Abstract
GTF2I and GTF2IRD1 encode a family of closely related transcription factors TFII-I and BEN critical in embryonic development. Both genes are deleted in Williams-Beuren syndrome, a complex genetic disorder associated with neurocognitive, craniofacial, dental and skeletal abnormalities. Although genome-wide promoter analysis has revealed the existence of multiple TFII-I binding sites in embryonic stem cells (ESCs), there was no correlation between TFII-I occupancy and gene expression. Surprisingly, TFII-I recognizes the promoter sequences enriched for H3K4me3/K27me3 bivalent domain, an epigenetic signature of developmentally important genes. Moreover, we discovered significant differences in the association between TFII-I and BEN with the cis-regulatory elements in ESCs and embryonic craniofacial tissues. Our data indicate that in embryonic tissues BEN, but not the highly homologous TFII-I, is primarily recruited to target gene promoters. We propose a “feed-forward model” of gene regulation to explain the specificity of promoter recognition by TFII-I factors in eukaryotic cells.
Introduction
TFII-I and BEN, products of paralogous genes Gtf2i and Gtf2ird1, display a dynamic expression pattern during mouse embryo development [1] [2]. At the blastocyst stage (E3.5–4.5) both transcription factors are localized to the cytoplasm and nuclei of the mouse inner cell mass (ICM) and trophectoderm. Consistently, in situ hybridization of mouse pre-implantation blastocysts revealed high levels of TFII-I mRNA in the ICM [3]. It is not surprising that the expression of TFII-I and BEN target genes was also detected in the early mouse embryo and ESCs [4] [5].
Although the TFII-I family members are expressed very early in development, they are not required for maintenance of pluripotency, because ablation of either Gtf2i or Gtf2ird1 in mouse embryos does not lead to peri-implantation lethality [6]. The characteristic feature of TFII-I factors is a presence of multiple helix-loop-helix domains (I-repeats) that can serve as independent DNA-binding modules, although their chromatin recognition properties are still not fully understood. The SELEX procedure performed with a set of isolated I-repeats identified the core RGATTR sequence as a common DNA-binding motif for repeats 4 and 5 of BEN and for repeats 4 and 6 of TFII-I [7]. This core consensus sequence corresponds to the bona fide BEN-binding sites located in the upstream regulatory regions of Hoxc8, Tnn1 Gsc, Scand3, Cfl, Shrm and Ezh2 genes [4] [8] [9] [10] [11] [12]. TFII-I and BEN bind to the DICE element TRTYBTCTHYACMR in the VH promoters of IgH genes [13]. Furthermore, SELEX with the full-length BEN delineated a binding motif GGGRSCWGCGAYAGCCSSH that bears no sequence similarity to the DICE or RGATTR core consensus sequence [14]. Although TFII-I and BEN recognize the same or similar motifs, only TFII-I, together with USF1/USF2 heterodimer, binds to the upstream cis element RBEIII (ACTGCTGA) necessary for transcription of Human Immunodeficiency Virus Type 1 [15]. In addition, TFII-I interacts with the E-boxes (CANNTG) and the pyrimidine-rich Initiator element (YYANWYY) [16]. It was speculated that TFII-I regulates c-Fos as well as the set of estrogen-responsive genes by recognizing the Initiator sequence [17] [18]. The regulation of VEGFR-2 and β-globin genes, for example, occurs through the recruitment of TFII-I to the Initiator and E-box elements, respectively [19] [20]. We reported that in mouse ESCs TFII-I binds to the canonical R4 consensus in the promoters of Cfdp1, Sec23a and Nsd1 [21].
Despite these findings, the direct in vivo targets of TFII-I factors are poorly defined. In the present work, we report genome-wide promoter mapping in mouse ESCs and embryonic craniofacial tissues. Chromatin immunoprecipitation (ChIP)-coupled DNA microarray analysis (ChIP-chip) revealed multiple TFII-I and BEN binding sites across the upstream regulatory regions of many developmental regulators. In addition to recognizing the previously defined consensus sequences, these proteins associate with the novel cis-regulatory elements. Collectively, our data indicate that chromatin recognition by TFII-I transcription factors is more complicated and diverse than previously considered.
Results
A Genome-wide Promoter Screen for TFII-I and BEN Binding Sites in Mouse ESCs and Embryonic Craniofacial Tissues
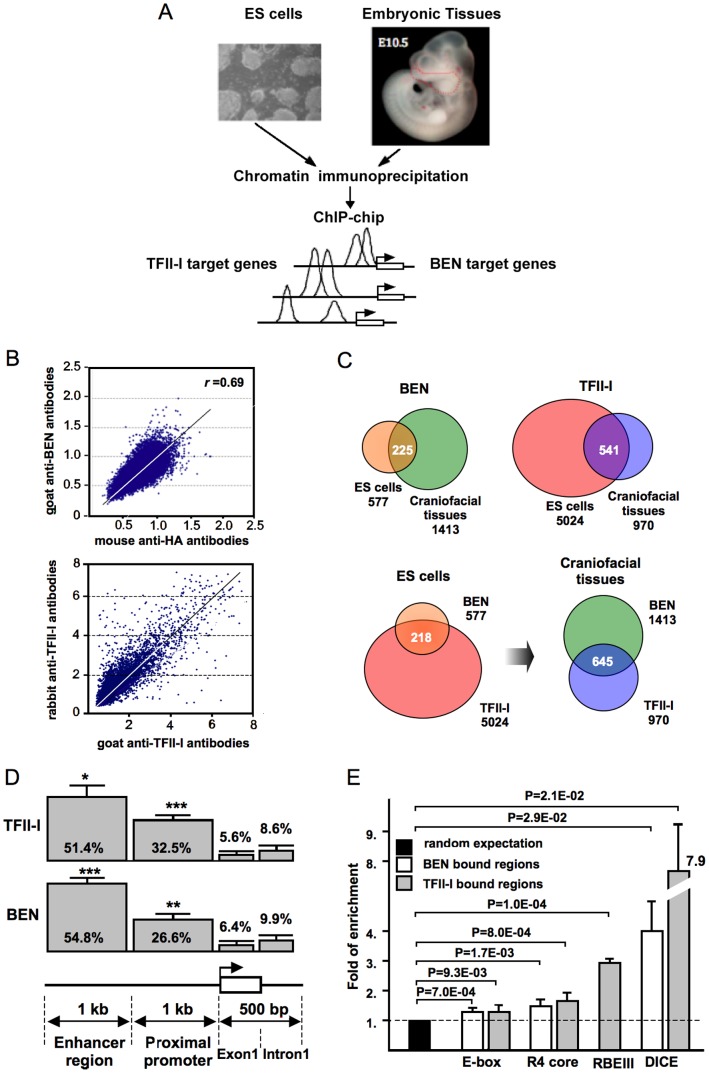
The promoter ChIP-chip analysis was used to identify regions bound by TFII-I and BEN in the mouse ESC lines E14tg2a and Ainv15 and embryonic craniofacial tissues (ETs) derived from E10.5 mouse embryos (Fig. 1A). The chromatin immunoprecipitation (IP) was performed with two types of antibodies for each transcription factor: rabbit and goat polyclonal antibodies for TFII-I-bound chromatin IP and goat anti-BEN and mouse anti-HA antibodies BEN-bound chromatin IP. First, we have checked the consistency of ChIP-chip obtained with two different antibodies for each transcription factor. Spearman’s rank correlation coefficient (r = 0.7–0.9, Fig. 1B) determined for each antibody pair is comparable with technical replicates (r = 0.8–0.9, not shown). A positive correlation between experiments using different antibodies provides credibility to the TFII-I and BEN binding peaks.
