing conditions). HA-Rad18 was readily detectible in extracts immunoprecipitated for Myc-Nse1 and assayed by Western blotting (Fig. 7C), since it immunoprecipitated with >50% efficiency compared to the results seen with HA-Rad18 immunoprecipitates. However, Myc-Nse1 was not detectable in extracts immunoprecipitated for HA-Rad18. This is likely to have been due to masking of the Myc tag in HA-Rad18 immunoprecipitates or the displacement of Nse1 from Rad18 following the binding of the 12CA5 (anti-HA) antibody.
FIG. 7.
Nse1 is an essential non-SMC component of the Smc5/6 complex. (A) The nse1 gene contains one intron (indicated as a grey-shaded panel [left panel]). This was confirmed by comparing the results of PCR analysis using a cDNA template and primers 1 and 2 (lane 1) or 1 and 3 (lane 2) to the results obtained with the same primer combinations and a genomic DNA template (lanes 3 and 4) (right panel). (B) Alignment of human, mouse, and yeast Nse1 proteins. Nse1 is conserved across species, with the human, mouse, and S. pombe Nse1 proteins showing several regions of high homology, while S. cerevisiae (CEREV) Nse1 contains several inserts that are not homologous with its eukaryotic counterparts. (C) S. pombe cell extracts were prepared from wild-type cells or from cells in which Rad18 was expressed from the endogenous locus as an N-terminal triple-HA-tagged protein. Cells carrying either a vector or pREP42-Myc-Nse1 were assayed by IP and probed with the indicated antibodies. HA-Rad18 was detectable in extracts immunoprecipitated with the anti-Myc antibody, 9E10 (lane 4, bottom panel). IB, immunoblot. (D) Human Smc6 coimmunoprecipitates with Nse1. HEK293T cells were transfected with either empty vector or HA-tagged human Nse1. Where indicated, cells were treated with nocodazole to arrest cells in mitosis, a point at which Smc6 is no longer associated with chromatin and is more readily extracted. Cell extracts were transferred to nitrocellulose, the top portion of the filter was assayed (using anti-Smc6 polyclonal antibodies) for Smc6 expression in whole-cell extracts, and the lower portion was probed with anti-HA (12CA5) monoclonal antibodies to detect Nse1-HA (left panels). Anti-HA immunoprecipitates were similarly analyzed by Western blotting (right panels).
We extended this analysis to human Nse1. Either empty vector or a HA-tagged Nse1 construct was transfected into HEK293T cells. Endogenous Smc6 was detectable using an anti-Smc6 polyclonal antibody in extracts immunoprecipitated for HA-Nse1 by Western blot analysis (Fig. 7D). Cells were also treated with nocodazole to arrest cells in mitosis prior to harvesting, as Smc6 has been reported to dissociate from chromosomes in mitosis. These data confirm that both fission yeast and human Nse1 interact with Rad18/Smc6 and that Nse1 is one of the non-SMC components of the Smc5/6 complex.
Cells lacking nse1 do not show normal cell cycle arrest in response to DNA damage.
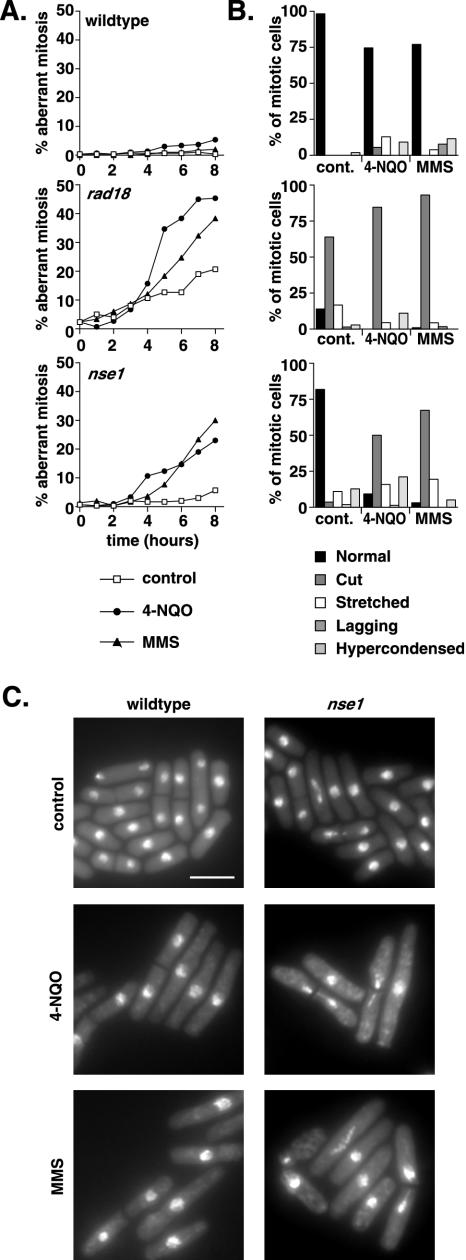
To address whether cells lacking nse1 fail to maintain a checkpoint arrest in the presence of DNA damage, we constructed an nse1Δ strain. Nse1 is essential in fission yeast. When heterozygous diploid cells were sporulated to allow dissection of haploid spores, only two viable spores, corresponding to the nse1+cells, were produced. Analysis of the inviable nse1Δ spores shows that these cells divide approximately four to five times prior to arrest.
