Abstract
New pharmacologic targets are needed for lung cancer. One candidate pathway to target is composed of the E1-like ubiquitin-activating enzyme (UBE1L) that associates with interferon-stimulated gene 15 (ISG15), which complexes with and destabilizes cyclin D1. Ubiquitin protease 43 (UBP43/USP18) removes ISG15 from conjugated proteins. This study reports that gain of UBP43 stabilized cyclin D1, but not other D-type cyclins or cyclin E. This depended on UBP43 enzymatic activity; an enzymatically inactive UBP43 did not affect cyclin D1 stability. As expected, small interfering RNAs (siRNAs) that reduced UBP43 expression also decreased cyclin D1 levels and increased apoptosis in a panel of lung cancer cell lines. Forced cyclin D1 expression rescued UBP43 apoptotic effects, which highlighted the importance of cyclin D1 in conferring this. Short hairpin RNA (shRNA)-mediated reduction of UBP43 significantly increased apoptosis and reduced murine lung cancer growth in vitro and in vivo after transplantation of these cells into syngeneic mice. These cells also exhibited increased response to all-trans-retinoic acid (RA), interferon (IFN), or cisplatin treatments. Notably, gain of UBP43 expression antagonized these effects. Normal-malignant human lung tissue arrays were examined independently for UBP43, cyclin D1, and cyclin E immunohistochemical expression. UBP43 was significantly (P < 0.01) increased in the malignant versus normal lung. A direct relationship was found between UBP43 and cyclin D1 (but not cyclin E) expression. Differential UBP43 expression was independently detected in a normal-malignant tissue array with diverse human cancers. Taken together, these findings uncovered UBP43 as a previously unrecognized anti-neoplastic target.
Keywords: UBP43/USP18, cyclin D1, lung cancer
Introduction
Lung cancer is the leading cause of cancer mortality for men and women in the United States (1). The 5 year survival rate for lung cancer patients is only 16% (1). Innovative ways to combat lung cancer are needed.
Carcinogenesis is a multistep process. Frequent genetic changes arise in human lung carcinogenesis and some drive this process (2). Prior work highlighted cyclin D1 as a chemopreventive or therapeutic target, as reviewed (3,4). Increased cyclin D1 expression occurs early during lung carcinogenesis (5,6) through gene amplification (7) or allele-specific expression imbalance (6), among other mechanisms. These and other findings implicated cyclin D1 as an antineoplastic target in the lung.
Lung carcinogenesis is inhibited by induced cyclin D1 proteasomal degradation. This occurred after treatment with retinoid X receptor (RXR, rexinoid) or retinoic acid receptor (RAR, retinoid) agonists (2,8,9). Cyclin D1 protein is also repressed by activating a pathway constituted by these retinoid regulated species: ubiquitin activating enzyme E1-like (UBE1L), interferon-stimulated gene 15 (ISG15) and ubiquitin protease UBP43 (UBP43/USP18) (10,11). This study examined whether UBP43 was an antineoplastic target.
UBE1L associates with ISG15, the ubiquitin-like protein family member; ISG15 conjugation is specifically removed by UBP43, the ubiquitin-specific protease (USP) family member (12,13). Deregulation of the UBE1L-ISG15-UBP43 pathway occurs in diverse tumor types (14–18). All-trans-retinoic acid (RA)-treated human bronchial epithelial (HBE) and acute promyelocytic leukemia (APL) cells increased UBE1L, ISG15, and UBP43 expression (11,15,16 and data not shown). Gain of UBP43 expression stabilized PML/RARα in APL cells (11) and augmented cyclin D1 expression in HBE cells (10). These findings implicated UBP43 as directly regulating the stability of cyclin D1 and other growth-regulatory proteins. Evidence for cyclin D1 as a chemopreventive target also came from mouse models and clinical studies (3,5,8,9,19–23).
