Abstract
The rate of exchange of peptide group NH hydrogens with the hydrogens of aqueous solvent is sensitive to neighboring side chains. To evaluate the effects of protein side chains, all 20 naturally occurring amino acids were studied using dipeptide models. Both inductive and steric blocking effects are apparent. The additivity of nearest-neighbor blocking and inductive effects was tested in oligo-and polypeptides and, surprisingly, confirmed. Reference rates for alanine-containing peptides were determined and effects of temperature considered. These results provide the information necessary to evaluate measured protein NH to ND exchange rates by comparing them with rates to be expected for the same amino acid sequence is unstructured oligo- and polypeptides. The application of this approach to protein studies is discussed.
Keywords: hydrogen exchange, side chain effects, steric hindrance, inductive effects, protein structure
INTRODUCTION
Information about the structure and conformational dynamics of proteins, resolved to the level of individual amino acid residues, is contained in their hydrogen exchange (HX) behavior. The protection against exchange imposed by protein structure is often expressed by the factor, P = krc/kprot, derived by comparing the HX rate measured in a protein (kprot) with the rate expected in a random coil model (krc).1 To extract structural information from measured protection factors, one must first know krc and understand the non-structural effects on which it is based. In aqueous solutions, peptide group HX is catalyzed by hydroxide and hydronium ions2 in pH-dependent reactions and by water in a pH-independent way.3,4 The contribution of all other potential catalysts is negligible.5 HX rates are, however, significantly influenced by neighboring side chains even in the absence of folded structure. Thus, krc depends not only on solution pH and temperature, but also on local amino acid sequence.
Inductive effects of polar side chains on the acid-and base-catalyzed HX behavior of peptide groups were demonstrated and calibrated by Molday et al.6 These workers also demonstrated an additivity rule for predicting the summed inductive effects on a peptide NH due to its two neighboring side chains. This knowledge, although incomplete, has been widely used in protein studies to correct measured HX data for side chain inductive effects. Over the 20 years since the work of Molday and coworkers,6 various issues have arisen. For example, recent work points to a previously unsuspected effect of some apolar side chains.7–9 The increasing use of HX approaches for protein studies emphasizes the need for a more complete analysis of primary structure effects. For this purpose we have examined the effects on peptide group HX due to all 20 naturally occurring amino acid side chains, determined reference rates pertinent for peptide group NH to ND exchange in proteins and oligopeptides, and considered the dependence of exchange rates on temperature. Hydrogen isotope effects are dealt with in the following paper.10
MATERIALS AND METHODS
Materials
Model dipeptide derivatives (N-acetyl amino acid N′-methyl amide; see Fig. 1b, inset) were purchased from Bachem Bioscience (Philadelphia, PA) or synthesized from the corresponding N-acetyl-amino acid-methylester by reaction with methylamine at 0°C. Dipeptides of glutamine and asparagine were synthesized from N-acetylated amino acids using methylamine with dicyclohexyl-carbodiimide coupling. The cysteine dipeptide was obtained by reducing the cystine model with dithiothreitol.11 Oligopeptides were purchased from Bachem Bioscience or made by solid state synthesis. Oxidized ribonuclease, a random chain model, was prepared from ribonuclease A (Sigma Chemical Co., St. Louis, MO) by treatment with performic acid12 to oxidize the four cystine cross bridges to cysteic acid. Poly-DL-alanine (PDLA), a randomly synthesized polymer (dp ~ 28) of racemic alanine, was obtained from Sigma Chemical Co. The purity and integrity of all the molecules used were checked by nuclear magnetic resonance (NMR). The NH and CαH resonances of the PDLA sample we used showed some substructure, apparently intrinsic to interactions of the D and L residues. The NH components all appeared to exchange at the same rate and did not change in separable fractions obtained by gel filtration high performance liquid chromatography (HPLC).
Fig. 1.
H to D exchange of peptide NHs in the alanine dipeptide reference molecule (a–c) and for oligo- and polypeptide reference molecules (d) at 5°C in 0.5 M KCl. a: NMR spectra after various times of exchange at pDread 4.05; b: exponential decay of the NH resonances; c: dependence of H-D exchange rates on pDread; d: H-D exchange behavior of alanine oligo- and polypeptide reference molecules. The data in c and d are fit with rate constants for acid, base, and water catalysis (solid curves) to obtain the alanine reference rate constants listed in Table I. Results for acid catalysis of PDLA exchange in low salt concentration (○) were treated separately (rates 20% slower); results for base catalysis at high (0.5 M KCl) and low salt concentrations agree within 7% and were merged.
Two-dimensional (2D) COSY,13 TOCSY,14,15 and/or NOESY16,17 spectra were used as necessary to assign proton resonances. For all the dipeptide models used, the peptide group NH to the right of the side chain, the N′H-methyl resonance which we will call R, resonates relatively upfield. The left NH on the N-acetyl group, called L, resonates downfield, often as a resolved doublet split by the CαH. For the separable protons in primary amide side chains (Asn, Gln), assignment of HE and Hz assumed that HE was represented by the downfield resonance.18
Methods
Kinetic HX data were collected by 1D 1H-NMR using a 500 MHz spectrometer (Bruker AM500). Exchange rates slower than 1 min−1 were measured by H-D exchange. For peptides with good solubility, the H-D exchange reaction was initiated by diluting the peptide 10-fold into D2O. Less soluble peptides were initially lyophilized from saturated solutions and dissolved directly into a larger volume (1.7 ×) of D2O. Samples were then pipetted into NMR tubes, loaded into the spectrometer, and a series of 1D spectra was recorded to follow the time-dependent loss of the NH resonances as H to D exchange proceeded (e.g., Fig. 1a,b).