Figure 1. Chromatin isolation and ChIP-chip analysis.
(A) The ChIP-chip strategy. Chromatin was isolated from mouse ESCs and the craniofacial region (CF) of E10.5 mouse embryos. Chromatin immunoprecipitation (ChIP) was performed with TFII-I and BEN-specific antibodies. CF region is marked in red with dashed lines. (B) The correlation between the genome-wide promoter binding using goat anti-BEN and mouse anti-HA antibodies to BEN (left) and rabbit and goat polyclonal antibodies to TFII-I (right). Spearman’s rank correlation coefficient (r) is calculated for each antibody pair. (C) The overall statistics of TFII-I and BEN target genes in ESCs and embryonic craniofacial tissues. (D) Distribution of the TFII-I and BEN-bound genomic sites with respect to the gene structure. The 2 kb region upstream of the transcription start site (TSS) is arbitrarily divided into two 1 kb segments (‘enhancer region’ and ‘proximal promoter’). 0.5 kb region downstream from TSS is split into exon and intron sequences. Bars represent the standard deviation calculated from four (TFII-I) or six (BEN) chip hybridizations, *p<0.1, **p<0.05, ***p<0.01.
5,744 TFII-I and 625 BEN binding peaks (FDR <0.19) were identified within 5,024 (Table S1) and 577 promoters (Table S2), respectively. 221 promoters are recognized by both transcription factors (Table S3A). Interestingly, TFII-I and BEN recognize the same sequence in 57 promoters (Table S3B).
We next investigated the occupancy of TFII-I factors in embryonic craniofacial tissues isolated from E10.5 mouse embryos. This experiment yielded 1,181 TFII-I and 1,520 BEN binding peaks (FDR <0.19) in 970 (Table S4) and 1,413 promoters (Table S5), respectively. Some peaks were detected in the promoter regions of the two closely located but oppositely directed genes and in such cases it was impossible to assign the binding peak to a corresponding gene. In contrast to the ESCs binding profile the promoter occupancy in ETs showed a 2.5-fold increase of BEN binding and a 5-fold decrease of TFII-I binding, respectively (Fig. 1C). Among 636 promoters occupied by both factors, 537 coincide with TFII-I and BEN binding to the same sequence (Table S6). Interestingly, the majority of genomic sites recognized by both proteins in ETs are also recognized by TFII-I in mouse ESCs. The density maps showed a high degree of TFII-I and BEN co-occupancy at the transcription start sites in ESCs and ETs (Fig. S1A).
The DNA-binding Motifs Recognized by the TFII-I Family of Transcription Factors
Our next goal was to investigate a distribution of DNA-binding sites with respect to gene structure. CEAS software provides summary statistics including the percentages of peaks that reside in the proximal promoters (1 kb upstream from RefSeq 5′ start), 1st exon, 1st intron, and enhancer regions (>1 kb from RefSeq) [22]. A significant enrichment was detected in the proximal enhancers occupied by TFII-I and BEN in ESCs and in embryonic craniofacial tissues (Fig. 1D ). Enrichment relative to random expectation (Fig. 1E) revealed a statistically significant abundance of the R4 consensus in the TFII-I (57.1%) and BEN-bound sequences (51.7%), respectively. E-boxes were the most enriched motifs and counted more than once among all known DNA-binding sequences. DICE is another sequence frequently bound by TFII-I and BEN. In contrast, RBEIII is a rare sequence recognized by TFII-I only.
Because we cannot rule out a probability that the chromatin recognition by the TFII-I family requires cooperation with additional transcription factors, DNA sequences around TFII-I and BEN bound sites were searched to identify potential protein partners listed in TRANSFAC and JASPAR databases. CEAS identified a number of hits for each motif both within the ChIP-regions and in the whole genome (p value <1.0E-5) (Tables S7 and S8). The genomic regions occupied by TFII-I and BEN showed significant enrichment for AP-2α, AP-2γ, ETF, Spz1, and E2F binding sequences. The BEN-bound sequences are also enriched with the recognition sites for SMADs, p300 and STATs. The relative abundance of these motifs suggests the existence of a specific protein interaction network.
The TFII-I and BEN bound sequences were also analyzed for the presence of new cis-regulatory elements [23]. We found three novel motifs with statistically significant enrichment (E-value less than 5E-21) (Fig. S1B and Table S9). TFII-I consensus sequence list is indicated in Fig. S1C.
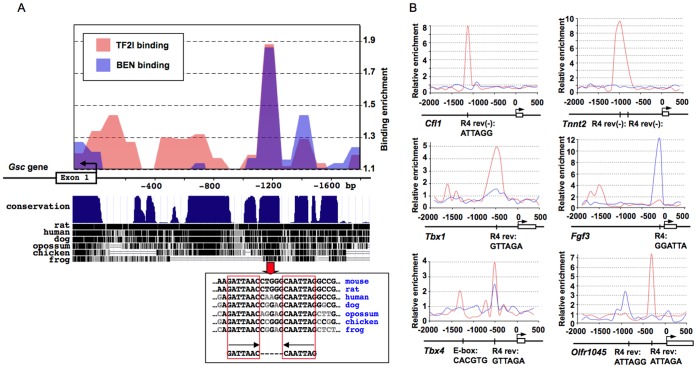
It was previously reported that the TFII-I family interacts with the distal promoter element of Gsc [10] [11]. ChIP-chip confirmed TFII-I and BEN occupancy in the Gsc distal element corresponding to a stereotypical configuration of the two conserved R4 sequences (Fig. 2A). Multiple hits for E-boxes, R4 consensus, DICE and RBEIII elements were detected within the TFII-I and BEN occupied sites across a large set of developmentally regulated genes (Fig. 2B and Table S9). We noticed that each protein displayed a unique chromatin recognition mode. For example, TFII-I binding occurs at the more distal promoter sites of Cfl1, Tnnt2 and Tbx1 while BEN recognizes the more proximal promoter of Fgf3 (Fig. 2B). Interestingly, TFII-I and BEN occupy the same sequence in the Tbx4 promoter although they bind different cis-regulatory elements in the Olfr1045 gene (Fig. 2B).
Figure 2. Genome-wide promoter recognition by TFII-I transcription factors in mouse ESCs. (.
A) TFII-I and BEN bind to the distal element of the Gsc promoter. The conservation plot is downloaded from the UCSC genome browser (http://genome.ucsc.edu/). (B) TFII-I binds to the promoters of Cfl1, Tnnt2 and Tbx1 while BEN occupies the promoter of Fgf3. TFII-I and BEN recognize the same sequence in the Tbx4 promoter although they bind to different cis-regulatory motifs within the promoter of Olfr1045. Red – TFII-I binding, blue – BEN binding.
Biological Functions of TFII-I and BEN Target Genes in Mouse ESCs
To gain insights into the TFII-I controlled biological processes we carried out the gene ontology (GO) analysis using the DAVID software [24]. The GO classification indicated significant enrichment in the ‘chromatin assembly’ (Fig. S2A and Table S10) and the ‘cell fate commitment’ categories amongst the TFII-I and BEN target genes in mouse ESCs (Fig. S2B and Table S10). Other over-represented cellular functions include ‘signal transduction’ for genes bound by TFII-I and ‘cell-cell signaling’ and ‘cytoskeleton organization’ for BEN targets. Multiple over-represented signaling pathways were found among TFII-I bound genes. BEN, on the other hand, primes mainly genes linked to the WNT signaling pathway (Fig. S2 C and Table S10).
