Abstract
Invasive species are of great interest to evolutionary biologists and ecologists because they represent historical examples of dramatic evolutionary and ecological change. Likewise, they are increasingly important economically and environmentally as pests. Obtaining generalizations about the tiny fraction of immigrant taxa that become successful invaders has been frustrated by two enigmatic phenomena. Many of those species that become successful only do so (i) after an unusually long lag time after initial arrival, and/or (ii) after multiple introductions. We propose an evolutionary mechanism that may account for these observations. Hybridization between species or between disparate source populations may serve as a stimulus for the evolution of invasiveness. We present and review a remarkable number of cases in which hybridization preceded the emergence of successful invasive populations. Progeny with a history of hybridization may enjoy one or more potential genetic benefits relative to their progenitors. The observed lag times and multiple introductions that seem a prerequisite for certain species to evolve invasiveness may be a correlate of the time necessary for previously isolated populations to come into contact and for hybridization to occur. Our examples demonstrate that invasiveness can evolve. Our model does not represent the only evolutionary pathway to invasiveness, but is clearly an underappreciated mechanism worthy of more consideration in explaining the evolution of invasiveness in plants.
Invasive species have always held a special place for ecologists and evolutionary biologists. Successful invaders that have colonized new regions within historical time provide real-life examples of ecological and evolutionary change. The demographic change from a small number of colonists to a sweeping wave of invaders is a dramatic ecological event. Likewise, those demographic changes—a founder event followed by a massive increase in numbers—may have dramatic evolutionary consequences. Not surprisingly, whole books have been dedicated to the basic science of invasive species (for example, see refs. 1 and 2).
Also, the applied biology of invasive species has become increasingly important as intentional and unintentional anthropogenic dispersal moves species from continent to continent at unprecedented rates. Invasive plants and animals are often thought of as agricultural pests, but they also pose a hazard for a variety of human concerns, including health, transportation, and conservation (3). Invasive species not only directly impact human well being, but they also are recognized as agents that alter community structure and ecosystem function (for example, see ref. 4). In the United States alone, the damage wrought by invasive species totals approximately $122 billion per year (5).
Only a tiny fraction of introduced species become successful invasives (6). Given that invasives are important for so many reasons, considerable effort has been spent trying to develop generalizations to determine which species are likely to become successful. In particular, ecological, taxonomic, and physiological correlates of invasive success have been sought to predict which introduced species might become successful (for example, see refs. 7–11). Less frequently, possible genetic correlates have been sought (for example, see ref. 12). Very little attention has been given to the possibility of the evolution of invasiveness after colonization.
Are invasives “born” (that is, are they released from fitness constraints) or are they “made” (that is, do they evolve invasiveness after colonization)? The fact that certain correlates of invasive success have been identified suggests that invasives are born. Also, Darwin's (13) observation that non-native genera are more likely to be successful invaders than are native genera supports the view that successful invasives are preadapted and do not evolve invasiveness in situ. Certain specific cases of invasives fit this model well. For example, the fact that invasiveness can sometimes be reversed by a biological control agent [(e.g., prickly pear in Australia (14) and Klamath weed in the American Pacific Northwest (15)] suggests that invasiveness can appear simply once an organism is released from its primary biological enemies. Also, it has been observed that “a strong predictor of invasiveness . . . is whether the organism has been invasive . . . elsewhere” (ref. 16, p. 627). Although such correlates may be statistically strong, they are typically weak in predicting invasions, leading one reviewer of the field to assert, “serendipity is often an important element in successful invasions” (ref. 12, p. 655) and another to lament, “It could be that invasions . . . are intrinsically unpredictable” (ref. 17, p. 10).
But for some successful invasive species, it may well be that a series of events after colonization is more important than intrinsic “colonizing ability.” In fact, two enigmatic phenomena associated with successful invasives suggest that many species are not preadapted to become successful invasives and that the right circumstances must transpire for invasiveness to occur (and perhaps evolve). The first is the observation that there is often a considerable lag phase between the establishment of local populations and their aggressive spread (16, 18). For example, Kowarik (19) reviewed 184 invasive woody species with known dates of first cultivation in Brandenburg, Germany. The mean delay in invasion was 131 years for shrubs and 170 years for trees. Delays on the order of decades may occur for herbaceous invasives as well (20). If these species were simply preadapted, then we would expect evidence of invasiveness relatively quickly. Second, multiple introductions often are correlated with the eventual success of non-native species establishment and invasiveness (21). For example, North America's most successful invasive birds, the European Starling and the House Sparrow, both became invasive only after repeated introduction (22). Collectively considered, these observations suggest genetic change and adaptive response play a role in the ultimate establishment of some invasive species.
We contend that hybridization may result in critical evolutionary changes that create an opportunity for increased invasiveness. As Anderson and Stebbins (23) pointed out, “hybridization between populations having very different genetic systems of adaptation may lead to . . . new adaptive systems, adapted to new ecological niches” (ref. 23, p. 378). Stebbins further examined what he came to call “the catalytic effects of such hybridization” (24) in subsequent articles (25, 26). Although Anderson and Stebbins did not consider the case of invasive species, they did acknowledge that human activities could be a powerful agent for bringing together cross-compatible species that had been previously isolated by ecology or geography.
Indeed, Abbott (27) observed that interspecific hybridization involving non-native plant species has often served as a stimulus for the evolution of entirely new, and sometimes invasive, species. Specifically, he noted that hybridization involving a non-native species and another (either native or non-native) has led to a number of new sexually reproducing plant species. The 10 examples he gives are either stabilized introgressants or allopolyploids. Some of these species have remained localized, but most have spread successfully far beyond their sites of origin. The latter group of his examples, plus many more we have accumulated, are listed in Table 1.
Table 1.