To study the fully alkaline form of dipeptides with titratable side chains, exchange was measured at neutral pH and higher. These rates, faster than 1 sec−1, were measured in H2O (H-H exchange) by solvent saturation transfer, using the Fourier transform analogue of the Forsen and Hoffman19 experiment or by selective saturation recovery20 interpreted according to the equation, 1/T1,meas = 1/T1 + kB[OH−], where pH covered a range through which the exchange term (kb[OH−]) varied from negligible to large. To place the H-H exchange data onto the same scale of log(kex) vs. pDread used for the H-D results (see Figs. 3, 6), the H-H results must be corrected for the kinetic isotope effect. To do this, the H-H rate data were plotted as if pHread equals pDread and 0.39 logarithmic units were subtracted from the value of log(kex) [see Fig. 3a of Ref. 10].
Fig. 3.
H-D exchange behavior of titratable amino acids in dipeptide models. The dashed curves show the alanine dipeptide reference rates. a,b: Lys and arg were fit by the usual three parameters for acid, base, and water catalysis. c,d: His, Asp, and Glu require additional rate constants for both the protonated and unprotonated forms. Values for pKa,read in D2O at 5°C are: His, 7.02; Glu, 4.53; Asp, 4.08. To shield coulombic effects these solutions contained 0.50 M KCl, as did all other solutions used here.
Fig. 6.
H-D exchange curves for NHs in some test oligopeptides. The dashed curve shows the exchange behavior of an internal NH in the reference trialanine peptide (from Fig. 1d). For each residue NH underlined, the solid line is the curve predicted using Eq. (2) with the reference rates in Table I and the side chain-specific factors in Table II. The success of the predictions demonstrates the additivity of inductive and blocking effects. The unblocked oligopeptides (free amino and carboxyl termini) are: Lys-Val-Ile-Leu-Phe (KVILF); Lys-Ala-Ile-Val-Asn-Lys (KAIVNK); Gly-Ala-Val-Ser-Thr-Ala (GAVSTA). Exchange behavior of the last NH of each oligopeptide was affected by titration of the terminal carboxyl group in the pD region measured and is not shown.
All sample handling and NMR experiments were at 5°C. Solutions contained 0.50 M KCl to shield possible charge effects.21 Citric acid (below pD 4) or succinic acid (above pD 4) at 50 mM concentrations was used for pH buffering in D2O solutions between pD 1 and 6.5, and 50 mM phosphate was used at pH values above 6 in H2O. Solution pH was measured at room temperature but the experiments were done at 5°C. Over this temperature range, citrate and succinate experience an 0.06 unit increase in pKa and phosphate pKa increases 0.1 units. The solution pH used was changed accordingly.
Data Analysis
HX rates were obtained from the time-dependent decrease in area of NH resonance peaks (Fig. 1). Processing used the program FELIX (Hare Research, Bothell, WA) and was partially automated. The results reported here involved the recording and processing of ~10,000 NMR spectra. The first NMR spectrum in each kinetic experiment was transformed, phase corrected, appropriate baseline regions were chosen, and integration intervals were picked. The same values were used to analyze all the sequential spectra in a given kinetic experiment. The output file of peak area vs. exchange time was imported into a non-linear least squares routine and fit to the equation, H = Hoexp(−kt) + baseline, to define the exchange rate at a given pD. The non-zero baseline level was due to water remaining in the diluted peptide solution.
Data were plotted to produce V-shaped curves of log(kex) vs. pD (see figures), and these were fit by Eq. (1) to obtain second-order rate constants for catalysis by specific acid (kA), specific base (kB), and water (kw), where KD is the D2O dissociation constant.
| (1) |
Data in the figures are plotted as obtained against pDread, the uncorrected pH meter reading using a glass pH electrode. However, calculated rate constants listed in the tables include the proper corrections to take account of the pH meter anomaly in D2O (pDcorr = pDread + 0.4),22 the pertinent solvent dissociation constants,10 and buffer temperature dependence. Measured exchange rates for the dipeptides of glutamic acid, aspartic acid, and histidine were fit using additional terms including the side chain pKa and rate constants for both the charged and uncharged forms.6 To reduce the number of fitting parameters, side chain pKas were measured independently, either by direct pD titration at 5°C or from the pD-dependent chemical shift of side chain protons observed in the experimental HX solutions.
RESULTS
Alanine Reference Rates
Figure 1 illustrates the determination of HX rate constants. Figure 1a follows the exchange of H to D at left (L) and right (R) peptide groups of the alanine dipeptide (N-Ac-Ala-N′MA). Figure 1b shows kinetic plots of the exchange data fit with a mono-exponential decay. Results for the alanine dipeptide model are summarized in Figure 1c. Figure 1d shows similar results for an internal NH of a blocked alanine tripeptide (N-Ac-Ala-Ala-Ala-N′MA) and for PDLA. The limbs of the curves of log rate against pD have slopes of −1 and +1 due to catalysis by D+ and OD− ions. Data were fit according to Eq. (1) to obtain rate constants for the acid-and base-catalyzed reactions and also for a water-catalyzed reaction that becomes significant near the pD of minimum rate. Results for the three alanine-based peptides studied are listed in Table I.
TABLE I.
H to D Exchange Rate Constants for Alanine-Based Reference Molecules at 278°K in 0.5 M KCl*
| logkA (M−1 min−1) | logkB (M−1 min−1) | logkw (min −1) | |
|---|---|---|---|
| N-Ac-Ala-N′MA (L) | 2.87 | 9.71 | −2.3 |
| N-Ac-Ala-N′MA (R) | 2.81 | 9.01 | −3.2 |
| N-Ac-A-A-A-N′MA (A) | 1.56 | 10.20 | −2.3 |
| PDLA | 1.19 | 9.90 | −2.5 |
These values were obtained from the data in Figure 1, are specific for NH to ND exchange in high salt concentration (0.5 M KCl), use pDcorr (= pDread + 0.4),22 and take the D2O molar ionization constant at 5°C as 10−15.65.36 (The figures in this paper show data measured at 0.5 M KCl on an uncorrected pDread scale.) For reference values at lower salt concentration, see Table III.