Biological Functions of TFII-I and BEN Target Genes in Embryonic Tissues
We also observed dramatic differences in the composition of genes targeted by TFII-I in mouse ESCs and ETs with regard to their biological functions. The most enriched cellular function among TFII-I bound genes in embryonic tissues is ‘signal transduction’ owing to a large group of olfactory, vomeronasal and taste receptor genes (Fig. S3B and Table S11). At the same time, BEN targets a broad spectrum of genes associated with ‘cell division’, ‘regulation of apoptosis’, ‘cell fate commitment’, ‘cell differentiation’, ‘cell motility’, ‘regulation of transcription’, ‘RNA processing’ and ‘regulation of translation’ (Supplemental Fig. S3A and Table S11). Many of these genes are linked to specific developmental processes, especially brain and skeletal development (Fig. S3C and Table S11).
Genome-wide Promoter Occupancy by TFII-I and BEN does not Correlate with the Expression Levels of Target Genes in Mouse ESCs
We addressed the question whether the genomic sites occupied by TFII-I exert transcriptional regulatory activity. We analyzed the Affymetrix expression data (GEO database: GDS1616 record) derived from the same E14tg2a mouse ESC line used for ChIP-chip. The default P-value cut-offs (0.04 and 0.06) provide boundaries for defining Present, Marginal, or Absent calls. Absent indicates that the expression level is below the threshold of detection and close to zero. Marginal call indicates the cases of an uncertainty. The distribution of gene targets marked as absent, marginal and present is 49.1%:2.4%:48.5% for BEN and 48.7%:2.6%:48.7% for TFII-I, respectively. This observation clearly indicates that binding of TFII-I factors does not correlate with the transcriptional activity of the corresponding genes. Quantitative mRNA expression analysis from mouse ESCs [25] was used to compare transcription of target genes. Similarly, we failed to detect significant differences in the overall level of expression except the modest decrease in transcription among BEN bound genes.
Correlation between TFII-I Target Genes and Embryonic Developmental Pathways
We analyzed genes bound by TFII-I in mouse ESCs and embryonic craniofacial tissues for various developmental processes. For this purpose the ChIP-chip gene list was compared with four different gene groups: 366 human genes associated with neural tube closure defects [26], 497 mouse genes linked to craniofacial development, 739 mouse genes associated with skeletal development and 976 mouse genes involved in brain development, respectively (MGI, http://www.informatics.jax.org/phenotypes.shtml). A chi-squared test was performed to estimate the average deviation of actually observed genes from the expected value of random variables (Table S12). We discovered that TFII-I binding sites are significantly enriched in ESCs, which is contrary to the enrichment of BEN in embryonic craniofacial tissues.
Abrogation of TFII-I and BEN by siRNA Knockdown in Mouse Neural Crest Cells
To find a correlation between TFII-I and BEN promoter binding in ESCs and the expression of key developmental genes at later stages of differentiation, we investigated siRNA-mediated silencing of Gtf2i and Gtf2ird1 in mouse neural crest cells. For this purpose we used the neural crest-derived JoMa1.3 cell line, which retains some features of stemness and the ability to differentiate into neurons, glia, smooth muscle cells, melanocytes and chondrocytes [27].
Three independent Gtf2i and Gtf2ird1 siRNA-mediated knockdowns have been performed with JoMa1.3 cells. The knockdown effect was monitored at three different time intervals (9 h, 24 h and 72 h) post transfection. Average 9.8-fold decrease in Gtf2i expression (>90% knockdown) and 5.8-fold decrease in Gtf2ird1 expression (∼80% knockdown) was observed 9 h post transfection. This silencing effect persisted through day 3 with the remaining 2.5-fold decrease of Gtf2i and 5-fold decrease of Gtf2ird1 expression, respectively (Fig. S4). We observed no notable morphological changes in JoMa1.3 cells, although expression of neural crest markers Twist and Snail1 as well as histone methyltransferases Ezh2 and Nsd1 was affected after TFII-I knockdown but not after BEN inactivation (Fig. 3E). ChIP-chip has also confirmed our previous observation that TFII-I occupies Ezh2 and Nsd1 proximal promoters in mouse ESCs [21]. Collectively, these results indicate that during ESC differentiation TFII-I regulates genes involved in cell fate commitment and lineage specification.
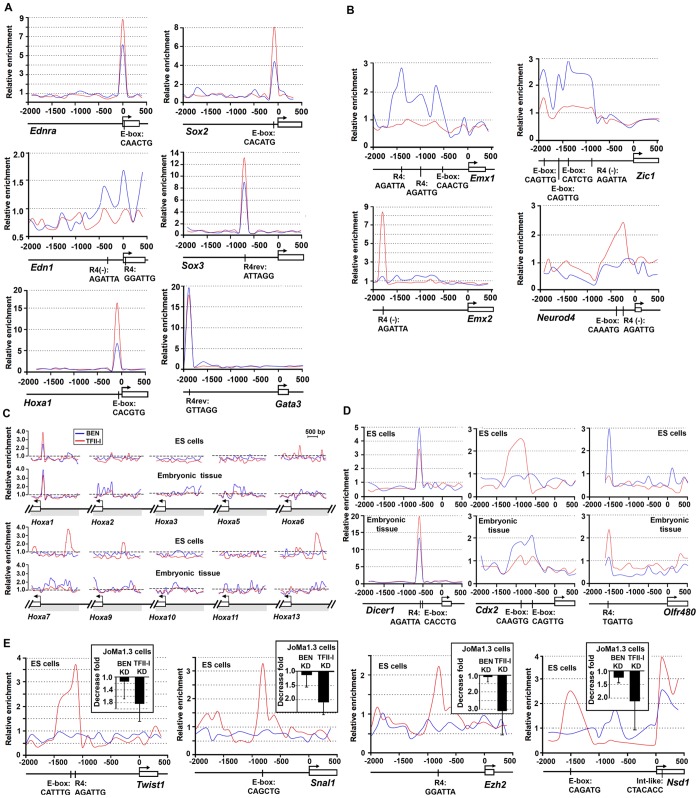
Figure 3. TFII-I factors occupy the promoters of key developmental regulators in ESCs and embryonic craniofacial tissues (ETs).
(A) TFII-I and BEN bind to the promoters of Ednra, Edn1, Sox2, Sox3, Hoxa1 and Gata3 implicated in neural crest and craniofacial development. (B) TFII-I occupies the promoters of Emx1, Emx2, Zic1 and Neurod4 involved in brain development. The notation and labeling are as in Figure 2B. (C) TFII-I and BEN occupy the promoter regions of the HoxA cluster (Hoxa1, Hoxa7 and Hoxa13) in mouse ESCs and ETs. (D) TFII-I and BEN recognize the same cis-regulatory element in the promoters of Dicer, Cdx2 and Olfr480 in stem cells and embryonic tissues. (E) TFII-I binds to the promoters of Twist1, Snail2, Ezh2 and Nsd1 (red lines) in ESCs, although BEN does not bind to these promoters (blue lines). siRNA-mediated knockdown of TFII-I down-regulates expression of Twist, Snail1, Ezh2 and Nsd1 in embryonic neural crest cells (JoMa1.3 line). Error bars represent the standard deviation calculated from three independent knockdown experiments.