Invasive taxa that evolved after intertaxon hybridization
| Derived taxon | Parent taxa | Family | Habit of hybrid lineage | Site of taxon's origin | Evidence beyond morphology | Ref. | How stabilized? | Invasiveness | Occurs in human-disturbed areas? |
|---|---|---|---|---|---|---|---|---|---|
| Amelanchier erecta | A. humulis × A. “clade B” | Rosaceae | Shrub | North America | n | 28 | Agamospermy | Highly invasive relative to congeners | Yes |
| Bromus hordeaceus | B. arvensis and B. scoparius | Poaceae | Annual herb | Europe | c, i, n | † | Allopolyploid | Aggressive ruderal | Yes |
| Cardamine insueta | C. rivularis × C. amara | Brassicaceae | Perennial herb | Europe | c, n, o | 30 | Allopolyploid | Successfully colonizing disturbed sites | Yes |
| Cardamine schulzii | C. rivularis × C. amara | Brassicaceae | Perennial herb | Europe | c, n, o | 30 | Allopolyploid | Successfully colonizing disturbed sites | Yes |
| Circaea × intermedia | C. alpina × C. lutetiana | Onagraceae | Perennial herb | Europe | as, s | 31 | Clonal growth | Sometimes a weed, often occurs in absence of one or both parents | Yes |
| Fallopia × bohemica | F. japonica* × F. sachalinensis* | Polygonaceae | Shrub | Europe | c, n, s | 32 | Clonal growth | Noxious weed | Yes |
| Glyceria × pedicillata | G. fluitans × G. notata | Poaceae | Perennial herb | Europe | s | 31, 33 | Clonal growth | “Example of a successful … sterile hybrid” | Yes |
| Helianthus annuus spp. texanus | H. annuus* × H. debilis spp. cucumerifolius | Asteraceae | Annual herb | North America | c, n, o | 34 | Recombinant | Weed of disturbed areas | Yes |
| Mentha × verticillata | M. aquatica × M. arvensis | Lamiaceae | Perennial herb | Europe | s | 33 | Clonal growth | Often in the absence of either parent | Yes |
| Nasturtium sterile | N. microphyllum × N. officinale | Brassicaceae | Perennial herb | Europe | c | 35 | Recombinant | Disturbed area weeds | Yes |
| Oenothera glazioviana (O. erythrosepala, O. lamarckiana) | O. hookeri* × O. biennis* | Onagraceae | Biennial herb | Europe | as, c | 36 | Permanent translocation heterozygosity | Weed | Yes |
| Senecio squalidus | S. aethensis* × S. chrysanthemumifolius* | Asteraceae | Perennial herb | Europe | i, o | 37, 38 | Recombinant | Rapidly spreading | Yes |
| Senecio vulgaris var. hibernicus | S. v. var. vulgaris × S. squalidus* | Asteraceae | Annual herb | Europe | as, c, i | 27 | Recombinant | Rapidly becoming ubiquitous | Yes |
| Sorghum almum | S. propinquum* × S. bicolor* | Poaceae | Perennial herb | South America | c, n | 39 | Allopolyploid | Weed | Yes |
| Spartina anglica | S. alterniflora* × S. maritima | Poaceae | Perennial herb | Europe | c, i | 40 | Allopolyploid, clonal growth | Noxious weed | Yes |
| Stachys × ambigua | S. palustris × S. sylvatica | Lamiaceae | Perennial herb | Europe | c, s | 31 | Clonal growth | Weed | Yes |
| Tragopogon mirus | T. dubius* × T. porrifolius* | Asteracae | Biennial herb | North America | c, i, n, o | 41 | Allopolyploid | Substantial increase in range and numbers | Yes |
| Tragopogon miscellus | T. dubius* × T. pratensis* | Asteracae | Biennial herb | North America | c, i, n, o | 41 | Allopolyploid | Substantial increase in range and numbers | Yes |
as, Artificial synthesis; c, cytological; i, isozymes; n, nuclear DNA; o, organelle DNA; s, full or partial sterility.
Signifies non-natives.
† Ainouche, M. L. & Bayer, R. J. (1996) Am. J. Bot. 83, Suppl., 135 (abstr.).
Abbott, Anderson, and Stebbins focused on interspecific hybridization. But their ideas should work equally well for hybridization among previously isolated populations of the same species. Therefore, we proceed below with a broad perspective.
We extend the ideas of Stebbins, Anderson, and Abbott to specifically address hybridization as a stimulus for the evolution of invasiveness. We restrict our examples to plants, but the model we develop may apply to other organisms as well. Below, we first provide many examples in which hybridization seems to have served as a stimulus for the evolution of a new invasive line. Second, we explain why plants with a history of hybridization may have a fitness advantage relative to those without such a history. Third, we discuss some scenarios that might lead to such hybridization. Finally, we examine how our model for interspecific hybridization could work equally well for hybridization between previously isolated populations of the same species.
Materials and Methods
We sought at least 25 well documented examples of the evolution of invasiveness in plants after a spontaneous hybridization event. We did not intend our review to be exhaustive, but instead concentrated on finding the most convincing examples.
We used four criteria for choosing our examples:
(i) More evidence than intermediate morphology must be available to support the hybrid origin of the invasive lineage. Intermediate morphology does not necessarily support the hypothesis of hybridity (42). Species-specific genetically based traits such as chromosomes, isozymes, and/or DNA-based markers provide more reliable evidence for hybrid parentage. The hypothesis also can receive support from comparison of artificially synthesized hybrids with the putative spontaneous hybrids and from the relative sterility of the putative hybrids compared with that of the parental species.
(ii) The hybridization event preceding the evolution of invasiveness must be spontaneous. Many artificial hybrids have escaped from cultivation to become naturalized invasives (e.g., certain mints, comfrey, poplars, and watercress; cf. ref. 31).