Table I provides reference NH to ND exchange rate constants for the following measurements on a series of di-, oligo-, and polypeptides under these same conditions. The results in Table I were obtained at 5°C to bring rates onto an easily measured time scale, and solutions contained 0.5 M KCl in order to suppress coulombic effects in subsequent comparisons with charged molecules. The use of reference rate constants for protein studies under a wide range of conditions discussed below. Reference rate constants pertinent to other isotope combinations are derived in the following paper.10
Side Chain Effects in Dipeptide Models
We want to evaluate the changes in peptide group HX rate imposed by nonalanine side chains. Figures 2–5 show HX curves for various amino acid dipeptides. The data were fit to Eq. (1) to obtain rate constants for acid, base, and water catalysis of NH exchange in D2O. Fits obtained are shown by the solid curves drawn in the figures. The dashed curve in each panel provides a visual reference for the exchange behavior of the alanine dipeptide model, taken from Figure 1c. HX rate increments relative to alanine are listed in logarithmic form in Table II.
Fig. 2.
a–d: H-D exchange behavior of neutral polar amino acids in dipeptide models. The leftward shift relative to the alanine dipeptide reference curve (dashed line) results from an inductive effect in which electron density is withdrawn from the peptide group, increasing the base-catalyzed rate and decreasing the acid-catalyzed rate. The side chain parameters in Table II quantify the changes in acid and base rate constants for the L and R peptide NHs in these residues.
Fig. 5.
H-D exchange curves for the peptide NHs of Asn and Gln dipeptide models (a,b) and for the R peptide of the proline dipeptide model in cis and trans forms (c). The dashed curve is the alanine reference.
TABLE II.
Effects of Amino Acid Side Chains on the HX Rates of Neighboring Peptides*
| Side chain (X) | Logkex(X) − Logkex(Ala)
|
|||
|---|---|---|---|---|
| Acid catalysis
|
Base catalysis
|
|||
| L | R | L | R | |
| Ala | 0.00 | 0.00 | 0.00 | 0.00 |
| Arg | −0.59 | −0.32 | 0.08 | 0.22 |
| Asn | −0.58 | −0.13 | 0.49 | 0.32 |
| Asp(COO−) | (0.9) | 0.58 | −0.30 | −0.18 |
| Asp(COOH) | (−0.9) | −0.12 | 0.69 | (0.6) |
| Cys | −0.54 | −0.46 | 0.62 | 0.55 |
| Cys2 | −0.74 | −0.58 | 0.55 | 0.46 |
| Gly | −0.22 | 0.22 | 0.27 | 0.17 |
| Gln | −0.47 | −0.27 | 0.06 | 0.20 |
| Glu(COO−) | (−0.9) | 0.31 | −0.51 | −0.15 |
| Glu(COOH) | (−0.6) | −0.27 | 0.24 | 0.39 |
| His | −0.10 | 0.14 | ||
| His+ | (−0.8) | −0.51 | (0.8) | 0.83 |
| Ile | −0.91 | −0.59 | −0.73 | −0.23 |
| Leu | −0.57 | −0.13 | −0.58 | −0.21 |
| Lys | −0.56 | −0.29 | −0.04 | 0.12 |
| Met | −0.64 | −0.28 | −0.01 | 0.11 |
| Phe | −0.52 | −0.43 | −0.24 | 0.06 |
| Pro(trans) | −0.19 | −0.24 | ||
| Pro(cis) | −0.85 | 0.60 | ||
| Ser | −0.44 | −0.39 | 0.37 | 0.30 |
| Thr | −0.79 | −0.47 | −0.07 | 0.20 |
| Trp | −0.40 | −0.44 | −0.41 | −0.11 |
| Tyr | −0.41 | −0.37 | −0.27 | 0.05 |
| Val | −0.74 | −0.30 | −0.70 | −0.14 |
| N-term (NH3+) | −1.32 | 1.62 | ||
| C-term (COO−) | 0.96 | (−1.8) | ||
| C-term (COOH) | (0.05) | |||
Results for the polar residues in Figures 2 and 3 exhibit inductive effects, marked by a leftward shift on the pD axis. Upward shifts in kmin for Ser and Cys L peptides are due to a large inductive effect on the water-catalyzed rate. Titration effects are apparent in Figure 3 even though the 0.50 M KCl present minimizes direct coulombic effects. Figure 4 shows that aliphatic and aromatic side chains impose steric blocking effects in which both acid and base rates are down shifted. The aromatic side chains impose both blocking and inductive effects as do some of the polar side chains. Steric blocking is larger for L than for R peptide NHs, presumably due to geometric proximity to the side chain. Results for the peptide group NHs of Asn and Gln are shown in Figure 5a,b. Figure 5c shows HX results for the R peptide of proline. The cis and trans proline isomers differ, apparently due to an inductive effect. The clearly resolved resonances of the cis and trans proline isomers (trans isomer upfield) appear in a ratio of 1.0:4.0. The data show that the cis-trans isomerization lifetime in the proline dipeptide must be larger than the HX time scale in Figure 5c (>1 hr at 5°C). Factors that affect the isomerization rate have been discussed.23,24
Fig. 4.
a–d: H-D exchange curves of aliphatic and aromatic amino acids in dipeptide models. The downward shifts relative to the alanine curve (dashed line) reflect side chain-dependent steric hindrance effects that slow both acid and base rates. The aromatics show both steric and inductive effects.