This study uncovered UBP43-dependent mechanisms that regulated lung cancer growth and tumorigenesis. Prior work (10) was extended by reporting gain of UBP43 expression preferentially stabilized cyclin D1 protein. UBP43 knock-down destabilized cyclin D1, but not other examined G1 cyclin proteins. This was confirmed by UBP43 transfection and treatment with cycloheximide (CHX), the protein synthesis inhibitor, which increased cyclin D1 protein stability versus controls. UBP43 transduction reduced apoptosis and increased cyclin D1 expression as well as growth of HBE and lung cancer cells. In contrast, UBP43 knock-down decreased cyclin D1 expression and inhibited growth and increased apoptosis in murine and human lung cancer cells. This significantly decreased lung cancer formation in FVB mice transplanted with syngeneic lung cancer cells that were engineered with reduced UBP43 expression. To explore directly whether UBP43 enzymatic activity mediated this, an enzymatically-inactive UBP43 species was engineered and used in these experiments.
To elucidate therapeutic implications, growth and apoptosis assays were performed after individual treatments with RA, interferon (IFN), or cisplatin in lung cancer cells independently engineered with loss or gain of UBP43 expression. To ascertain clinical relevance, UBP43 immunohistochemical expression profiles were studied using new assays and a normal-malignant human lung tissue array. UBP43 was significantly increased in the malignant versus normal lung tissues. A direct relationship was found between UBP43 and cyclin D1. Results were confirmed and extended using a normal-malignant tissue array with diverse tumor types. Taken together, these findings implicate UBP43 as a tractable target for lung and other cancers.
Materials and Methods
Cell Culture
The ED-1 murine lung cancer cell line was cultured in RPMI 1640 (Invitrogen, Carlsbad, CA) medium with 10% fetal bovine serum (FBS, Gemini, Calabasas, CA), 4mM L-glutamine (Invitrogen, Carlsbad, CA), 100units/ml penicillin (Invitrogen), and 100μg/ml streptomycin (Invitrogen) (24). ED-1L cells were isolated as a single cell subclone of ED-1 cells. H23, HOP62, and A549 lung cancer cell lines were cultured as before (25). BEAS-2B immortalized HBE cells were cultured in serum-free LHC-9 medium (Biofluids, Akron, OH) (10,11). PT67 and 293 cells were cultured in DMEM (Invitrogen) medium with 10% FBS (Gemini), 4mM L-glutamine (Invitrogen), 100units/ml penicillin (Invitrogen), and 100μg/ml streptomycin (Invitrogen). Cells were cultured at 37°C in a humidified incubator with 5% CO2. Cell lines were obtained from and authenticated (using genotypic and phenotypic assays) by American Type Culture Collection (ATCC, Manassas, VA) except for ED-1 cells that were previously authenticated (24).
Expression Plasmids and Transient Transfection
The pcDNA4-UBP43, pSG5-UBE1L, pCMV-hemagglutinin (HA)-ISG15, pCMV-HA-cyclin D1, lysine-less pCMV-HA-cyclinD1 33–269 (mut-cyclin D1), pCMV-HA-cyclin D2, pCMV-HA-cyclin D3, pCMV-HA-cyclin E, enhanced green fluorescent protein (EGFP) expression plasmid, and pRetroX-IRES-ZsGreen1-UBP43 retroviruses were previously described (10,11,26). Flag-tagged-UbcH8 was purchased (Addgene, Cambridge, MA). Short hairpin RNA (shRNA) retroviruses and lentiviruses were used to knock-down human or mouse UBP43 species (Open Biosystems, Huntsville, AL). Empty vectors (Open Biosystems) were controls (11). Indicated UBP43 cysteine (C) residues were transversed to serine (S) residues (to render UBP43 enzymatically inactive) using the QuickChange site-directed mutagenesis kit (Strategene, La Jolla, CA). DNA sequence analyses confirmed engineered species.
Transient transfection of logarithmically growing cells was accomplished using the Effectene Transfection Reagent (Qiagen, Valencia, CA) (11,26). EGFP-expressing plasmids assessed transfection efficiencies. Transfections were in triplicate. Each experiment was independently replicated three times.