(iii) The hybrid derivatives must be established as a novel, stabilized lineage and not simply as transient, localized hybrid swarms. In some cases, genetic or reproductive mechanisms may stabilize hybridity (e.g., allopolyploidy, permanent translocation heterozygosity, agamospermy, and clonal spread; cf. ref. 43). Some have become new, reproductively isolated, recombinant species. In other cases, introgression may be so extensive that the hybrid lineage swamps out one or both of its parents, becoming a coalescent complex.
(iv) The new lineage must exhibit some degree of invasiveness. We define invasive populations as those that are capable of colonizing and persisting in one or more ecosystems in which they were previously absent. The minimal criterion of invasiveness for our hybrid derivative is that it must replace at least one of its parental taxa or invade a habitat in which neither parent is present. We hold to this criterion for those few cases in which one parent is itself invasive.
We did not restrict ourselves to examples of hybridization involving one or more non-natives, because the evolution of invasiveness by hybridization should be independent of the geographical source of the parental material.
Results and Discussion
We found 28 examples representing 12 families where invasiveness was preceded by hybridization; these examples are detailed in Tables 1 and 2. We encountered another 2 dozen or so examples of invasive lineages thought to have a hybrid origin (e.g., Lonicera × bella, Oenothera wolfii × Oenothera glazioviana, and Platanus racemosa × Platanus acerifolia). The latter did not sufficiently meet our criteria, mostly because only morphology was offered to support their putative hybrid origin.
Table 2.
Invasive lineages that evolved after intertaxon hybridization
| Parent taxa | Family | Habit of hybrid lineage | Site of new lineage's origin | Evidence beyond morphology | Ref. | How stabilized? | Invasiveness | Occurs in human-disturbed areas? |
|---|---|---|---|---|---|---|---|---|
| Avena barbata* × A. strigosa* | Poaceae | Annual herb | North America | i | M. Blumler, personal communication | Selfing genotype | Spreading rapidly | Yes |
| Beta vulgaris spp. vulgaris* × B. v. spp. maritima | Chenopodiaceae | Annual herb | Europe | n, o, b | 44 | Coalescent complex | Noxious weed | Yes |
| Carpobrotus edulis* × C. chilense | Aizoaceae | Perennial herb | North America | as, i | 45, 46 | Clonal growth | Replacing one parent | Yes |
| Lythrum salicaria* × L. alatum | Lythraceae | Perennial herb | North America | d, s | 47 | Clonal growth | Noxious weed | Yes |
| Onopordum acanthium* × O. illyricum* | Asteraceae | Perennial herb | Australia | n | 48 | Coalescent complex | Weed | Yes |
| Raphanus raphanistrum* × R. sativus* | Brassicaceae | Annual herb | North America | as, c, s | 49 | Coalescent complex | Weed | Yes |
| Rhododendron ponticum* × R. catawbiense* | Ericaceae | Shrub | Europe | n, o | 37, 50 | Coalescent complex | Noxious weed | Yes |
| Secale cereale* × S. montanum* | Poaceae | Perennial herb | North America | i, s | 51 | Coalescent complex | Weed | Yes |
| Spartina alterniflora* × S. foliosa | Poaceae | Perennial herb | North America | as, n, s | 52, 53 | Clonal growth | Replacing one parent | Yes |
| Viola riviniana × V. reichenbachiana | Violaceae | Perennial herb | Europe | c, n | 54 | Coalescent complex | Invading polluted forests | Yes |
as, Artificial synthesis; c, cytological; i, isozymes; n, nuclear DNA; o, organelle DNA; s, full or partial sterility.
Signifies non-natives.
In some of our examples, the hybrid-derived lineages have already achieved a taxonomic epithet (detailed in Table 1). In other cases, a new invasive lineage has been identified and studied but not yet named, to our knowledge (detailed in Table 2). In each case, we give the parental species, plant family, habit of the hybrid derivative, its site of origin, and the evidence supporting a history of hybridization for the new lineage. We cite one or two good comprehensive references for each example. In many cases, the best reference is an article or review that cites many supporting sources of empirical research. To list each of those is beyond the scope of this paper. Finally, we present how the novel lineage is maintained and indicate the scope of its invasiveness, including whether the lineage is known to grow, at least in some instances, in human-disturbed areas.
Some characteristics of our sample seem to be quite broad; many diverse families are represented. Hybridity is stabilized by a variety of mechanisms, from cytological (polyploidy and permanent translocation heterozygosity) to apomictic (agamospermy and clonal growth). In many cases, the new hybrid lineage is a coalescent complex that absorbs one or both parental types, especially among the unnamed cases in Table 2. Likewise, invasiveness runs the gamut from cases in which the new hybrid lineage is displacing a parent or spreading into a new community to cases in which the hybrid lineage is an established noxious weed.
But we also note some interesting trends in our sample. Life history traits tend to be concentrated within a narrow subset of those traits possible. Almost all of our examples are herbaceous (24 of 28). However, the majority of the cases involve perennial species (19 of 28). Interestingly, these characteristics also are found to be frequent among cases of spontaneous hybridization. For example, Ellstrand et al. (55) examined the 10 genera in the British flora with the highest number of different spontaneous hybrids. They found that most were perennial herbs.
These trends make sense. Perennial hybrids will persist longer than will annuals, giving more time for stabilization opportunities to occur, especially if clonal reproduction is available (as it is in about half of our examples). The predominance of herbaceous over woody examples in our Tables is consistent with Harper's (56) prediction that colonizing plants allocate more resources to reproductive rather than to vegetative growth. Iteroparous perennial herbs appear to maximize fitness by investing in sexual structures and vegetative spread instead of investing in permanent structures (57).
It has been suggested that Old World or temperate ecosystems may be less susceptible to invasives than are New World or tropical ecosystems, and that most successful plant invaders have Mediterranean or Central European origins (58). The rationale for this view is that Old World species have had a much longer evolutionary history with human disturbance, particularly agricultural disturbance. These views have been modified by the recognition that historical patterns of plant invasions simply may have followed paths of commerce; indeed, numbers of invasive species in the Old World have increased as New- to Old World commerce has increased (59). Interestingly, most of our examples come from the Old World, not the New. Finally, all but two examples (Sorghum almum and the Onopordum hybrids) are Holarctic, and all are temperate. These latter patterns may have more to do with the geographic distribution of evolutionary biologists than with any biological phenomenon.