We used spore germination in the presence of DNA-damaging agents to assess the ability of cells lacking Nse1 to arrest in response to DNA damage. Analogous spore germination experiments have been previously reported for rad18Δ. After germination, the alkylating agent MMS or the UV-mimetic 4-NQO was added to cultures and monitored for a subsequent 8 h. As assessed by microscopy, wild-type spores exhibited cell cycle arrest and minimal mitotic abnormalities in the presence of these drugs. Conversely, nse1Δ spores accumulated mitotic abnormalities in the presence of DNA damage (Fig. 8A). The most common abnormal phenotype, which corresponded to approximately 50% of mitotic figures after 8 h in 4-NQO or 70% of mitotic figures after 8 h in MMS, was one in which the nuclear material was bisected by a septum. The appearance of nuclear abnormalities in both the presence and absence of DNA damage was delayed in nse1Δ cells compared to the results seen for rad18Δ cells. This is consistent with the observed viability of nse1Δ spores for several cell cycles longer than rad18Δ spores. nse1Δ cells continued to accumulate mitotic abnormalities after 8 h in the presence or absence of DNA damage and, moreover, showed aberrant mitoses at frequencies similar to those for rad18Δ cells when the drugs were added several hours later, presumably when Nse1 was more completely depleted (unpublished data). These data show that (like cells deficient in rad18) cells lacking Nse1 fail to arrest normally in response to DNA damage, suggesting a function for the Smc5/6 complex, rather than for Rad18 alone, in the DNA damage checkpoint.
FIG.8.
The Smc5/6 complex is essential for cell cycle arrest following DNA damage. (A) To assess the kinetics of aberrant mitosis in nse1Δ cells, spores from an nse1+/nse1::ura4 heterozygous diploid were germinated until the appearance of polar growth (10 h) at 30°C in medium lacking uracil to select for nse1::ura4. rad18::ura4 and rad18+ nse1+ spores were used as controls. DNA-damaging agents (MMS and 4-NQO) were then added, or cultures were left untreated (control); the results were monitored for a further 8 h. (B) Samples were taken, fixed, and stained with DAPI to assess the proportion and type of abnormal mitoses. nse1::ura4 cells manifested abnormalities 1 to 2 cell cycles later than rad18::ura4 cells. cont., control. (C) Examples of untreated (control) and 4-NQO- and MMS-treated nse1::ura4 and nse1+ cells after 8 h.
DISCUSSION
To successfully recover from DNA damage, cells mount a coordinated response in which the damage is repaired by lesion-specific enzymes; checkpoints ensure a halt to cell cycle progression to allow time for repair to be completed. We have an advanced understanding of both repair mechanisms and the processes by which a checkpoint delay is initiated (8, 40). Relatively little is known about how these responses are coordinated and how a checkpoint arrest is relieved on completion of repair.