In prior work, Molday et al.6 studied rate changes due to inductive effects of polar side chains. Present results for neutral polar side chains generally agree with the earlier rates to within 25%. Results for the charged side chains are more divergent due in part to uncertainties introduced by titration effects and by a significant water contribution not considered in the earlier work. Apolar side chains were not previously studied.
Additivity of Side Chain Effects
Molday et al.6 showed that the inductive effects of two neighboring side chains on an included peptide NH act independently. A given side chain multiplies the exchange rate of a neighboring peptide NH by a characteristic factor independently of the effect of the other neighboring side chain. This makes it possible to easily predict the exchange rate of any peptide NH when its two neighboring side chains are known. In order to exert steric blocking effects, however, neighboring side chains may have to compete for the same space and thus may not act independently.
To test the interactive nature of side chain blocking, we synthesized and studied oligopeptides that place in sequential positions residues with large apolar blocking effects and also polar inductive effects. HX rates measured were then compared to rates predicted on the basis of nearest-neighbor additivity. In Figure 6 each panel shows measured HX data for a particular NH in the oligopeptide specified. The solid line is the curve predicted by using the factors in Table II and the nearest-neighbor additivity rule, computed as in Eq. (2), presented later. In these oligopeptide predictions, the alanine reference behavior used was that of the middle residue in the trialanine peptide shown in Figure 1d. The other NHs in the trialanine peptide showed somewhat different rates presumably due to end effects and/or differential blocking effects.
Figure 6 shows that predicted rate curves for peptide NHs in the oligopeptides are successful even when the summed blocking effects are larger than 10-fold and when both blocking and inductive effects interact. Significant errors are found only for the peptide group NH neighboring the oligopeptide amino terminus. In this case, an estimate of the large correction required for the presence of a neighboring unblocked N-terminus was obtained by comparison of the alanine dipeptide with alanine-N-methylamide (ala-NHCH3). The discrepancies seen at the near-terminal position are fairly small relative to the large correction factor used (Table II), appear disparate, and were not pursued further.
Water-Catalyzed Rate
To fit the rate-pD curves, a pD-independent water-catalyzed rate term [kw in Eq. (1)] is often required to account for data points close to the pD minimum. This small rate contribution accounts for roughly 1/4 of the rate at the pD minimum and becomes negligible as the acid or the base rate increases. Water catalysis has been measured before.3,4,25
The water rate found for NHs adjacent to apolar blocking side chains is smaller than for alanine and increases when the peptide group basicity is increased due to neighboring polar side chains (see below). Figure 7a shows that water rates found for the dipeptide NHs (L NHs) correlate with the side chain inductive effect measured by the shift in pDmin. Since ΔpDmin increases with basicity of the peptide group, this result suggests that the pH-independent water reaction involves the water molecule acting as a base to directly extract the NH proton. If so, the water rate for a given peptide NH might be predicted by using the base factors of the neighboring side chains, as written in the last term of Eq. (2) (see below). Figure 7b provides a partial test for this approach by comparing water rates predicted in this way with water rates measured by fitting the HX behavior of the oligopeptide NHs that have reasonably well-determined water rates (data in Fig. 6).
Fig. 7.
Water-catalyzed HX rate: a: Water rates measured for the dipeptide NHs are correlated with the relative basicity of the peptide group, expressed in terms of the shift in pDmin relative to the alanine dipeptide. Data are shown for the 17 dipeptides (including Cys2) with well-determined water rates (Pro and the titratable residues—Asp, Glu, and His—are excluded), b: To test predictive schemes for the water rate, water rates predicted as in the last term in Eq. (2a) (using only the base factors) are compared with water rates measured by fitting the oligopeptide data in Figure 6. The data points represent the eight moderately well-determined NHs in Figure 6 (left-most panels excluded). The diagonal at unit slope indicates the position for a perfect prediction of the rates.
Protein Reference Rates
Measured reference rates are significantly different for the alanine dipeptide, oligopeptide, and polypeptide models (Fig. 1c,d, Table I). The alanine dipeptide provides a proper reference for the dipeptides studied, and the internal NH of the blocked trialanine peptide succeeds for oligopeptide predictions (Fig. 6). What is the correct reference for protein predictions?
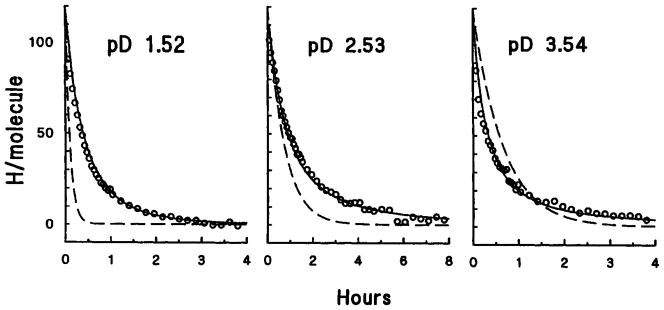
Experiments were done to test the predictability of HX rates in a polypeptide. The overall H to D exchange behavior of random chain oxidized ribonuclease is shown in Figure 8. Rate data were taken near the pD minimum at pDread 2.5, and at pD values on both sides of the minimum where exchange is dominated by either acid or base catalysis. The solid lines are the overall HX curves predicted using the PDLA reference rates, the known ribonuclease sequence, the side chain factors in Table II, and a nearest-neighbor additivity rule [Eq. (2) page 84]. At pD 1.52 acid catalysis dominates, side chain inductive and steric effects both act to slow HX rates, and the HX curve for oxidized ribonuclease is generally slower than for PDLA. At pD 3.54 base catalysis dominates, some NHs are slowed by steric effects while others are accelerated by inductive effects, so that the measured HX curve crosses over the reference PDLA curve.
Fig. 8.