More than half the cases (18 of 28) involve at least one non-native parental taxon. This correlate may be an artifact of how difficult it is to reconstruct evolutionary events; observed changes in the distribution of non-natives provide a historical context for identifying a hybridization event. “Frequently, the history of these events is known, allowing examination of the factors which may have favoured the spread of a new taxon following its origin” (ref. 27, p. 402). On the other hand, the correlate may have real evolutionary significance. Human-mediated dispersal may magnify the potential for hybridization by increasing the migration distances and the number of independent colonization events severalfold as compared with other processes.
All of our invasives grow in habitats characterized by human disturbance, at least in part of their range. Anderson and Stebbins (23) predicted that human disturbance should both mix previously isolated floras as well as create novel niches well suited to novel hybrid-derived genotypes, that is, to create niches better suited to intermediates or segregants than to the parental species. We caution that human-disturbed habitats may be much better studied and visited more frequently than those isolated from human activity.
How Can Hybridization Stimulate the Evolution of Invasiveness?
We are well aware that not all hybridization leads to increased fitness or adaptive evolution (60). But hybridization can lead to adaptive evolution in a number of ways. We examine some hypotheses that describe how hybridization can catalyze the evolution of invasiveness, gaining support from our examples in Tables 1 and 2 when appropriate. The following hypotheses are not likely to be exhaustive nor are they necessarily mutually exclusive.
Evolutionary novelty.
The generation of novel genotypes is the most common hypothesis for hybridization's role in adaptive evolution (for example, see refs. 23, 25, 26, 27, 60, 61, and 62). Stebbins (26) explains it succinctly: “. . . recombination which inevitably takes place in . . . fertile progeny of hybrids gives rise to a large quantitative increase in . . . the gene pool. . . . Although this recombination gives rise to a great preponderance of genotypes which are not well adapted to any environment, nevertheless a minority of them may represent better adaptations to certain environments than do any of the genotypes present in the parental species populations” (ref. 26, p. 26).
One of our examples seems to fit this model perfectly. When sugar beets (which are biennials) are grown for seed production near the Mediterranean Sea, some of their seed is sired by nearby populations of wild beets (which are annuals). Therefore, sugar beet seed grown for commercial purposes in northern Europe has a fraction of hybrid seed. The resulting hybrid plants are morphologically similar to the crop but are annuals, bolting, flowering, and setting seed, leaving a woody root that cannot be sold, that in fact damages harvesting and processing machinery (44). These beet hybrids have given rise to weedy lineages, whose evolutionary novelty of annuality preadapts them for invasive success in cultivated beet fields.
Additional support for this hypothesis comes from invasive hybrid lineages that colonize well defined communities that have not been colonized by either parent. Our Tables supply at least three such examples. Viola riviniana and Viola reichenbachiana hybridize occasionally throughout Europe (31). But in central Germany, a hybrid lineage has successfully colonized pine forests affected by calcareous pollutants (54). Our second example involves Rhododendron ponticum in Britain, which colonizes areas much colder than those of its native range in Iberia. This wider ecological tolerance is correlated with its history of hybridization in Britain with the cold-tolerant Rhododendron catawbiense from North America (50). Our final example is Spartina anglica of the British Isles, an allopolyploid derivative of the native Spartina maritima and Spartina alterniflora, introduced from the east coast of North America. “After initial colonization of an estuary, the species characteristically becomes a dominant component of the marsh, producing extensive and dense monospecific swards. In contrast, the progenitor species have retained a limited distribution” (ref. 63, p. 393).
We have numerous examples in our Tables of invasive hybrid derivatives that either occur in the absence of either parent or are outcompeting one or both parents. It is not clear that those examples (and even the three detailed above) are necessarily cases of evolutionary novelty or just cases of superior fitness attributable to fixed heterosis (see Fixed heterosis below). It is always possible that both novelty and heterosis may occur simultaneously. Further support for the hypothesis at hand could come from experimental studies that specifically compare the fitness of hybrid-derived lines to their parental types under a variety of different environmental parameters.
Evolutionary novelty may result from the fixation of intermediate traits, from the recombination of traits from both parents, or from traits that transgress the phenotype of both parents. Although transgressive traits are well known to occur in plant hybrids and their derivatives (64), recently they have been found to be so frequent that it has been posited that “transgression is the rule rather than the exception” (ref. 65, p. 363). Of the cases mentioned above, it seems that novelty in Beta is caused by the recombination of traits from both parents and that novelty in Viola involves a trait that transgresses the niche of the parent taxa.
Genetic variation.
Recombination in hybrids generates both novelty and variation. A hypothesis related to the one just discussed is that the increase in genetic variation produced in a hybrid lineage can, in itself, be responsible for the evolutionary success of that lineage (26). We recognize that this argument falls within the category of “group selection.” But we also recognize that invasiveness is itself a group trait, one that is defined by the spread and persistence of groups of individuals, one that cannot be measured from a single individual.
Overall, at the population level, early successional plant species have about the same level of genetic variation as those occurring later in succession (for example, see ref. 66). Nonetheless, in our examples of Raphanus in California (49), of Secale in California (ref. 51 and references therein), and of Viola in Germany (54), the hybrid-derived populations were found to have much more genetic variation than were those of the parental species. Not surprisingly, all of those examples involve freely recombining “coalescent complexes” as opposed to our examples in which the genotype is tightly restrained from recombination. Thus, although these examples are compatible with the genetic variation hypothesis, rigorous experimental work with such systems would be a better test of this idea.
Fixed heterosis.