We have shown that the Smc5/6 complex is required to maintain cell cycle arrest following DNA damage. Both dominant-negative and depletion mutants of rad18 fail to maintain a checkpoint arrest in response to DNA damage. Despite these mutants exhibiting proficient Rad3-dependent Chk1 activation, after an initial delay they reenter the cell cycle, resulting in highly aberrant mitoses. This is consistent with results obtained with the previously characterized hypomorphic rad18-74 mutant, confirming that (in the absence of fully functional Rad18) cells are unable to maintain a Chk1-imposed checkpoint arrest and that such cells enter mitosis, presumably with unrepaired DNA. It was important to assess this with additional rad18 alleles, as rad18-X, the only other allele available prior to this study, behaves like most other DNA repair mutants in that irradiation induces a sustained and terminal cell cycle arrest (25, 54). We have isolated human and fission yeast homologs of Nse1, a non-SMC component of the Smc5/6 complex. During the revision of the manuscript, another group also confirmed the presence of Nse1 and a second subunit, Nse2, in the fission yeast Smc5/6 complex (an electronic preprint is available at http://www.jbc.org/cgi/reprint/M308828200v1) (32). In the presence of DNA damage, analyses of cells lacking Nse1 showed that these cells fail to arrest normally in response to DNA damage and that they undergo lethal mitoses. Taken together, these data suggest a role for the Smc5/6 complex in coordination of an activated checkpoint (which we propose is necessary for the monitoring of DNA repair) prior to mitotic entry.
Cdc25-22 block and release is a common protocol used to synchronize fission yeast cells in G2. We have not used this protocol in our study as we found that in all the rad18 mutant backgrounds except that of rad18-X, the cdc25-22 block-and-release process potentiated a checkpoint defect. Moreover, a significant proportion (∼5%) of abnormal mitoses were observed in these mutants in the absence of irradiation. These were not present prior to block and release or in rad18− mutants at increased temperatures, though the percentage of cells showing mitotic defects was increased severalfold following irradiation. Indeed, the effectiveness of this method of synchronization with otherwise wild-type cells is dependent on the duration of the block. When cells are blocked at 36°C for 3 h before release, a normal checkpoint response is observed. When this block is extended to 4.5 h, no checkpoint arrest is seen (data not shown); presumably, the checkpoint cannot regulate increased pools of cyclin B/Cdc2 that accumulate during the arrest. This likely explains the differential checkpoint responses of cdc25-22-synchronized UV-C-irradiated rad18-74 cells, which failed to arrest, and the lactose gradient-synchronized cells treated with ionizing radiation, which behaved similarly to the mutants described in this paper (54).
Overexpression of Rad18 leads to gross structural changes to the nuclear architecture, leading to extrusion of the chromatin to the margins of the nucleus. Surprisingly, the vast majority of Rad18 under these conditions was not chromatin associated. This is unlikely to be a nonspecific effect of protein accumulation, as this result was not seen with similar overexpression of Cut14 and Cut3, the fission yeast Smc2 and Smc4 homologs, or with Nse1 overexpression (M. Yanagida, personal communication; our unpublished data). Further, it is not likely that this reflects a positive gain-of-function phenotype, as Rad18 mutants lacking a Walker A motif behave similarly and the hollow-sphere-like nuclei were produced by overexpression of just the first coiled-coil domain. This phenotype somewhat resembles that of cells lacking both topoisomerase I and II (51). It is possible that the overexpression of Rad18 could interfere with the function of topoisomerases, resulting in this nuclear phenotype. We were not able to reverse the effects of Rad18 overexpression by co-overexpression of Top2.
While cells lacking rad18 divide two to four times, only ∼20% of these cells arrest in mitosis. Those arrested in interphase show only a modest cellular elongation, indicating growth arrest rather than a block to the nuclear cycle, during which cells continue to elongate. A similar result is seen in nse1Δ cells, though these manage to divide four to six times. The mitotically arrested cells could arise from an interphase defect. rad18 mutants, with the topoisomerase II mutation top2-191, are synthetically lethal. Together with the condensin, topoisomerase II is a major determinant of mitotic chromosome condensation (10, 28, 50, 54). The major mitotic defect (with or without irradiation) seen in rad18− and nse1− mutants is the stretching of chromatin along the plane of division, frequently for the length of the cell. This may be the result of segregation of incompletely condensed chromatin, as it is a more modest version of what is seen in the fission yeast Smc2 and Smc4 mutants, in which φ-shaped chromosomes are produced (45). While Smc6 comes off chromatin at mitosis, it is possible that at the G2-M boundary the Smc5/6 complex aids in the loading of condensin and/or topoisomerase II (49). The arrest may be due to a more global defect in chromosomal organization, impacting on gene expression to the point of pleiotropically affecting many biochemical pathways.