Overall H-D exchange behavior of the NHs in random chain oxidized ribonuclease near the pDmin (center) and where dominated by acid (left) and base (right) catalysis (5°C). The solid line is the H-D exchange curve obtained by summing the predicted exchange behavior of all the individual NHs in the polymer. The dashed line shows the reference HX behavior of PDLA drawn with an amplitude of 119 NHs, the number of NHs in ribonuclease.
Good agreement is found between the measured data and the predicted curves (solid lines in Fig. 8) when the HX rate of PDLA is used as a reference. Use of the trialanine peptide reference rate (Fig. 1d), which provides good predictions for the oligopeptide data (Fig. 6), gives predicted curves that are uniformly too fast (~ 2-fold) for the oxidized ribonuclease data, in part due to their different translational diffusion coefficients.
Salt Effects
In this work, coulombic effects due to titratable side chains were intentionally suppressed by the use of 0.50 M KCl so that other side chain effects could more easily be identified and evaluated. The presence of 0.5 M KCl alters exchange rates of the charged residues by only about 30%, estimated by comparison of dipeptide rates measured here with similar results of Molday et al.6 at lower salt. Salt effects on uncharged molecules were directly studied by H-D exchange (Fig. 1d) and also by H-H exchange using saturation transfer methods in the acid and base-catalyzed regions (data not shown). Rates due to acid catalysis were found to be increased in 0.5 M KCl by a factor of 1.2 and base catalysis was slowed by about 1.07.
These results together with the temperature corrections discussed below were used to derive the alanine oligo- and polypeptide reference rates in Table III. Table III lists reference rates pertinent for 20°C at the low salt conditions normally used in protein studies.
TABLE III.
H to D Exchange Rate Constants for Alanine-Based Reference Molecules at 293°K*
| logkA (M−1 min−1) | logkB (M−1 min−1) | logkw (min−1) | |
|---|---|---|---|
| N-Ac-A-A-A-N′MA | 2.04 | 10.36 | −1.5 |
| PDLA | 1.62 | 10.05 | −1.5 |
These values are specific for NH to ND exchange under normal low salt conditions, use pDcorr. (= pDread + 0.4),22 and take the D2O molar ionization constant at 20°C as 10−15.05 36 To calculate the HX rate of a random coil peptide NH between any two side chains at 20°C and normal low salt conditions, one can use Eq. (2) with the reference rate constants listed here and the side chain factors in Table II. To obtain HX rates at other temperatures, one can apply Eq. (3) with the following activation energies: Ea(kA) = 14 kcal/mol, Ea(kB) = 17 kcal/mol, Ea(kw) = 19 kcal/mol (see text). Reference rate constants listed here are specific for NH to ND exchange. For other isotope combinations, see Connelly et al.10
Some Side Chain NH Rates
Figure 9 shows HX results measured in the dipeptide models for the indole NH of Trp, the NδH side chain proton of Arg, and some side chain amide protons. Values for Asn Hz and Gln HE were poorly defined due to overlap with a contaminant, and HE of Asn could not be measured. Rate constants for the side chain NHs measured are listed in Table IV. Some previous results have been reported for Trp indole NH20,26–28 and for Asn and Gln6,29 using various conditions and isotope combinations.
Fig. 9.

H-D exchange curves for some side chain NHs in dipeptide models in D2O at 5°C. Rate constants are listed in Table IV.
TABLE IV.
H to D Exchange Rates of Some Side Chains in Dipeptides at 278°K
| logkA (M−1 min−1) | logkB (M−1 min−1) | logkw (min−1) | |
|---|---|---|---|
| Arg(NδH) | 2.0 | 11.3 | — |
| Asn(Hz) | 4.5 | 9.4 | — |
| Gln(HE) | 5.2 | 9.5 | — |
| Gln(Hz) | 4.8 | 9.1 | −0.9 |
| Trp(N1H) | 5.1 | 9.4 | −1.0 |
DISCUSSION
As we have seen, the exchange with D2O of the peptide group NH is pH-dependent and describes a V-shaped curve of log (kex) vs. pD, exhibiting catalysis by specific acid, specific base, and water. The rate constants are sensitive to inductive and steric blocking effects imposed by neighboring side chains. The understanding and proper use of this information are important in studies of protein structure and dynamics.
Side Chain Effects
The rate of peptide NH abstraction by OD− is less than diffusion-limited by two orders of magnitude due to the strongly basic pKa of the peptide group.30,31 Polar side chains tend to withdraw electron density from neighboring peptide groups, increasing their acidity. This inductive effect increases the rate for base-catalyzed removal of the NH proton and reduces the rate for deposition of an additional proton, the rate-limiting step in the acid-catalyzed HX pathway. Thus, polar side chains tend to raise the right limb of the V-shaped rate vs. pD curve and lower the left limb, moving pDmin to the left with little effect on kmin. Ser and Cys in Figure 2 provide good examples, and all the polar residues in Figures 2 and 3 show an inductive component. Inductive effects on peptide NH exchange were noted by Leichtling and Klotz32 and were calibrated by Molday et al.6 for a number of polar amino acid side chains.
Figure 4 documents a different kind of behavior. A number of side chains slow both acid- and base-catalyzed rates of their neighboring peptides more or less equally, and thus shift the rate-pD curve downward, so that kmin is reduced with little change in pDmin. These results indicate that some conformers of the side chains themselves or side chain-induced conformers of the main chain sterically interfere with the encounter complex [peptide group + (D + or OD−) + associated waters]. Initial molecular dynamics calculations lend credence to this mechanism.