Genetic or reproductive mechanisms that stabilize hybridity (e.g., allopolyploidy, permanent translocation heterozygosity, agamospermy, and clonal spread) also will fix heterotic genotypes. It may well be that the fitness boost afforded by fixed heterozygosity is all that is necessary to make a hybrid lineage invasive. Given the ubiquity of heterosis in both agricultural and natural systems, we are surprised how rarely fixed heterosis is posited as a role of hybridization in adaptive evolution (but see ref. 43). The majority of our examples (especially in Table 1) are capable of fixing heterotic genotypes by agamospermy (e.g., Amelanchier), by allopolyploidy (e.g., Bromus, Cardamine, Sorghum, and Tragopogon), by permanent translocation heterozygosity (Oenothera), and by clonal spread (e.g., Circaea, Fallopia, Glyceria, Mentha, and Stachys).
The case of the invasive S. anglica in the British Isles is perhaps our most notorious example (40, 63). This species originated by chromosome doubling of the sterile hybrid between the Old World S. maritima and the New World S. alterniflora. Genetic analysis found fixed heterozygosity at many of this species' loci, but also showed that S. anglica is almost totally lacking in genetic variation among individuals. Despite its relatively narrow ecological amplitude, it has invaded intertidal flats, replacing more diverse native plant communities, altering succession, and limiting the availability of food to wading birds.
But note that we also were able to use S. anglica as a possible example of invasive success attributable to evolutionary novelty (see Evolutionary novelty above). It is not clear whether invasive success in S. anglica and in our other examples is caused by (i) the fitness benefits conferred by heterosis, (ii) the fixation of an evolutionarily novel genotype by a mode of reproduction that frustrates recombination, or (iii) both. Common garden experiments could test these hypotheses by asking whether hybrids have superior fitness to one or both parental types under specific environmental conditions. We are aware of one such study among our examples, involving Carpobrotus and demonstrating heterosis in the hybrids (46).
Dumping genetic load.
Populations with a history of isolation and a small population size may accumulate detrimental mutations. In such populations, mildly deleterious alleles become fixed, leading to slow erosion of average fitness (see examples in refs. 67 and 68). Hybridization between such populations can afford an opportunity to escape from this mutational load, particularly if recombination permits selection to act to reduce the frequency of detrimental alleles. If recombination creates genotypes with reduced load, then they and their descendants will enjoy increased fitness relative to their progenitors, even without fixed heterozygosity. In fact, certain stabilized diploid hybrid segregates have been shown to maintain higher viability and fecundity than do their parental taxa (L. Rieseberg, unpublished data). We are not aware of prior discussions suggesting that hybridization might stimulate adaptive evolution through dumping genetic load. Nonetheless, the fitness gained might in itself be sufficient to account for invasiveness, especially if invasiveness comes at the expense of the replacement of one or both of the parental species.
Measuring genetic load is a challenging area of experimental quantitative genetics. Presently, it would be difficult to test this hypothesis without being able to assess the relative load of the hybrid derivative versus that of the parental species. We are not aware of any experimental work that has attempted such a comparison.
Human Activities and Some Hybridization Scenarios.
The following anthropogenic activities could enhance both the likelihood of hybridization and the likelihood of forming new niches that favor hybrid derivatives.
(i) Bringing together previously isolated populations.
Humans have become an ecologically significant vector of dispersal, often moving species at high rates and over long distances (for example, see ref. 69). Modern transportation has accelerated that process, including bringing together cross-compatible species that previously were geographically isolated. More than one-third of our invasive hybrid derivatives involves cases in which both parental species were introduced to the location where the initial hybridization event occurred. Another 25% involve cases in which one parent was introduced and the other was native. In most cases in which at least one parental species is introduced, the dispersal involved was on the order of thousands of kilometers. In fact, in all but 3 of the 18 cases, the introduced parental species were native to another continent.
(ii) Opening new “hybrid” zones.
Human activities often result in ecological disturbance. Anderson (70) noted that disturbance, human or otherwise, opens an array of niches that might be better suited for hybrids than for their parents. Furthermore, Stebbins (25) pointed out that, with disturbance, “the initial occurrences of hybridization [will] be in many instances, much more frequent” (ref. 25, p. 248). Although all of our examples of invasive hybrid derivatives occur at least partially in disturbed sites, some of them are found almost exclusively in human-disturbed sites (Amelanchier, Bromus, Cardamine, Helianthus, Nasturtium, and Viola). It is interesting to note that these examples more frequently involve cases in which long-distance dispersal is not a factor.
We hypothesize, then, that human activities can encourage hybridization through (i) long-distance dispersal that brings together previously isolated but closely related taxa, (ii) disturbance that provides habitat suitable for hybrid progeny, or (iii) a combination of dispersal and disturbance. Once hybridization has occurred, if invasiveness evolves, it may do so instantly, for example, as a genotype fixed by a mode of reproduction that restricts recombination, or more slowly, for example, if selection works to sieve out the best adapted genotypes among an array of recombinants.
Can Hybridization Within Taxa Lead to Invasiveness?
There is no reason why the observations above should be restricted to interspecific hybridization. We hypothesize that a hybridization event among well differentiated populations of the same species may act in the same way as does hybridization among species to serve as a stimulus for the evolution of invasiveness. Introduction of distantly related individuals of the same species from different parts of its range may yield an evolutionary stimulus that is essentially the same as is the introduction of different species.