What role does the Smc5/6 complex play in checkpoint maintenance? We have no evidence that the complex is a signaling component of the checkpoint per se; rather, the data suggest that the defects in checkpoint maintenance in rad18 and nse1 mutants reflect a defect in the organization of the interphase nucleus. Smc6 is present on chromatin only during interphase in human cells, and preliminary data suggest that this is also the case for fission yeast (reference 49 and our unpublished data). By analogy to findings for the condensin and cohesin complexes, we propose that the Smc5/6 complex facilitates various DNA repair processes via organization of chromatin in the vicinity of lesions to allow repair complexes to have access to DNA. This would explain why rad18 mutants are sensitive to a wide range of DNA-damaging agents that are repaired by different pathways (25, 54). Although rad18 mutants are profoundly defective in the repair of double-stranded DNA breaks, they are more modestly defective in the excision of cyclobutane dimers and of four to six photoproducts induced by exposure to 200 J of UV-C/m2 (25, 54). We have not found a significant retention of thymidine dimers (detected with the TDM-2 monoclonal antibody) in rad18-dn or rad18-so cells following exposure to 100 J of UV-C/m2 (data not shown). However, the excision of these dimers from chromosomal DNA does not necessarily reflect the completion of the repair process and they are not the only lesions induced by UV-C. Epistasis analysis suggests that Rad18 participates in recombinational events (25), which are also required following UV-C irradiation in S. pombe. By analogy to findings involving cut5 and pol1 mutants in S. pombe, in which the absence of replication factors on chromatin fails to generate a checkpoint whereas stalled replication is a strong checkpoint inducer, the detection of repair complexes on chromatin may be the factor that signals maintenance of checkpoint arrest (11, 33, 44, 46). We favor a model in which defects in the Smc5/6 complex can result in a reduced ability to establish repair at all sites of lesions or to maintain repair complexes on damaged templates, leading to premature termination of the arrested signal. We cannot rule out, however, that DNA repair might be completed in the absence of functional Rad18, as we observed for thymidine dimer excision. It is also possible that the defects we have observed are due to some other defect in organizing chromosomes in a manner appropriate for mitosis.
Our proposal that the Smc5/6 complex plays a role in establishing or facilitating the access of repair complexes to damaged sites rather than directly signaling from chromatin to maintain Chk1 activity leaves the signal an unknown entity. The failure to resume cell cycle progression after the establishment of a checkpoint would lead to permanent cell cycle arrest, thus making it difficult to identify the genes required to attenuate a checkpoint. Nevertheless, delineating this signaling pathway is as important as identifying the pathway leading to Chk1 activation and remains a significant challenge in efforts to define cellular response to DNA damage.
Acknowledgments
We are grateful to Alan Lehmann for the provision of anti-Rad18 sera, Mitsuhiro Yanagida for top2 plasmids, Tsukasa Matsunaga for the TDM-2 antibody, and Michael Krien and Doris Germain for critical reading of the manuscript.
S.H. is the recipient of a University of Melbourne Research Scholarship, and M.O. is a Scholar of the Leukemia and Lymphoma Society. This research was also supported by grants from the Australian Research Council (A00103857) and the NIH/NCI (CA100076-01).