The blocking effect is always larger for the L than the R peptide, presumably because L is geometrically more accessible to the side chain. Blocking is most obvious for but not limited to beta-branched side chains. The gamma-branched leucine side chain is a quite effective blocker, especially for its L peptide (4-fold slowing), as are the three aromatics (~2.5-fold slowing). Earlier observations of HX slowing in polymeric amides33,34 may have been due to similar effects. The rank order of side chain blocking ability appears to be similar to that seen for other peptide-directed chemical reactions, e.g., the coupling reaction in polypeptide synthesis.35 The fact that side chains sterically hinder the interaction of peptide groups with aqueous species may have implications for a number of issues in addition to HX behavior including peptide synthesis, solvent partitioning equilibria, and apparent hydrogen bond strength in proteins.
Additivity of Side Chain Effects
It is surprising that side chain blocking effects are simply additive. For example, consider the Ile-(NH)-Val sequence in Figure 6 (KAIVNK). This NH is R to the Ile side chain and L to Val. In the dipeptide models the Ile side chain slows acid catalysis of its R peptide NH by 4-fold. Val slows acid catalysis of its L peptide by 5-fold. What degree of slowing should one expect for the Ile-NH-Val sequence? In one view the peptide group is blocked all but 1/4 of the time due to Ile and, independently, all but 1/5 of the time due to Val. The two side chains acting together might exhibit a positive, negative, or null interaction effect. The measured result is that Ile and Val together cause a simply combinatoric 20-fold slowing (4 × 5) in the acid-catalyzed exchange of the included NH. A similar result occurs for the base-catalyzed reaction; the Ile and Val side chains together produce a measured 7-fold slowing, which compares with the factor of 8.5 expected for a combinatoric effect (1.7 × 5). The other determinations in Figure 6 exhibit similar predictability.
The detailed bases of these effects are not clear. In light of proton transfer theory30 [see eq. (4) and accompanying discussion in ref. 10], the blocking factors seem most likely to reflect a decrease in the rate of formation of the peptide-catalyst encounter complex. The additive behavior found and the ability of a side chain to simultaneously block both its neighboring peptides indicate that the steric effect does not involve spatial blocking of the incoming catalyst but rather represents an energetic disfavoring of the encounter complex. A likely possibility is that the side chains interfere with the ordering of the solvation shell that accompanies formation of the charged complex. Independently imposed blocking factors would then be logarithmically additive because their free energy effects are additive.
Prediction of Peptide Group HX Rates
In protein HX studies, the comparison of measured HX rates (kprot) with calculated random chain rates [krc in Eq. (2)] has been widely used to quantify structural protection or slowing factors, defined as P = krc/kprot. Till now, the random chain rate has been calculated by using PDLA reference rates3,5 together with side chain inductive effects and the nearest-neighbor rule found by Molday et al.6 The present work updates this analysis and extends it to include the newly discovered steric hindrance effects.
Eq. (2) can be used to predict the HX rate of an unstructured peptide NH neighbored by particular side chains in a random chain conformation (krc).
| (2a) |
Eq. (2a) replaces the side chain-specific acid, base, and water rate constants in Eq. (1) (kA, kB, kw) with standard rate constants for the pertinent alanine reference peptide (kA,ref, kB,ref, kw,ref; Table III) multiplied by the side chain-specific acid (AL,AR) or base (BL, BR) factors (Table II). Eq. (2b) shows that the predicted first-order rate constant for acid catalysis—k(acid)—can be obtained by adding the logarithms of the appropriate terms (listed in Tables I, II) and then taking the antilogarithm.
| (2b) |
Analogous exercises yield the rates for base and for water catalysis. The expected exchange rate for any NH is the sum of its independently computed acid, base, and water terms [Eq. (2a)]. Different solution conditions (temperature, etc.) are accounted for by suitably adjusting the reference rates and catalyst concentrations. The A and B factors are insensitive to solution conditions (except for salt-sensitive charge effects). Note that these factors are written from the perspective of the side chain. R refers to the peptide group to the right of the side chain in question and L to its left.
As an example let us calculate the expected HX rate of the oligopeptide Ile-NH-Val proton, considered just above, at pD 2.0 and 5°C in 0.5 M KCl (as measured in Fig. 6). For these conditions, log[OD−] = 2.00 − 15.65 = −13.65. From the oligopeptide reference rates in Table I pertinent for the high salt condition, log(kA) = 1.56, log(kB) = 10.20, and log(kw) = −2.3. The NH is R from Ile and L from Val (Table II). Therefore, the acid rate for Ile-NH-Val [Eq. (2b)] is antilog (1.56 − 0.74 − 0.59 − 2.00) = 1.70 × 10 −2 min−1. The base-catalyzed rate is antilog (10.20 −0.70 −0.23 −13.65) = 4.16 × 10−5 min−1. The water rate is antilog (−2.30 −0.70 −0.23) = 5.9 × 10−4 min−1. The predicted HX rate is the sum of the acid, base, and water rates.
For a protein at 20°C under normal low salt conditions, the PDLA reference rates in Table III should be used.
Temperature Dependence
Reference rate constants pertinent for alanine peptides in normal low salt conditions and specific for 20°C are listed in Table III. To predict HX rates at other temperatures, each rate or reference rate constant in Eq. (2) should be modified according to Eq. (3).
| (3) |
We recommend use of the activation energies specified below Table III, obtained from available results3,4,25 and the present work weighted to account for different levels of accuracy in the various data sets. These are 14, 17, and 19 kcal/mol for kA, kB, and kw, respectively.
The apparent activation energy for the base-catalyzed reaction (17 kcal/mol) is the sum of the rather small Ea value for the second-order, OD−-catalyzed rate constant (4 kcal/mol) plus the relatively large equilibrium enthalpy for D2O or H2O ionization (~14 kcal/mol).1,36 Use of an apparent Ea of 17 kcal/mol in Eq. (3), when applied to k(base) in Eq. (2a), eliminates the need to separately compute [OD−] as a function of temperature. Alternatively, one can use 3 kcal/mol for the activation energy of kB,ref and separately calculate specific base concentration (OD− or OH−) from known pH and the equations provided by Covington et al.36 for solvent pKa. The two approaches give very similar results over a wide temperature range.