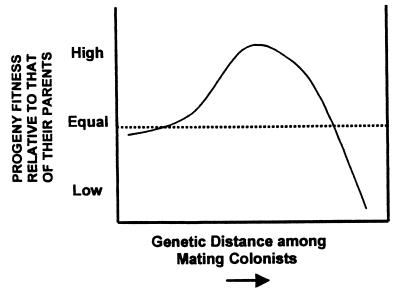
Just as with interspecific hybridization, we do not expect all intraspecific hybridization events to lead to invasiveness. One can posit an optimal level of relatedness yielding the genotypes most likely to become invasive (Fig. 1). Our arguments are similar to those developed to explain an optimal outcrossing distance (71). Hybridization among very closely related populations should not result in any evolutionary changes different from matings within a population. Likewise, very distantly related populations may have evolved cross-incompatibility or produce sterile or otherwise unfit progeny. Thus, we would expect that only a small fraction of interpopulation combinations would yield progeny with superior fitness as compared with their parents. Still, those progeny might not become invasive in an environment that was limiting abiotically (e.g., too saline or xeric) or biotically (e.g., by predators or parasites).
Figure 1.
As genetic distance between mating colonists increases, so too should heterosis in their progeny—up to a point—then, progeny fitness declines as outbreeding depression becomes important.
Nonetheless, if hybridization among populations of the same taxa played an important role in the evolution of invasiveness, then we might expect certain correlates for the appearance of invasiveness. First, we would expect that invasiveness would occur after multiple introductions of a species, because multiple introductions would be necessary for providing genotypes from disparate sources. In fact, species that are intentionally introduced would have an advantage in this regard. Second, we would expect that invasiveness would occur after a lag time, during which hybridization and selection would act to create and increase invasive genotypes. As noted in our introduction, both of these phenomena have occurred so frequently that they have attracted the attention of students of invasive species. In fact, invasive species often originate from multiple foci, each with an independent origin (for example, see refs. 72 and 73). If these foci spread and coalesce, there is an opportunity for hybridization among these independent lineages.
Finally, we might expect that if the evolution of invasiveness followed a bout of hybridization between well differentiated populations, then the resulting populations should likely be more genetically diverse than were their progenitors. This suggestion may seem surprising because of the commonly held view that invasives should be relatively genetically depauperate as a result of the bottlenecks associated with their colonization dynamics (21). On the other hand, hybridization between well differentiated populations resulting from introductions from different sources ought to leave relatively high levels of within-population polymorphism as a “signature.”
We have found two such examples. Echium plantagineum is a noxious weed of Australia. The average population there was found to be more diverse than were those genetically analyzed in its native range in Europe (74). This species has been introduced more than once to Australia, both intentionally and unintentionally (75). Similarly, North American populations of the introduced weed cheatgrass, Bromus tectorum, were found to have increased within-population genetic variation as compared with populations from its source range in Europe and northern Africa (76). Again, there is ample evidence of multiple introductions (77).
Conclusions
Discussions of the population biology of invasives have focused largely on their ecology and on the evolutionary consequences of the invasive process. The evolution of invasiveness as an adaptive trait has been largely neglected. We have extended—and, indeed, hybridized—the ideas of Stebbins, Anderson, and Abbott concerning the evolutionary significance of hybridization to offer one model for the evolution of invasiveness. That is, hybridization can, through one or more mechanisms, catalyze the evolution of invasiveness. Human dispersal and human disturbance both act to accelerate the process and increase the opportunities for hybrid lineages to take hold. The process is not unique to plants. In fact, evidence recently has emerged that “a new, aggressive Phytophthora pathogen of alder trees in Europe” seems to have arisen through interspecific hybridization (ref. 78, p. 5878). Likewise, hybridization between different honeybee subspecies has given rise to the infamous Africanized bees of the New World (79).
Certain caveats are in order. We recognize that only a fraction of hybridization events will lead to the evolution of invasiveness. We do not claim that all invasive species have evolved invasiveness. As we note in our introduction, sometimes certain ecological explanations appear to be the most parsimonious, such as encountering an unfilled niche, competitive superiority, or ecological release. Nor do we claim that hybridization is the sole evolutionary pathway to invasiveness. Other evolutionary pathways to invasiveness already have received some attention. For example, weeds have evolved to mimic unrelated crops and have become successful invaders of agroecosystems (80). Also, Jain and Martins (29) observed that a single gene mutation apparently is responsible for the appearance of invasiveness of rose clover in California.
At the moment, evolution of invasiveness remains an underappreciated area of research on a topic of great applied and basic importance. We have shown that one way to get a handle on studying such evolution is to use examples that have a genetic signature for reconstructing past events. Any other pathways in which past events can be reconstructed should be equally valuable for study. We anticipate that the study of the evolution of invasiveness should be able to provide answers for why invasiveness occurs in some cases and does not occur in others.
Acknowledgments
We thank V. Symonds, K. Gallagher, and N. Sherman for discussions early in the development of the manuscript and also thank the following for their comments on draft versions of the manuscript: R. Abbott, D. Crawford, J. Hamrick, I. Parker, L. Rieseberg, A. Snow, P. Soltis, and R. Whitkus. We are grateful for feedback from the 1998 Ecological Genetics Group Meeting and the first workshop of the Collaboratory on the Population Biology of Invasive Species (funded by a grant from the National Science Foundation through a grant to the University of California at Irvine). This work was supported by funding from a University of California Competitive grant (1997–980069) and an Environmental Protection Agency grant (R-826102–01-0) to N.C.E., and by National Science Foundation Grants 9973734 to K.A.S. and 9322795 to K.A.S. and C. D'Antonio.
Footnotes
This paper was presented at the National Academy of Sciences colloquium “Variation and Evolution in Plants and Microorganisms: Toward a New Synthesis 50 Years After Stebbins,” held January 27–29, 2000, at the Arnold and Mabel Beckman Center in Irvine, CA.