REFERENCES
- 1.al-Khodairy, F., and A. M. Carr. 1992. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 11:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aono, N., T. Sutani, T. Tomonaga, S. Mochida, and M. Yanagida. 2002. Cnd2 has dual roles in mitotic condensation and interphase. Nature 417:197-202. [DOI] [PubMed] [Google Scholar]
- 3.Barr, S. M., C. G. Leung, E. E. Chang, and K. A. Cimprich. 2003. ATR kinase activity regulates the intranuclear translocation of ATR and RPA following ionizing radiation. Curr. Biol. 13:1047-1051. [DOI] [PubMed] [Google Scholar]
- 4.Basi, G., E. Schmid, and K. Maundrell. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123:131-136. [DOI] [PubMed] [Google Scholar]
- 5.Bhat, M. A., A. V. Philp, D. M. Glover, and H. J. Bellen. 1996. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with topoisomerase II. Cell 87:1103-1114. [DOI] [PubMed] [Google Scholar]
- 6.Birkenbihl, R. P., and S. Subramani. 1992. Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 20:6605-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boddy, M. N., P. Shanahan, W. H. McDonald, A. Lopez-Girona, E. Noguchi, J. R. Yates III, and P. Russell. 2003. Replication checkpoint kinase Cds1 regulates recombinational repair protein Rad60. Mol. Cell. Biol. 23:5939-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carr, A. M. 2002. DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair 1:983-994. [DOI] [PubMed] [Google Scholar]
- 9.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 10.Cuvier, O., and T. Hirano. 2003. A role of topoisomerase II in linking DNA replication to chromosome condensation. J. Cell Biol. 160:645-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Urso, G., B. Grallert, and P. Nurse. 1995. DNA polymerase alpha, a component of the replication initiation complex, is essential for the checkpoint coupling S phase to mitosis in fission yeast. J. Cell Sci. 108:3109-3118. [DOI] [PubMed] [Google Scholar]
- 12.Edwards, R. J., N. J. Bentley, and A. M. Carr. 1999. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat. Cell Biol. 1:393-398. [DOI] [PubMed] [Google Scholar]
- 13.Fousteri, M. I., and A. R. Lehmann. 2000. A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J. 19:1691-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujioka, Y., Y. Kimata, K. Nomaguchi, K. Watanabe, and K. Kohno. 2002. Identification of a novel non-structural maintenance of chromosomes (SMC) component of the SMC5-SMC6 complex involved in DNA repair. J. Biol. Chem. 277:21585-21591. [DOI] [PubMed] [Google Scholar]
- 15.Furnari, B., N. Rhind, and P. Russell. 1997. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science 277:1495-1497. [DOI] [PubMed] [Google Scholar]
- 16.Haering, C. H., J. Lowe, A. Hochwagen, and K. Nasmyth. 2002. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9:773-788. [DOI] [PubMed] [Google Scholar]
- 17.Harvey, S. H., M. J. Krien, and M. J. O'Connell. 30 January 2002, posting date. Structural maintenance of chromosomes (SMC) proteins, a family of conserved ATPases. Genome Biol. 3:REVIEWS3003. [Online.] [DOI] [PMC free article] [PubMed]
- 18.Hauf, S., I. C. Waizenegger, and J. M. Peters. 2001. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293:1320-1323. [DOI] [PubMed] [Google Scholar]
- 19.Hirano, T. 2002. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16:399-414. [DOI] [PubMed] [Google Scholar]
- 20.Hirano, T., R. Kobayashi, and M. Hirano. 1997. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89:511-521. [DOI] [PubMed] [Google Scholar]
- 21.Hirano, T., and T. J. Mitchison. 1994. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell 79:449-458. [DOI] [PubMed] [Google Scholar]
- 22.Kim, S. T., B. Xu, and M. B. Kastan. 2002. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 16:560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo, T., T. Wakayama, T. Naiki, K. Matsumoto, and K. Sugimoto. 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294:867-870. [DOI] [PubMed] [Google Scholar]
- 24.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 25.Lehmann, A. R., M. Walicka, D. J. Griffiths, J. M. Murray, F. Z. Watts, S. McCready, and A. M. Carr. 1995. The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol. Cell. Biol. 15:7067-7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Losada, A., M. Hirano, and T. Hirano. 1998. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 12:1986-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowe, J., S. C. Cordell, and F. van den Ent. 2001. Crystal structure of the SMC head domain: an ABC ATPase with 900 residues antiparallel coiled-coil inserted. J. Mol. Biol. 306:25-35. [DOI] [PubMed] [Google Scholar]
- 28.Maeshima, K., and U. K. Laemmli. 2003. A two-step scaffolding model for mitotic chromosome assembly. Dev. Cell 4:467-480. [DOI] [PubMed] [Google Scholar]
- 29.Martinho, R. G., H. D. Lindsay, G. Flaggs, A. J. DeMaggio, M. F. Hoekstra, A. M. Carr, and N. J. Bentley. 1998. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 17:7239-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattingly, R. R., A. Sorisky, M. R. Brann, and I. G. Macara. 1994. Muscarinic receptors transform NIH 3T3 cells through a Ras-dependent signalling pathway inhibited by the Ras-GTPase-activating protein SH3 domain. Mol. Cell. Biol. 14:7943-7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127-130. [DOI] [PubMed] [Google Scholar]