Genome-wide Promoter Occupancy by TFII-I and BEN in ESCs and Embryonic Craniofacial Tissues
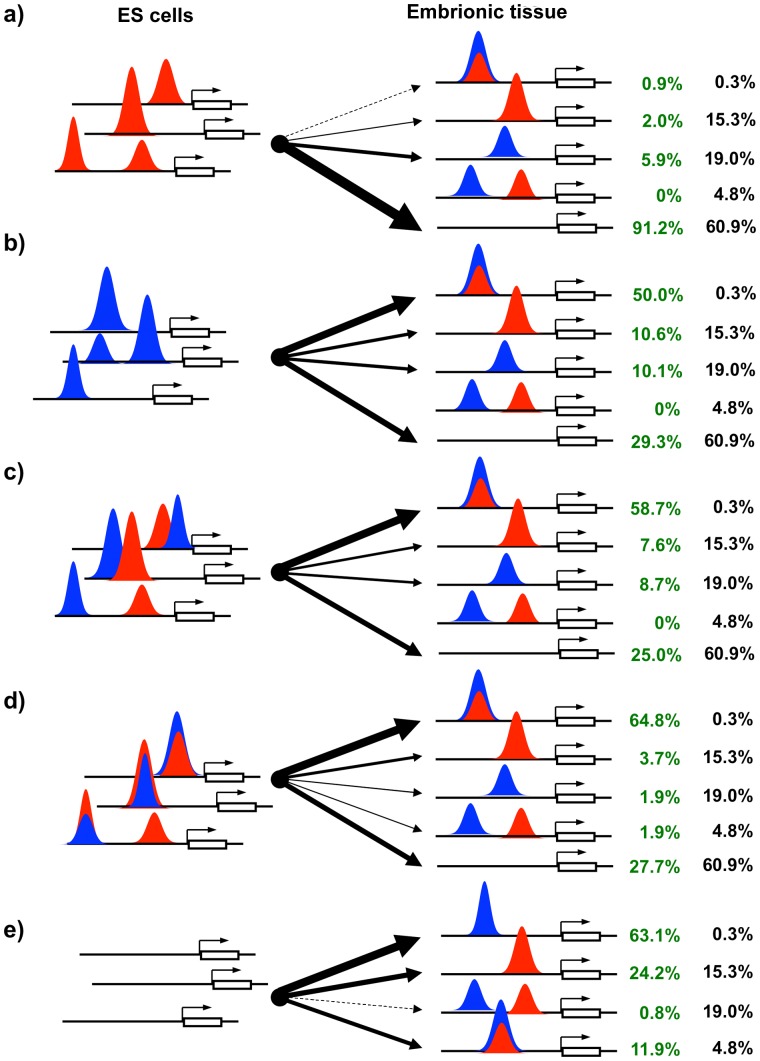
We established that promoter recognition by TFII-I and BEN shows significant difference in ESCs and embryonic tissues. First, the majority of promoters occupied by TFII-I in ESCs become free in ETs (Fig. 4,a); second, a large number of ESC promoters bound by BEN recruit TFII-I and BEN to the same site in ET promoters (Fig. 4,b); third, ESC promoters occupied by TFII-I and BEN are still recognized by both transcription factors in ETs, predominantly at the same sites, although in some cis-regulatory sites TFII-I and BEN binding is lost completely (Fig. 4,c,d); and fourth, promoters active only in ETs recruit more BEN than TFII-I (Fig. 4,e). For example, both proteins occupy the same sequence in the Hoxa1 and Dicer promoters, while BEN either replaces or is being replaced with TFII-I in the Cdx2 and Olfr480 promoters, respectively (Fig. 3C,D).
Figure 4. Promoter recognition by the TFII-I family.
TFII-I (in red) and BEN (in blue) possess distinct promoter recognition properties in ESCs and embryonic craniofacial tissues (ETs). First, the majority of ESC promoters occupied by TFII-I become vacant in ETs (a); second, a large number of ESC promoters recognized by BEN recruit both transcription factors to the same site in ETs (b); third, the ESC promoters occupied by TFII-I and BEN are still recognized by both transcription factors in ETs, predominantly in the same sequence, although some sites lost their binding completely (d); and fourth, the promoters active in ETs recruit more BEN than TFII-I (e). The black numbers on the right indicate percentage expected from the random distribution of TFII-I or BEN binding. The green numbers indicate the observed distribution significantly deviated from the random distribution (chi-squared test).
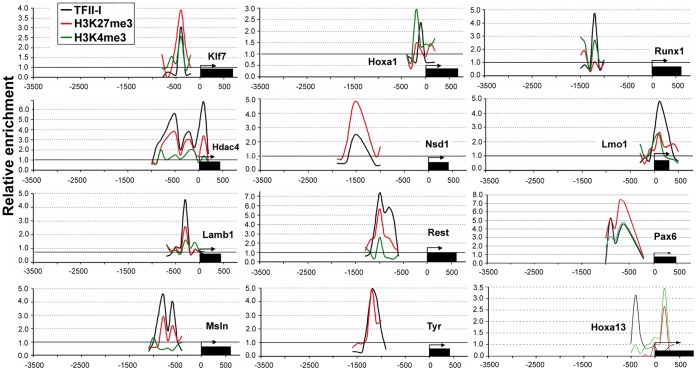
Correlation between TFII-I and BEN Promoter Occupancy and Genome-wide Distribution of H3K27me3 and H3K4me3 Bivalent Marks
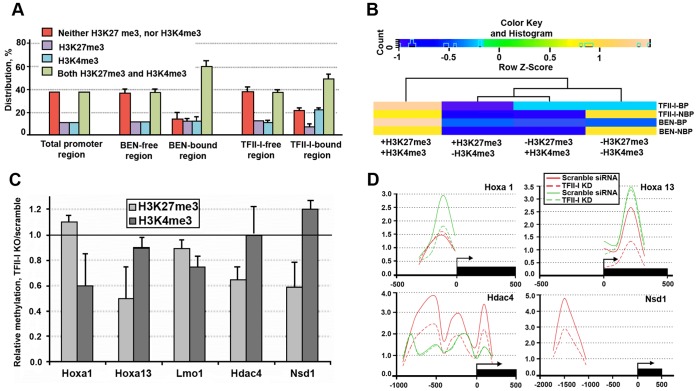
The lack of correlation between promoter recognition by TFII-I and BEN and gene expression in mouse ESCs prompted us to examine the status of histone methylation in the genomic regions occupied by these factors. For this purpose we used H3K27me3 and H3K4me3 promoter-binding profile (NCBI GEO database: record GSE17387) performed on the same GPL8943 platform (mouse 385 K RefSeq Promoter Array) in B6D2F2 mouse ESCs [28]. We observed co-occupancy of H3K4me3/K27me3 bivalent marks and TFII-I factors in the promoters of many developmental regulators. The promoter occupancy for a few genes is illustrated in Fig. 5. Correlation analysis showed enrichment of bivalent marks (P<0.05) in the regulatory regions occupied by TFII-I or BEN (Fig. 6A). To confirm this observation, we used previously published ChIP-seq data sets for H3K4me3 and H3K27me3 (GSM307618 and GSM307619). The peak finding program MACS produced 26,339 H3K4me3 and 5,529 H3K27me3 peaks, respectively. Colocalization of bivalent domains with the genomic sites bound TFII-I factors is represented as a heat-map (Fig. 6B). We found a statistically significant (P<0.05) association between bivalent domain and promoter sites occupied by TFII-I and BEN. H3K27 tri-methylation at the promoters of Hoxa13, Hdac4 and Nsd1 (Fig. 6C,D) is significantly affected in cells depleted of TFII-I. We noticed that H3K4 tri-methylation at the Hoxa1 promoter was also reduced in ESCs.