References
- 1.Elton C S. The Ecology of Invasion by Animals and Plants. London: Chapman & Hall; 1958. [Google Scholar]
- 2.Mooney H A, Drake J A, editors. Ecology of Biological Invasions of North America and Hawaii. New York: Springer; 1986. [Google Scholar]
- 3.United States Congress, Office of Technology Assessment. Harmful Non-Indigenous Species in the United States. Washington, D.C.: U.S. Government Printing Office; 1993. [Google Scholar]
- 4.Horvitz C C, Pacarella J B, McMann S, Freedman A, Hofstetter R H. Ecol Appl. 1998;8:947–974. [Google Scholar]
- 5.Pimentel D, Lach L, Zuniga R, Morrison D. BioScience. 2000;50:53–65. [Google Scholar]
- 6.Williamson M. Experientia. 1993;49:219–224. [Google Scholar]
- 7.Bazzaz F A. In: Ecology of Biological Invasions of North America and Hawaii. Mooney H A, Drake J A, editors. New York: Springer; 1986. pp. 96–110. [Google Scholar]
- 8.Daehler C C. Biol Conserv. 1998;84:167–180. [Google Scholar]
- 9.Pyšek P. In: The Ecology and Evolution of Clonal Plants. De Kroon H, van Groenendael J, editors. Leiden, The Netherlands: Backhuys; 1997. pp. 405–427. [Google Scholar]
- 10.Pyšek P. Oikos. 1998;82:282–294. [Google Scholar]
- 11.Rejmanek M. Biol Conserv. 1996;78:171–181. [Google Scholar]
- 12.Gray A J. Philos Trans R Soc London B. 1986;314:655–674. [Google Scholar]
- 13.Darwin C. On the Origin of Species. London: Murray; 1859. [Google Scholar]
- 14.Dodd A P. In: Biogeography and Ecology in Australia. Keast A, Crocker R L, Christian C S, editors. The Hague: W. Junk; 1959. , Monographiae Biologicae no. 8. [Google Scholar]
- 15.Huffaker C B, Kennett C E. J Range Manage. 1959;12:69–82. [Google Scholar]
- 16.Ewel J J, O'Dowd D J, Bergelson J, Daehler C C, D'Antonio C M, Gomez D, Gordon D R, Hobbs R J, Holt A, Hopper K R, et al. BioScience. 1999;49:619–630. [Google Scholar]
- 17.Williamson M. Ecography. 1999;22:5–12. [Google Scholar]
- 18.Mack R N. In: Studies on Plant Demography: John L. Harper Festschrift. White J, editor. London: Academic; 1985. pp. 127–142. [Google Scholar]
- 19.Kowarik I. In: Plant Invasions: General Aspects and Special Problems. Pyšek P, Prach K, Rejmanek M, Wade P M, editors. The Hague: SPB Academic; 1995. pp. 15–38. [Google Scholar]
- 20.Pyšek P, Prach K. J Biogeogr. 1993;204:413–420. [Google Scholar]
- 21.Barrett S C H, Husband B C. In: Plant Population Genetics, Breeding, and Genetic Resources. Brown A H D, Clegg M T, Kahler A L, Weir B S, editors. Sunderland, MA: Sinauer; 1990. pp. 254–278. [Google Scholar]
- 22.Ehrlich P R, Dobkin D S, Wheye D. The Birder's Handbook: A Field Guide to the Natural History of North American Birds. New York: Simon & Schuster; 1988. [Google Scholar]
- 23.Anderson E, Stebbins G L. Evolution. 1954;8:378–388. [Google Scholar]
- 24.Stebbins G L. Flowering Plants: Evolution Above the Species Level. Cambridge, MA: Belknap; 1974. [Google Scholar]
- 25.Stebbins G L. Proc Am Philos Soc. 1959;103:231–251. [PubMed] [Google Scholar]
- 26.Stebbins G L. Taxon. 1969;18:26–35. [Google Scholar]
- 27.Abbott R. Trends Ecol Evol. 1992;7:401–405. doi: 10.1016/0169-5347(92)90020-C. [DOI] [PubMed] [Google Scholar]
- 28.Campbell C S, Wojciechowski M F, Baldwin B G, Alice L A, Donoghue M J. Mol Biol Evol. 1997;14:81–90. doi: 10.1093/oxfordjournals.molbev.a025705. [DOI] [PubMed] [Google Scholar]
- 29.Jain S, Martins P. Am J Bot. 1979;66:361–366. [Google Scholar]
- 30.Urbanska K M, Hurka H, Landolt E, Neuffer B, Mummenhoff K. Plant Syst Evol. 1997;204:233–256. [Google Scholar]
- 31.Stace C A. Hybridization and the Flora of the British Isles. London: Academic; 1975. [Google Scholar]
- 32.Bailey J P, Child L E, Wade M. In: Plant Invasions: General Aspects and Special Problems. Pyšek P, Prach K, Rejmanek M, Wade P M, editors. The Hague: SPB Academic; 1995. pp. 141–150. [Google Scholar]
- 33.Stace C A. New Flora of the British Isles. Cambridge, U.K.: Cambridge Univ. Press; 1991. [Google Scholar]
- 34.Rieseberg L H, Beckstrom-Sternberg S, Doan K. Proc Natl Acad Sci USA. 1990;87:593–597. doi: 10.1073/pnas.87.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bleeker W, Hurka H, Koch M. Florist Rundbriefe. 1997;31:1–8. [Google Scholar]
- 36.Cleland R E. Oenothera: Cytogenetics and Evolution. New York: Academic; 1972. [Google Scholar]
- 37.Abbott R J, Milne R I. BCPC Symp Proc. 1995;64:53–64. [Google Scholar]
- 38.Abbott R J, James J K, Irwin J A, Comes H P. Watsonia. 2000;23:123–138. [Google Scholar]