- 32.McDonald, W. H., Y. Pavlova, J. R. Yates III, and M. N. Boddy. 8 September 2003. Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J. Biol. Chem. 10.1074/jbc.M308828200. [DOI] [PubMed]
- 33.McFarlane, R. J., A. M. Carr, and C. Price. 1997. Characterisation of the Schizosaccharomyces pombe rad4/cut5 mutant phenotypes: dissection of DNA replication and G2 checkpoint control function. Mol. Gen. Genet. 255:332-340. [DOI] [PubMed] [Google Scholar]
- 34.Melby, T. E., C. N. Ciampaglio, G. Briscoe, and H. P. Erickson. 1998. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol. 142:1595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melo, J. A., J. Cohen, and D. P. Toczyski. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15:2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 37.Morishita, T., Y. Tsutsui, H. Iwasaki, and H. Shinagawa. 2002. The Schizosaccharomyces pombe rad60 gene is essential for repairing double-strand DNA breaks spontaneously occurring during replication and induced by DNA-damaging agents. Mol. Cell. Biol. 22:3537-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Connell, M. J., C. Norbury, and P. Nurse. 1994. Premature chromatin condensation upon accumulation of NIMA. EMBO J. 13:4926-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Connell, M. J., J. M. Raleigh, H. M. Verkade, and P. Nurse. 1997. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 16:545-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Connell, M. J., N. C. Walworth, and A. M. Carr. 2000. The G2-phase DNA-damage checkpoint. Trends Cell Biol. 10:296-303. [DOI] [PubMed] [Google Scholar]
- 41.Raleigh, J. M., and M. J. O'Connell. 2000. The G2 DNA damage checkpoint targets both wee1 and cdc25. J. Cell Sci. 113:1727-1736. [DOI] [PubMed] [Google Scholar]
- 42.Rhind, N., B. Furnari, and P. Russell. 1997. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev. 11:504-511. [DOI] [PubMed] [Google Scholar]
- 43.Saka, Y., F. Esashi, T. Matsusaka, S. Mochida, and M. Yanagida. 1997. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 11:3387-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saka, Y., P. Fantes, T. Sutani, C. McInerny, J. Creanor, and M. Yanagida. 1994. Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 13:5319-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saka, Y., T. Sutani, Y. Yamashita, S. Saitoh, M. Takeuchi, Y. Nakaseko, and M. Yanagida. 1994. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 13:4938-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saka, Y., and M. Yanagida. 1993. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+. Cell 74:383-393. [DOI] [PubMed] [Google Scholar]
- 47.Sutani, T., T. Yuasa, T. Tomonaga, N. Dohmae, K. Takio, and M. Yanagida. 1999. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 13:2271-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tatebayashi, K., J. Kato, and H. Ikeda. 1998. Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics 148:49-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor, E. M., J. S. Moghraby, J. H. Lees, B. Smit, P. B. Moens, and A. R. Lehmann. 2001. Characterization of a novel human SMC heterodimer homologous to the Schizosaccharomyces pombe Rad18/Spr18 complex. Mol. Biol. Cell 12:1583-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uemura, T., H. Ohkura, Y. Adachi, K. Morino, K. Shiozaki, and M. Yanagida. 1987. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell 50:917-925. [DOI] [PubMed] [Google Scholar]
- 51.Uemura, T., and M. Yanagida. 1984. Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 3:1737-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uhlmann, F., F. Lottspeich, and K. Nasmyth. 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400:37-42. [DOI] [PubMed] [Google Scholar]
- 53.Uhlmann, F., and K. Nasmyth. 1998. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 8:1095-1101. [DOI] [PubMed] [Google Scholar]
- 54.Verkade, H. M., S. J. Bugg, H. D. Lindsay, A. M. Carr, and M. J. O'Connell. 1999. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol. Biol. Cell 10:2905-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verkade, H. M., T. Teli, L. V. Laursen, J. M. Murray, and M. J. O'Connell. 2001. A homologue of the Rad18 postreplication repair gene is required for DNA damage responses throughout the fission yeast cell cycle. Mol. Genet. Genomics 265:993-1003. [DOI] [PubMed] [Google Scholar]
- 56.Walworth, N. C., and R. Bernards. 1996. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science 271:353-356. [DOI] [PubMed] [Google Scholar]
- 57.Yazdi, P. T., Y. Wang, S. Zhao, N. Patel, E. Y. Lee, and J. Qin. 2002. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 16:571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zou, L., D. Cortez, and S. J. Elledge. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16:198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]