Figure 5. Colocalization of bivalent chromatin with TFII-I bound sites.
TFII-I associates with the promoter regions of key developmental genes enriched for H3K4me3 and H3K27me3 marks.
Figure 6. TFII-I binding overlaps with bivalent domain in ESCs. (.
A) The promoter occupancy by TFII-I and BEN correlates with genome-wide distribution of H3K27me3 and H3K4me3 bivalent marks (green bars). (B) Heat map indicates the co-localization frequency of TFII-I and BEN with bivalent domain. TF2I-BP, TFII-I bound promoters; TF2I-NBP, promoters free of TFII-I; BEN-BP, BEN bound promoters; BEN-NBP, promoters free of BEN. (C) Depletion of TFII-I by siRNA knockdown reduces H3K4me3 at Hoxa1 and H3K27me3 at Hoxa13, Hdac4 and Nsd1. (D) ChIP revealed that TFII-I depletion affects H3K27me3 at the promoters of Hoxa13, Hdac3 and Nsd1 and H3K4me3 at the Hoxa1 promoter. H3K27m3 is in red; H3K4me3 is in green.
Discussion
Genome-wide Promoter Binding by TFII-I and BEN in ESCs and Embryonic Craniofacial Tissues
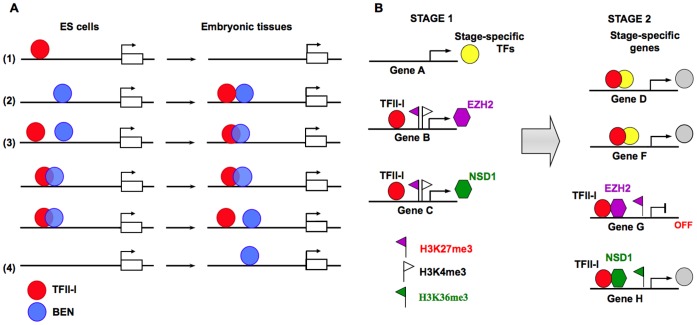
Our studies revealed that TFII-I and BEN recognize multiple distinct sequence motifs. In ESCs predominantly TFII-I primes the majority of genes associated with signal transduction and cell fate commitment. On a contrary, in embryonic craniofacial tissues BEN binds genes linked to tissue and organ development (Fig. S2, S3 and Table S10–11). We discovered that promoter occupancy by TFII-I and BEN is significantly different in ESCs and ETs. The following general rules were deduced from the promoter occupancy profiles (Fig. 7A). First, the majority of ESC promoters bound by TFII-I become vacant in ETs. Second, most ESC promoters recognized by BEN retain the ability to recruit both transcription factors in ETs. Third, the majority ESC promoters occupied by TFII-I and BEN recruit these factors to the same cis-regulatory sites in ETs. Fourth, BEN predominantly recognizes promoters active in ETs.
Figure 7. Target recognition by TFII-I factors. (.
A) Chromatin recognition by TFII-I transcription factors in ESCs and embryonic craniofacial tissues (ETs). (1), the majority of TFII-I-bound ESC promoters tend to lose TFII-I binding in ETs. (2), the majority of BEN-bound ESC promoters recruit TFII-I de novo in ETs. (3), most of TFII-I and BEN-bound ESC promoters continue to retain these factors in ETs. (4), the promoters active in ETs recruit more BEN than TFII-I. (B) The feed-forward model explains the lack of correlation between the promoter binding by TFII-I and expression of the corresponding genes (stage 1, ESCs). Transcription factors (TFs) activated by TFII-I at stage 2 (ETs) can also recognize the TFII-I target sites and together they could initiate expression of stage-specific genes and additionally activate chromatin-modifying genes Ezh2 and Nsd1. These epigenetic factors mark novel target promoters (genes F and G) for repression or activation (e.g. repressive mark H3K27me3 and active mark H3K36me3). Stage 1, embryonic stem cells; stage 2, embryonic tissues.
The under-representation of genes targeted by TFII-I in embryonic craniofacial tissues is in a strike contrast to the abundance of genes recognized by BEN (Table S4–12). In part, this could be explained by posttranslational modifications mediated by various signaling mechanisms that could alter the TFII-I binding ability as it was demonstrated for the TGFβ/activin-dependent induction of Gsc [11] [29].
Diversity of the TFII-I Recognition Elements
ChIP-chip revealed that TFII-I and BEN display a complex DNA binding mode. Although, in the majority of cases, TFII-I family members bind to the canonical R4 consensus sequence or E-boxes, not all the high-confident binding sites can be explained by the presence of these motifs. MEME analysis has revealed that TFII-I factors recognize additional sequences (Fig. S1B and Table S9), but even these novel motifs cannot explain the full complexity of promoter occupancy. Nevertheless, we were able to confirm TFII-I and BEN binding to a previously characterized distal element of the Gsc promoter (Fig. 2A). Both proteins co-occupy the same site in the Hoxa1 and Dicer1 promoters in ESCs and embryonic tissues (Fig. 3C,D). TFII-I factors can also exhibit mutually exclusive DNA-binding mode. In mouse ESCs TFII-I and BEN colocalize at the promoters of Olfr480 and Cdx2. In embryonic craniofacial tissues only TFII-I binds to the promoter of Olfr480 while BEN replaces TFII-I in the Cdx2 promoter (Fig. 3D).
Based on the sequence similarity the core consensus sequence RGATTR can be considered as a subset of the Initiator motif. Although the Initiator-like motifs were found among the TFII-I-bound sites, these sequences scatter frequently across the mammalian genome without specific protein binding capacity. The fact that not all the consensus sequences were identified at genomic loci bound by TFII-I and BEN suggests that promoter recognition is also determined by other transcription factors or the local chromatin structure. In this connection, it is notable that TFII-I or BEN binding sites coincide with the regions of extensive sequence conservation suggesting that additional regulatory proteins may influence their recruitment (Tables S7 and S8).
In conclusion, TFII-I factors display complex chromatin recognition, which differs from simple cooperative interaction or competition between TFII-I and BEN for a common binding site.
Diversity of the TFII-I and BEN Target Genes
TFII-I and BEN prime a large group of the olfactory, vomeronasal, taste receptor and histone genes (Tables S1–6). The odorant receptor genes with over thousand members constitute the largest gene family in the mouse genome [30]–[31]. A mature olfactory sensory neuron is thought to express just one olfactory receptor. The transcriptional mechanisms and the regulatory elements that mediate the expression of a single olfactory receptor gene per neuron remain poorly understood [32]. BEN was previously identified as a transcription factor involved in the regulation of the olfactory receptor gene 262 [33] Among other abundant TFII-I and BEN targets are histone 1H1, 1H2 and 2H clusters (Tables S1–S6).