- 39.Paterson A H, Schertz K F, Lin Y-R, Chang Y-L. Proc Natl Acad Sci USA. 1995;92:6127–6131. doi: 10.1073/pnas.92.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gray A J, Marshall D F, Raybould A F. Adv Ecol Res. 1991;21:1–61. [Google Scholar]
- 41.Novak S J, Soltis D E, Soltis P S. Am J Bot. 1991;78:1586–1600. [Google Scholar]
- 42.Rieseberg L H, Ellstrand N C. Crit Rev Plant Sci. 1993;12:213–241. [Google Scholar]
- 43.Grant V. Plant Speciation. 2nd Ed. New York: Columbia Univ. Press; 1981. [Google Scholar]
- 44.Parker I M, Bartsch D. In: Transgenic Organisms—Biological and Social Implications. Tomiuk J, Wöhrmann K, Sentker A, editors. Basel: Birkhauser; 1996. pp. 147–161. [Google Scholar]
- 45.Gallagher K G, Schierenbeck K A, D'Antonio C M. Am J Bot. 1997;84:905–911. [PubMed] [Google Scholar]
- 46.Vilà M, D'Antonio C M. Ecol Appl. 1998;8:1196–1205. [Google Scholar]
- 47.Strefeler M S, Darmo E, Becker R L, Katovich E J. Am J Bot. 1996;83:265–273. [Google Scholar]
- 48.O'Hanlon P C, Peakall R, Briese D T. Mol Ecol. 1999;8:1239–1246. doi: 10.1046/j.1365-294x.1999.00646.x. [DOI] [PubMed] [Google Scholar]
- 49.Panetsos C A, Baker H G. Genetica. 1967;38:243–274. [Google Scholar]
- 50.Milne, R. I. & Abbott, R. J. (2000) Mol. Ecol., in press. [DOI] [PubMed]
- 51.Sun M, Corke H. Theor Appl Genet. 1992;83:321–329. doi: 10.1007/BF00224278. [DOI] [PubMed] [Google Scholar]
- 52.Ayres D R, Garcia-Rossi D, Davis H G, Strong D R. Mol Ecol. 1999;8:1179–1186. [Google Scholar]
- 53.Daehler C C, Strong D R. Am J Bot. 1997;84:607–611. [PubMed] [Google Scholar]
- 54.Neuffer B, Auge H, Mesch H, Amarell U, Brandl R. Mol Ecol. 1999;8:365–377. [Google Scholar]
- 55.Ellstrand N C, Whitkus R W, Rieseberg L H. Proc Natl Acad Sci USA. 1996;93:5090–5093. doi: 10.1073/pnas.93.10.5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harper J. Population Biology of Plants. London: Academic; 1977. [Google Scholar]
- 57.Crawley M J. Philos Trans R Soc London B. 1986;314:711–731. [Google Scholar]
- 58.Di Castri F. In: Ecology of Biological Invasions: A Global Perspective. Drake J A, Mooney H, editors. New York: Wiley; 1989. pp. 3–16. [Google Scholar]
- 59.Binggeli P, Hall J B, Healey J R. A Review of Invasive Woody Plant Species in the Tropics. Bangor, U.K.: University of Wales; 1998. , School of Agriculture and Forestry Sciences Publ. No. 13. [Google Scholar]
- 60.Arnold M L. Natural Hybridization and Evolution. Oxford: Oxford Univ. Press; 1997. [Google Scholar]
- 61.Lewontin R C, Birch L C. Evolution. 1966;20:315–336. doi: 10.1111/j.1558-5646.1966.tb03369.x. [DOI] [PubMed] [Google Scholar]
- 62.Weiner J. The Beak of the Finch. New York: Vantage; 1994. [Google Scholar]
- 63.Thompson J D. BioScience. 1991;41:393–401. [Google Scholar]
- 64.Grant V. Genetics of Flowering Plants. New York: Columbia Univ. Press; 1975. [Google Scholar]
- 65.Rieseberg L H, Archer M A, Wayne R K. Heredity. 1999;83:363–372. doi: 10.1038/sj.hdy.6886170. [DOI] [PubMed] [Google Scholar]
- 66.Hamrick J L, Godt M J W. In: Plant Population Genetics: Breeding and Genetic Resources. Brown A H D, Clegg M T, Kahler A L, Weir B S, editors. Sunderland, MA: Sinauer; 1990. pp. 43–63. [Google Scholar]
- 67.Mills L S, Smouse P. Am Nat. 1994;144:412–431. [Google Scholar]
- 68.Lande R. Conserv Biol. 1995;9:782–791. [Google Scholar]
- 69.Sauer J D. Plant Migration: The Dynamics of Geographic Patterning in Seed Plant Species. Berkeley: Univ. of California Press; 1988. [Google Scholar]
- 70.Anderson E. Evolution. 1948;2:1–9. [Google Scholar]
- 71.Waser N M. In: The Natural History of Inbreeding and Outbreeding, Theoretical and Empirical Perspectives. Thornhill N W, editor. Chicago: Univ. of Chicago Press; 1993. pp. 173–199. [Google Scholar]
- 72.Cook L M, Soltis P S, Brunsfeld S J, Soltis D E. Mol Ecol. 1998;7:1293–1302. [Google Scholar]
- 73.Moody M E, Mack R N. J Appl Ecol. 1988;25:1009–1021. [Google Scholar]
- 74.Burdon J J, Brown A H D. Aust J Biol Sci. 1986;39:369–378. [Google Scholar]
- 75.Piggin C M, Sheppard A W. In: The Biology of Australian Weeds. Groves R H, Shepherd R C H, editors. Vol. 1. Melbourne: R. G. & F. J. Richardson; 1995. pp. 87–110. [Google Scholar]
- 76.Novak S J, Mack R N. Heredity. 1993;71:167–176. [Google Scholar]
- 77.Novak S J, Mack R N, Soltis P S. Can J Bot. 1993;71:1441–1448. [Google Scholar]
- 78.Brashier C M, Cooke D E L, Duncan J M. Proc Natl Acad Sci USA. 1999;96:5878–5883. doi: 10.1073/pnas.96.10.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Camazine S, Morse R A. Am Sci. 1988;76:465–471. [Google Scholar]
- 80.Barrett S C H. Econ Bot. 1983;37:255–282. [Google Scholar]