In principle, NHs flanked by non-alanine side chains may have somewhat different Ea values, dictated by the differing rate factors listed in Table II. Inductive effects (Table II) represent changes in peptide group pKa which will directly affect Ea and temperature dependence. Blocking effects may or may not alter Ea values, depending on whether they are enthalpic or entropically based. The inclusion of these effects has little practical significance; e.g., an error less than 2-fold is obtained for predicted rates up to 100°C when the side chain factor is as much as 10.
Note that when a pH buffer with significant temperature dependence is used, this must be separately considered to obtain the correct solution pH at the temperature of the experiment. Also, it should be clear that the temperature dependence just discussed refers specifically to the chemical exchange rate of the peptide group in random chain conformation. In a structured protein, HX rates may also respond to additional temperature-dependent effects on the stability of local or global structure.
Protein Studies
As we have seen, side chain inductive and blocking effects are both essentially additive in random chain polypeptides, and here the side chain factors listed in Table II appear sufficient to predict HX rates. The use of this approach implicitly assumes that a peptide NH in the act of exchange experiences the same averaged conformational manifold as a random chain. In applying these predictions to rates measured in structured proteins, the implications of this assumption need to be considered.
In proteins, peptide NHs may be held exposed to solvent or may be involved in hydrogen bonded structure. For an NH exposed at the surface of a native protein, main chain and neighboring side chain conformations are unlikely to be dynamically randomized. This may not change the inductive effects of neighboring side chains but may well alter the steric blocking effects calibrated here. A similar problem exists for NHs involved in structural hydrogen bonding, which are thought to require H-bond breakage and often some cooperative main chain unfolding in order to exchange.1,37,38 The conformational properties of these transient open states are not yet clear. Recent progress in structural studies shows that unstructured polypeptides tend to adopt a condensed rather than a random coil form.39 Therefore, unfolded forms may well experience constraints that increase or decrease HX blocking relative to the random chain conformation. The sizable steric blocking effects found here with small molecules warn that such effects in locally unfolded forms may be large. Structural slowing factors for individual NHs calculated as just described should be considered in this light.
Acknowledgments
We thank Zhan Deng for her participation in the early stages of this work and Gregory P. Connelly for his help in bringing this project to a successful conclusion. This work was supported by NIH grant DK11295.
Abbreviations
- HX
hydrogen exchange
- pDmin
the pD at the minimum rate in the pD-dependent HX curve
- kmin
exchange rate constant at the pDmin
- PDLA
poly-DL-alanine
- N-Ac-Ala-N′MA
the alanine dipeptide N-acetylalanine-N′-methylamide (CH3CONHCH(CH3)CONHCH3), and analogously for other amino acid dipeptides
References
- 1.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic-acids. Q Rev Biophys. 1984;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 2.Berger A, Loewenstein A, Meiboom S. NMR study of the protolysis and ionization of N-methylacetamide. J Am Chem Soc. 1959;81:61–67. [Google Scholar]
- 3.Englander JJ, Calhoun DB, Englander SW. Measurement and calibration of peptide group hydrogen-deuterium exchange by ultraviolet spectrophotometry. Anal Biochem. 1979;92:517–524. doi: 10.1016/0003-2697(79)90693-6. [DOI] [PubMed] [Google Scholar]
- 4.Gregory RB, Crabo L, Percy AJ, Rosenberg A. Water catalysis of peptide hydrogen isotope exchange. Biochemistry. 1983;22:910–917. doi: 10.1021/bi00273a031. [DOI] [PubMed] [Google Scholar]
- 5.Englander SW, Poulsen A. Hydrogen-tritium exchange of the random chain polypeptide. Biopolymers. 1969;7:329–339. [Google Scholar]
- 6.Molday RS, Englander SW, Kallen RG. Primary structure effects on peptide group hydrogen exchange. Biochemistry. 1972;11:150–158. doi: 10.1021/bi00752a003. [DOI] [PubMed] [Google Scholar]
- 7.Robertson AD, Baldwin RL. Hydrogen exchange in thermally denatured ribonuclease A. Biochemistry. 1991;30:9907–9914. doi: 10.1021/bi00105a014. [DOI] [PubMed] [Google Scholar]
- 8.Lu J, Dahlquist FW. Detection and characterization of an early folding intermediate of T4 lysozyme using pulsed hydrogen exchange and two-dimensional NMR. Biochemistry. 1992;31:4749–4756. doi: 10.1021/bi00135a002. [DOI] [PubMed] [Google Scholar]
- 9.Radquist SE, Buck M, Topping KD, Dobson CM, Evans PA. Hydrogen exchange in native and denatured states of hen egg white lysozyme. Proteins. 1992;15:237–248. doi: 10.1002/prot.340140210. [DOI] [PubMed] [Google Scholar]
- 10.Connelly GP, Bai Y, Jeng MF, Englander SW. Isotope effects in peptide group hydrogen exchange. Proteins. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 11.Cleland WW. Dithiothreitol, a new protective agent for SH groups. Biochemistry. 1964;3:480–482. doi: 10.1021/bi00892a002. [DOI] [PubMed] [Google Scholar]