The “Feed-forward Model” of TFII-I Regulation
Recruitment of TFII-I to a large number of developmental promoters does not correlate with the expression of the corresponding genes. Surprisingly, we discovered that TFII-I-bound regions co-localize with a bivalent chromatin structure associated with cell fate commitment and differentiation (Fig. 5,6). We speculate that TFII-I is involved in the formation or maintenance of the bivalent chromatin state, although we cannot exclude the possibility that TFII-I recognizes H3K4me3/K27me3-enriched genes and furthermore, the developmental dynamics of TFII-I binding depend on the availability of such bivalent marks. We recently found that during ESC differentiation TFII-I modulates expression of a large group of regulatory genes involved in chromatin modifying and epigenetic reprogramming [5]. Collectively, these data prompted us to adopt the “feed-forward model” of gene regulation (Fig. 7B) previously proposed to explain the ability of transcription factors to recognize multiple genomic loci [34]. We have previously proposed that TFII-I may exert a specific regulatory effect upon the target genes by changing the histone methylation status of bivalent chromatin [35].
According to this model TFII-I binds to the promoters of bivalent genes with delayed expression except of a few developmental regulators (stage 1, stem cells) that together with TFII-I, regulate other genes (stage 2, embryonic tissues) (Fig. 7B). Among TFII-I targets are genes implicated in lineage differentiation (Gsc, members of Hox and Dlx family) and epigenetic modulators (Ezh2 and Nsd1 histone methyltransferase and Hdac4 histone deacetylase).
Materials and Methods
ESC Maintenance and Differentiation
Mouse ESCs Ainv15 (SCRC-1029) and E14tg2a (CRL-1821) were obtained from ATCC, VA. Ainv15 was maintained on the feeder cells in DMEM (Invitrogen, CA) supplemented with 15% fetal calf serum (HyClone Laboratories, UT), 1000 U/ml LIF (Chemicon, CA), non-essential amino acids (Invitrogen, CA), penicillin-streptomycin-L-glutamine and 2-mercaptoethanol. The formation of embryoid bodies was initiated in E14tg2a cells by removal of LIF and 2-mercaptoethanol. After depletion of the feeder cells, 5×105 ESCs were allowed to aggregate in suspension culture in Petri dishes. E14tg2a cells were grown in a feeder-free environment in KD MEM (Gibco, CA) containing 15% knockout serum replacement (Invitrogen, CA), 1000 U/ml LIF (Chemicon, CA), 0.1 mM non-essential amino acids, penicillin-streptomycin-L-glutamine and 2-mercaptoethanol. The cell envelope-associated alkaline phosphatase (ALP) activity was determined using a commercially available kit ALP-10 (Sigma-Aldrich, MO). Briefly, a test cell population was washed twice in PBS, lysed in 0.1% Triton X for 5 min prior to addition of the p-nitrophenyl phosphate buffer.
Culture of JoMa1.3 Cells and siRNA Knockdown of TFII-I Proteins
Mouse neural crest cell line JoMa1.3 [27] was grown on cell culture dishes coated with fibronectin. For maintenance, DMEM:Ham’s F12 (1:1) medium (Invitrogen, CA) was supplemented with 1% N2-Supplement (Invitrogen, CA), 2% B27-Supplement (Invitrogen, CA), 10 ng/ml EGF (R&D Systems Inc., MN), 1 ng/ml FGF (R&D Systems Inc., MN), 100 U/ml Penicilin–Streptomycin (Invitrogen, CA), 200 nM Tamoxifen (Sigma-Aldrich, MO) and 10% chick embryo extract (Gemini Bio-Products, CA). siRNA-mediated knockdown of Gtf2i or Gtf2ird1 in JoMa1.3 cells was performed using ON-TARGETplus SMARTpool siRNA (Dharmacon, IL). Three independent transfections were performed for each gene. To ensure minimal non-specific effects on gene expression we used the control negative siRNA (ON-TARGETplus Non-targeting Pool).
Generation of Tet-inducible BEN Expressing ESC Line
Murine full-length cDNA for Gtf2ird1 (GenBank accession #AY030288) was amplified by PCR using Herculase II DNA polymerase (Stratagene, CA). The PCR product was cloned into pLox vector and resulting construct plox/Gtf2ird1 was co-electroporated with pSalk-Cre into the genetically modified for this purpose cell line Ainv15 [36]. After electroporation stable clones were obtained by selection with 350 µg/ml of G-418 (Gibco, CA), and screened for integration by PCR. Cells were induced for BEN expression with 1 mg/ml doxycycline (Sigma-Aldrich, MO) in media without LIF.
QRT-PCR Analysis
cDNA was synthesized from 0.1–2 µg of RNA with Omniscript Reverse transcription kit (Qiagen, CA) using oligo dT or random N12 primers. Quantitative RT-PCR was performed on Applied Biosystems 7300 or on Bio-Rad MiniOpticon MJ Real-Time PCR system using TaqMan expression assays (Applied Biosystems, CA). The list of assays used in this study is given in Supplemental table S1. PCR reactions were performed in duplicates for each cDNA sample. All transcript levels were normalized to that of Gapdh and the relative expression ratios were calculated using the delta-delta Ct method [37] or the Pfaffl method [38], when probe efficiency was lower than 100%. Statistical analyses were performed using the InStat 3.0b software (GraphPad Software Inc., CA).
Chromatin Immunoprecipitation (ChIP) and ChIP-chip
ChIP experiments were performed with mouse ESC lines E14tg2a and Ainv15 or embryonic tissues using ChIP-IT kit (Active Motif, CA). About 120 mouse embryos on gestational day 10.5 (E10.5) were dissected by a tungsten needle to produce craniofacial embryonic material for ChIP. Chromatin samples were sheared using Branson sonifier 150 (Heinemann, Germany). Chromatin-binding proteins were reverse cross-linked and digested with Proteinase K (Thermo Fisher Scientific, MA). 10 µl of the chromatin supernatant was saved as Input DNA before immunoprecipitation (IP). ChIP experiment was performed with goat anti-BEN polyclonal IgG M-19, goat anti-TFII-I polyclonal IgG V-18, mouse anti-HA monoclonal IgG2b 12CA5 (sc-14714X, sc-9943X and sc-57592, Santa Cruz Biotechnology, CA) and with rabbit anti-TFII-I polyclonal antibodies 4562 (Cell Signaling Technology, MA) and antibody-chromatin complexes were purified using protein G-coated magnetic beads (Active Motif, CA). After reverse cross-linking and purification on QIAquick spin columns (Quagen, CA), immunoaffinity-enriched DNA fragments (IP) and the input samples were amplified using whole-genome GenomePlex Complete WGA kit (Sigma-Aldrich, MO). IP and input samples were labeled in separate reactions with Cy5 and Cy3, respectively, and then were co-hybridized to Mouse 385 K RefSeq Promoter Arrays (Roche NimbleGen, WI). The data were extracted using NimbleScan software. Peaks were detected by searching for four or more probes with a signal above a cut-off value using a 500-bp sliding window. A log2 ratio of the IP versus input samples was calculated and the transcription start site mapping was performed using SignalMap software (Roche NimbleGen, WI). The promoter recruitment was validated experimentally by independent ChIP.