Transient transfection of cells with the desired small interfering RNAs (siRNAs) was accomplished using Lipofectamine 2000 (Invitrogen) reagent. Different siRNAs that targeted UBP43, cyclin D1 or a RISC-free control siRNA were synthesized (Dharmacon, Lafayette, CO). Two siRNAs that independently targeted UBP43 were: human UBP43 siRNA1 (5′-CTGCATATCTTCTGGTTTA-3′), human UBP43 siRNA2 (5′-GGAAGAAGACAGCAACATG-3′), murine UBP43 siRNA1 (5′-CGTTGTTTGTCCAGCACGA-3′) and murine UBP43 siRNA2 (5′-AGGAACTCGAGGACGGAAA-3′). Two siRNAs that independently targeted human cyclin D1 were: cyclin D1 (5′-ACAACTTCCTGTCCTACTA-3′, siRNA3) and (5′-GTTCGTGGCCTCTAAGATG-3′, siRNA4). Transfection efficiencies were monitored by co-transfecting the siGLO Green Transfection Indicator (Dharmacon).
Generation of Stable UBP43 Transfectants
The pRetroX-IRES-ZsGreen1-UBP43 retroviral vector or an empty retrovirus was transfected into the RetroPack PT67 Packaging Cell Line (Clontech) using Effectene Transfection Reagent (Qiagen). Viral supernatants transduced cells with polybrene (4μg/ml) (Sigma, Milwaukee, WI). Green Fluorescent Protein (GFP) positive cells were harvested 48 hours later using a FACStar Plus cytometer (Becton Dickinson, San Jose, CA). This was repeated a week later. Three independent experiments were performed, each in triplicate.
ShRNA retrovirus (for human cells) and lentivirus (for murine cells) for UBP43 knock-downs were generated in PT67 cells or 293T cells, respectively. Stable selection was achieved with puromycin (2μg/ml, Sigma) treatment. Five candidate shRNAs were individually examined. The two with the most efficient UBP43 knock-downs were studied.
Real-Time PCR Assays
For real-time reverse transcription (RT) assays, primers were: human UBP43 forward primer 5′-GAGGCTGGACGCTTGCAT-3′ and reverse primer 5′-AGCACGACTTCACTTCCAGGAA-3′ human cyclin D1 forward primer 5′-AACTACCTGGACCGCTTCCT-3′ and reverse primer 5′-CCACTTGAGCTTGTTCACCA-3′ human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward primer 5-ACCTTTGGCATTGTGGAGG-3′ and reverse primer 5′-ACACATTGGGGGTAGGAACA-3′. Assays were performed using established methods (11).
Immunoblot Analyses
Cells were lysed with ice-cold radioimmunoprecipitation (RIPA) buffer using sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) assays (10,11,26). Primary antibodies for immunoblot analyses were two independently developed polyclonal antibodies recognizing UBP43 (11), rabbit polyclonal antibody against cyclin D1 (Santa Cruz, Santa Cruz, CA), murine monoclonal antibody against HA-tagged proteins (Covance, Princeton, NJ), murine monoclonal antibody for Flag-tagged proteins (OriGene, Rockville, MD), and a goat polyclonal antibody for actin (Santa Cruz). Secondary anti-mouse and anti-rabbit antibodies were purchased (Amersham, Piscataway, NJ) as was an anti-goat (Santa Cruz) antibody. Quantifications of signals were as before (11). To assess cyclin D1 protein stability after UBP43 transfection, cells were treated with or without CHX (40μg/ml, Sigma).
Proliferation and Apoptosis Assays
Growth was measured using the CellTiter-Glo Assay Kit (Promega, Madison, WI) (19). Triplicate replicate experiments were performed. To assess for drug responses, independent stable transductants with loss or with gain of UBP43 expression were each treated with or without RA (Sigma), IFN (PBL, Piscataway, NJ), or cisplatin (Sigma). Empty retroviruses served as controls. Apoptosis was measured by Annexin V:FITC positivity and flow cytometry using the Annexin V Assay Kit (AbD Serotec, Raleigh, NC). Cell lines were assayed with the Caspase-Glo 3/7 Assay Kit (Promega).