- 12.Hirs CWH. The oxidation of ribonuclease with performic acid. J Biol Chem. 1956;219:611–621. [PubMed] [Google Scholar]
- 13.Nagayama K, Kumar A, Wüthrich K, Ernst RR. Experimental techniques of two-dimensional correlated spectroscopy. J Magn Reson. 1980;40:321–334. [Google Scholar]
- 14.Braunschweiler L, Ernst RR. Coherence transfer by isotropic mixing: Application to proton correlation spectroscopy. J Magn Reson. 1983;53:521–528. [Google Scholar]
- 15.Bax A, Davis DG. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J Magn Reson. 1985;65:355–360. [Google Scholar]
- 16.Macura S, Ernst RR. Elucidation of cross-relaxation in liquids by two-dimensional NMR spectroscopy. Mol Phys. 1980;41:95–117. [Google Scholar]
- 17.Kumar A, Ernst RR, Wüthrich K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem Biophys Res Commun. 1980;95:1–6. doi: 10.1016/0006-291x(80)90695-6. [DOI] [PubMed] [Google Scholar]
- 18.Perrin CL, Arrhenius GML. Mechanisms of acid-catalyzed proton exchange in N-methyl amides. J Am Chem Soc. 1982;104:6693–6696. [Google Scholar]
- 19.Forsen S, Hoffman RA. Study of moderately rapid chemical exchange reactions by means of nuclear magnetic double resonance. J Chem Phys. 1963;39:2892–2901. [Google Scholar]
- 20.Waelder S, Lee L, Redfield AG. Nuclear magnetic resonance studies of exchangeable protons. I Fourier transform saturation- recovery and transfer of saturation of the tryptophan indole nitrogen proton. J Am Chem Soc. 1975;97:2927–2928. doi: 10.1021/ja00843a066. [DOI] [PubMed] [Google Scholar]
- 21.Kim PS, Baldwin RL. Influence of charge on the rate of amide proton exchange. Biochemistry. 1982;21:1–5. doi: 10.1021/bi00530a001. [DOI] [PubMed] [Google Scholar]
- 22.Glasoe PF, Long FA. Use of glass electrodes to measure acidities in deuterium oxide. J Phys Chem. 1960;64:188–193. [Google Scholar]
- 23.Grathwohl C, Wüthrich K. NMR studies of the rates of proline cis-trans isomerization in oligopeptides. Biopolymers. 1981;20:2623–2633. [Google Scholar]
- 24.Nall B. Proline isomerization and folding. Comments Mol Cell Biophys. 1985;3:123–143. [Google Scholar]
- 25.Jeng MF, Englander SW. Stable submolecular folding units in a non-compact form of cytochrome c. J Mol Biol. 1991;221:1045–1061. doi: 10.1016/0022-2836(91)80191-v. [DOI] [PubMed] [Google Scholar]
- 26.Waelder SF, Redfield AG. Nuclear magnetic resonance studies of exchangeable protons. II The solvent exchange rate of the indole nitrogen proton of tryptophan derivatives. Biopolymers. 1977;16:623–629. doi: 10.1002/bip.1977.360160311. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi T, Nakanishi M, Tsuboi M. Hydrogen-deuterium exchange study of amino acids and proteins by 200 to 300 nm spectroscopy. Anal Biochem. 1981;110:242–249. doi: 10.1016/0003-2697(81)90142-1. [DOI] [PubMed] [Google Scholar]
- 28.O’Neil JD, Sykes BD. Side chain dynamics of detergent solubilized membrane protein: Measurement of tryptophan and glutamine hydrogen exchange rates in M13 coat protein by 1H NMR spectroscopy. Biochemistry. 1989;28:6736–6745. doi: 10.1021/bi00442a029. [DOI] [PubMed] [Google Scholar]
- 29.Krishna NR, Sarathy KP, Huang DH, Stephens RL, Glickson JD, Smith CW, Walter R. Primary amide hydrogen exchange in model amino acids: Asparagine, glutamine and glycine amides. J Am Chem Soc. 1982;104:5051–5053. [Google Scholar]
- 30.Eigen M. Proton transfer, acid-base catalysis, and enzymatic hydrolysis. Angew Chem Int Ed Engl. 1964;3:1–19. [Google Scholar]
- 31.Englander SW, Englander JJ. Hydrogen tritium exchange. Methods Enzymol. 1972;26c:406–413. doi: 10.1016/s0076-6879(72)26021-9. [DOI] [PubMed] [Google Scholar]
- 32.Leichtling BH, Klotz IM. Catalysis of hydrogen-deuterium exchange in peptides. Biochemistry. 1966;5:4026–4036. [Google Scholar]
- 33.Scarpa JS, Mueller DD, Klotz IM. Slow hydrogen-deuterium exchange in a non-α-helical polyamide. J Am Chem Soc. 1967;89:6024–6030. [Google Scholar]
- 34.Hvidt A, Corett E. Kinetics of hydrogen-deuterium exchange in poly(N-vinylacetamide) measured by infrared spectroscopy. J Am Chem Soc. 1970;92:5546–5550. [Google Scholar]
- 35.Ragnarsson U, Karlsson SM, Sandberg BEB. Studies on the coupling step in solid phase peptide synthesis. Further competition experiments and attempts to assess formation of ion pairs. J Org Chem. 1974;39:3837–3841. doi: 10.1021/jo00940a008. [DOI] [PubMed] [Google Scholar]
- 36.Covington AK, Robinson RA, Bates RG. The ionization constant of deuterium oxide from 5 to 50 degrees. J Phys Chem. 1966;70:3820–3824. [Google Scholar]
- 37.Englander SW. Measurement of structural and free energy changes in hemoglobin by hydrogen exchange methods. Ann NY Acad Sci. 1975;244:10–27. doi: 10.1111/j.1749-6632.1975.tb41518.x. [DOI] [PubMed] [Google Scholar]
- 38.Englander SW, Mayne L. Protein folding studied by hydrogen exchange labeling and 2D NMR. Annu Rev Biophys Biomol Struct. 1992;21:243–265. doi: 10.1146/annurev.bb.21.060192.001331. [DOI] [PubMed] [Google Scholar]
- 39.Ptitsyn OB. Protein folding: Hypotheses and experiments. J Protein Chem. 1987;6:273–293. [Google Scholar]