Histone Methylation Analysis
For analysis of histone methylation, siRNA-mediated knockdown of Gtf2i or Gtf2ird1 was performed in mouse ESCs E14tg2a using ON-TARGETplus SMARTpool siRNAs (Dharmacon, IL). Anti-H3K27me3 (39535, Active Motif, CA), anti-H3K4me3 (ab1012, Abcam, MA), and anti-H3 (39163, Active Motif, CA) antibodies were used for ChIP experiment. Quantitative data were obtained by qRT-PCR with SYBR Green and custom-made primers and expressed as a ratio of methylated H3 versus total H3 histone. For co-locaization analysis, H3K4me3 and H3K27me3 ChIP-Seq data were downloaded from NCBI GEO (GSM307618 and GSM307619).
Bioinformatics and Statistical Analysis
The putative TFII-I and BEN binding sites were searched as a pattern consensus on both strands using the MacVector 9.5 software (Oxford Molecular Group, UK). Histone peaks were determined using the peak finding program MACS (Model-based Analysis for ChIP-Seq) from Dana-Farber Cancer Institute and Harvard School of Public Health in Boston, MA (http://liulab.dfci.harvard.edu/MACS/). The TFII-I-binding peaks and methylated regions were considered co-localized if they overlapped or the distance between the closer ends of the two genomic coordinates was less than 200 bp apart from each other. The numbers for the co-localization between all different combinations were converted into percent ratios and used Heatmap2 (http://hosho.ees.hokudai.ac.jp/~kubo/Rdoc/library/gplots/html/heatmap.2.html) package in R to produce the heat-map visualization. Heatmap represents the numbers in color code chart with different colors used to represent different z-scores for the data. To compare DNA sequences the EMBOSS Pairwise Alignment Tools were used (www.ebi.ac.uk/tools/emboss/align/index.html). Statistical analysis was performed with InStat 3.0b (GraphPad Software Inc., CA).
Supporting Information
Density map and novel TFII-I binding consensus sequences. (A) Colocalization of TFII-I binding in mouse ESCs (a) and BEN binding in embryonic craniofacial tissues (b). For the density map, 5744 TFII-I-binding peaks from ESCs (a) and 1520 BEN-binding peaks from embryonic craniofacial tissues (b) were aligned relative to transcription start sites (left panels) and the corresponding BEN-binding peaks (a) or TFII-I-binding peaks (b) are shown in the right panel. Red dashed lines indicate position of the aligned “reference” peaks. (B) The novel consensus sequences recognized by TFII-I factors. (C) The canonical TFII-I and BEN binding motifs.
(DOC)
The gene ontology and KEGG pathway analysis in mouse ESCs. (A) The basic cellular functions. (B) The basic cellular processes. (C) The KEGG pathways. The red and blue bars represent fold enrichment of the TFII-I and BEN bound genes, respectively. The red stars indicate the statistically significant functional categories (p-value 1.5E-1).
(DOC)
Gene ontology and KEGG pathway analysis of TFII-I target genes in mouse embryonic craniofacial tissues. (A) The basic cellular functions. (B) The basic cellular processes. (C) The developmental categories.
(DOC)
siRNA knockdown efficiency of TFII-I and BEN in JoMa cells.
(DOC)
TFII-I binding sites in the ESC promoters.
(XLS)
BEN binding sites in the ESC promoters.
(XLS)
Promoters in ESCs recognized by TFII-I factors.
(XLS)
Promoters in embryonic craniofacial tissues bound by TFII-I.
(XLS)
Promoters in embryonic craniofacial tissues bound by BEN.
(XLS)
Promoters in ESCs recognized by TFII-I and BEN to the same sequence.
(XLS)
Enrichment for transcription factor binding motifs across the TFII-I and BEN bound promoter regions in ESCs.
(XLS)
Enrichment for transcription factor binding motifs in the TFII-I and BEN bound promoter regions in embryonic craniofacial tissues.
(XLS)
The consensus binding motifs within the TFII-I and BEN bound promoter regions.
(DOC)
Pathway analysis in mouse ESCs.
(DOC)
Pathway analysis in mouse embryonic craniofacial tissues.
(DOC)
TFII-I transcription factors target a large set of developmental regulators.
(DOC)
Acknowledgments
pSalk-Cre, pLox and mouse JoMa1.3 cell line are kind gifts from Dr. M. Kyba and Dr. H. Schorle. The authors would like to thank N. Chimge for the Tet-inducible BEN expressing cell line.
Funding Statement
This work was supported by the National Institutes of Health (NIH) grants K02 DE18412 and R01DE017205 and State of Connecticut Stem Cell Initiative 09-SCB-UCHC to DB. The work by SHH, PJ and DGS was supported in part by the funding from State of Connecticut Stem Cell Initiative (Project No. 06SCC04). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bayarsaihan D, Bitchevaia NK, Enkhmandakh B, Tussie-Luna MI, Leckman JF, et al. (2003) Expression of BEN, a member of TFII-I family of transcription factors, during mouse pre- and postimplantation development. Gene Expres Patterns 3: 577–587. [DOI] [PubMed] [Google Scholar]
- 2. Enkhmandakh B, Bitchevaia N, Ruddle F, Bayarsaihan D (2004) The early embryonic expression of TFII-I during mouse preimplantation development. Gene Expr Patterns 4: 25–28. [DOI] [PubMed] [Google Scholar]
- 3. Yoshikawa T, Piao Y, Zhong J, Matoba R, Carter MG, et al. (2006) High-throughput screen for genes predominantly expressed in the ICM of mouse blastocysts by whole mount in situ hybridization. Gene Expr Patterns 6: 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Makeyev AV, Bayarsaihan D (2009) New TFII-I family target genes involved in embryonic development. Biochem Biophys Res Commun 386: 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chimge N, Makeyev AV, Waigel SJ, Enkhmandakh B, Bayarsaihan D (2012) PI3K/Akt-dependent functions of TFII-I transcription factors in mouse embryonic stem cells. J Cell Biochem 113: 1122–1131. [DOI] [PubMed] [Google Scholar]
- 6. Enkhmandakh B, Makeyev AV, Erdenechimeg L, Ruddle FH, Chimge NO, et al. (2009) Essential functions of the Williams-Beuren syndrome-associated TFII-I genes in embryonic development. Proc Natl Acad Sci U S A 106: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vullhorst D, Buonanno A (2005) Multiple GTF2I-like repeats of general transcription factor 3 exhibit DNA binding properties. Evidence for a common origin as a sequence-specific DNA interaction module. J Biol Chem 280: 31722–31731. [DOI] [PubMed] [Google Scholar]
- 8. Bayarsaihan D, Ruddle FH (2000) Isolation and characterization of BEN, a member of the TFII-I family of DNA-binding proteins containing distinct helix-loop-helix domains. Proc Natl Acad Sci USA 97: 7342–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O’Mahoney JV, Guven KL, Lin J, Joya JE, Robinson CS, et al. (1998) Identification of a novel slow-muscle-fiber enhancer binding protein, MusTRD1. Mol Cell Biol 18: 6641–6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ring C, Ogata S, Meek L, Song J, Ohta T, et al. (2002) The role of a Williams-Beuren syndrome-associated helix-loop-helix domain-containing transcription factor in activin/nodal signaling. Genes Dev 16: 820–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ku M, Sokol SY, Wu J, Tussie-Luna MI, Roy AL, et al. (2005) Positive and negative regulation of the transforming growth factor beta/activin target gene goosecoid by the TFII-I family of transcription factors. Mol Cell Biol 25: 7144–7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chimge NO, Makeyev AV, Ruddle FH, Bayarsaihan D (2008) Identification of the TFII-I family target genes in the vertebrate genome. Proc Natl Acad Sci U S A 105: 9006–9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tantin D, Tussie-Luna MI, Roy AL, Sharp PA (2004) Regulation of immunoglobulin promoter activity by TFII-I class transcription factors. J Biol Chem 279: 5460–5469. [DOI] [PubMed] [Google Scholar]
- 14. Lazebnik MB, Tussie-Luna MI, Roy AL (2008) Determination and functional analysis of the consensus binding site for TFII-I family member BEN, implicated in Williams-Beuren syndrome. J Biol Chem 283: 11078–11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen J, Malcolm T, Estable MC, Roeder RG, Sadowski I (2005) TFII-I regulates induction of chromosomally integrated human immunodeficiency virus type 1 long terminal repeat in cooperation with USF. J Virol 79: 4396–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roy AL, Du H, Gregor PD, Novina CD, Martinez E, et al. (1997) Cloning of an inr- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. EMBO J 16: 7091–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ogura Y, Azuma M, Tsuboi Y, Kabe Y, Yamaguchi Y, et al. (2006) TFII-I down-regulates a subset of estrogen-responsive genes through its interaction with an initiator element and estrogen receptor alpha. Genes Cells 11: 373–381. [DOI] [PubMed] [Google Scholar]
- 18. Grueneberg DA, Henry RW, Brauer A, Novina CD, Cheriyath V, et al. (1997) A multifunctional DNA-binding protein that promotes the formation of serum response factor/homeodomain complexes: identity to TFII-I. Genes Dev 11: 2482–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jackson TA, Taylor HE, Sharma D, Desiderio S, Danoff SK (2005) Vascular endothelial growth factor receptor-2: counter-regulation by the transcription factors, TFII-I and TFII-IRD1. J Biol Chem 280: 29856–29863. [DOI] [PubMed] [Google Scholar]
- 20. Crusselle-Davis VJ, Vieira KF, Zhou Z, Anantharaman A, Bungert J (2006) Antagonistic regulation of beta-globin gene expression by helix-loop-helix proteins USF and TFII-I. Mol Cell Biol 26: 6832–6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Makeyev AV, Bayarsaihan D (2011) Molecular basis of Williams-Beuren syndrome: role of TFII-I in craniofacial development. Cleft Plate Craniof J 48: 109–116. [DOI] [PubMed] [Google Scholar]
- 22. Shin H, Liu T, Manrai AK, Liu XS (2009) CEAS: cis-regulatory element annotation system. Bioinformatics 25: 2605–2606. [DOI] [PubMed] [Google Scholar]
- 23.Bailey TL, Elkan C (1994) “Fitting a mixture model by expectation maximization to discover motifs in biopolymers”. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, 28–36, AAAI Press, Menlo Park, California. [PubMed]
- 24. Dennis GJ, Sherman BT, Hosack DA, Yang J, Gao W, et al. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biology 4: P3. [PubMed] [Google Scholar]
- 25. Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salbaum JM, Kappen C (2010) Neural tube defect genes and maternal diabetes during pregnancy. Birth Defects Res A Clin Mol Teratol 88: 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maurer J, Fuchs S, Jager R, Kurz B, Sommer L, et al. (2007) Establishment and controlled differentiation of neural crest stem cell lines using conditional transgenesis. Differentiation 75: 580–591. [DOI] [PubMed] [Google Scholar]
- 28. Dahl JA, Reiner AH, Klungland A, Wakayama T, Collas P (2010) Histone H3 lysine 27 methylation asymmetry on developmentally-regulated promoters distinguish the first two lineages in mouse preimplantation embryos. PLoS One 5: e9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li QR, Xing XB, Chen TT, Li RX, Dai J, et al. (2011) Large-scale phosphoproteome profiles comprehensive features of mouse embryonic stem cells. Mol Cell Proteomics 10: M110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kambere MB, Lane RP (2007) Co-regulation of a large and rapidly evolving repertoire of odorant receptor genes. BMC Neurosci Suppl 3: S2. [DOI] [PMC free article] [PubMed]
- 31. Shykind BM, Rohani SC, O’Donnell S, Nemes A, Mendelsohn M, et al. (2004) Gene switching and the stability of odorant receptor gene choice. Cell 117: 801–815. [DOI] [PubMed] [Google Scholar]
- 32. Vassalli A, Feinstein P, Mombaerts P (2011) Homeodomain binding motifs modulate the probability of odorant receptor gene choice in transgenic mice. Mol Cell Neurosci 46: 381–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hoppe R, Frank H, Breer H, Strotmann J (2003) The clustered olfactory receptor gene family 262: Genomic organization, promoter elements, and interacting transcription factors. Genome Res 13: 2674–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Macquarrie KL, Fong AP, Morse RH, Tapscott SJ (2011) Genome-wide transcription factor binding: beyond direct target regulation. Trends Genet 27: 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bayarsaihan D, Makeyev AV, Enkhmandakh B (2012) Epigenetic modulation by TFII-I during embryonic stem cell differentiation. J Cell Biochem 113: 3056–3060. [DOI] [PubMed] [Google Scholar]
- 36. Kyba M, Perlingeiro RC, Daley GQ (2002) HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109: 29–37. [DOI] [PubMed] [Google Scholar]
- 37. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 38. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Density map and novel TFII-I binding consensus sequences. (A) Colocalization of TFII-I binding in mouse ESCs (a) and BEN binding in embryonic craniofacial tissues (b). For the density map, 5744 TFII-I-binding peaks from ESCs (a) and 1520 BEN-binding peaks from embryonic craniofacial tissues (b) were aligned relative to transcription start sites (left panels) and the corresponding BEN-binding peaks (a) or TFII-I-binding peaks (b) are shown in the right panel. Red dashed lines indicate position of the aligned “reference” peaks. (B) The novel consensus sequences recognized by TFII-I factors. (C) The canonical TFII-I and BEN binding motifs.
(DOC)
The gene ontology and KEGG pathway analysis in mouse ESCs. (A) The basic cellular functions. (B) The basic cellular processes. (C) The KEGG pathways. The red and blue bars represent fold enrichment of the TFII-I and BEN bound genes, respectively. The red stars indicate the statistically significant functional categories (p-value 1.5E-1).
(DOC)
Gene ontology and KEGG pathway analysis of TFII-I target genes in mouse embryonic craniofacial tissues. (A) The basic cellular functions. (B) The basic cellular processes. (C) The developmental categories.
(DOC)
siRNA knockdown efficiency of TFII-I and BEN in JoMa cells.
(DOC)
TFII-I binding sites in the ESC promoters.
(XLS)
BEN binding sites in the ESC promoters.
(XLS)
Promoters in ESCs recognized by TFII-I factors.
(XLS)
Promoters in embryonic craniofacial tissues bound by TFII-I.
(XLS)
Promoters in embryonic craniofacial tissues bound by BEN.
(XLS)
Promoters in ESCs recognized by TFII-I and BEN to the same sequence.
(XLS)
Enrichment for transcription factor binding motifs across the TFII-I and BEN bound promoter regions in ESCs.
(XLS)
Enrichment for transcription factor binding motifs in the TFII-I and BEN bound promoter regions in embryonic craniofacial tissues.
(XLS)
The consensus binding motifs within the TFII-I and BEN bound promoter regions.
(DOC)
Pathway analysis in mouse ESCs.
(DOC)
Pathway analysis in mouse embryonic craniofacial tissues.
(DOC)
TFII-I transcription factors target a large set of developmental regulators.
(DOC)