In Vivo Tumorigenicity and Statistical Assays
Lentiviral-mediated UBP43 knock-down was achieved in ED-1 cells. Cells were harvested in phosphate buffered saline (PBS) supplemented with 10% mouse serum (Invitrogen); 8 × 105 cells from each transfectant were individually injected into tail veins of respective 8-week-old female FVB syngeneic mice. Ten mice were used in each experimental arm with replicate experiments performed. Mice were sacrificed 4 weeks after injections following an Institutional Animal Care and Use Committee (IACUC)-approved protocol. Tissues were formalin-fixed, processed and hematoxylin and eosin stained (27). Scoring of lung tumors was as in prior work (25).
Normal-Malignant Tissue Arrays
Resected non-small cell lung carcinomas (NSCLCs) with case-matched adjacent histopathologically normal lung tissues were examined using a normal-malignant lung tissue array from the New Hampshire State Cancer Registry and the Dartmouth-Hitchcock Tumor Registry (25). Studies were reviewed and approved by Dartmouth’s Institutional Review Board (IRB) for human subjects.
Normal and malignant lung tissues were each formalin-fixed and processed as before (27). Tissues were independently examined for cyclin D1 and cyclin E immunohistochemical expression profiles (10,27). Results were compared to those obtained with different polyclonal anti-UBP43 antibodies used to detect UBP43 immunohistochemically (11). Antibody specificities were confirmed using blocking peptides.
To extend analyses to other human cancers, a normal-malignant tissue array with lung and other cancers was purchased (Life Span Biosciences, Seattle, WA) and probed for UBP43 immunohistochemical expression. UBP43 levels were scored by the reference pathologist (V.M.), who quantified cytoplasmic staining using established methods (22).
Statistical Analyses
Two-tailed T-tests were used. Results were presented as means ± standard deviation (S.D.). Statistical significance was noted with these symbols: * P < 0.05 and ** P < 0.01.
Results
In Vitro Studies
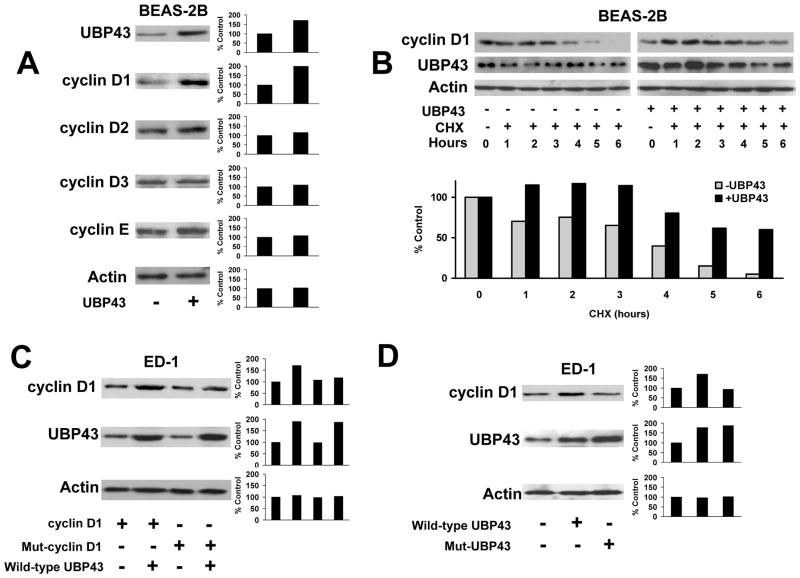
Prior work implicated UBP43 as an antineoplastic target that opposed UBE1L effects on cyclin D1 (10,16). To determine whether UBP43 affected stability of other G1 cyclins, HBE cells were transfected with UBP43 and independently probed for exogenously expressed cyclin D1, cyclin D2, cyclin D3, or cyclin E proteins. Only cyclin D1 was appreciably stabilized by gain of UBP43 expression (Fig. 1A). UBP43 transfection also increased endogenous cyclin D1 protein, but not cyclin D1 mRNA expression (Supplemental Fig. 1 and data not shown). To establish whether UBP43 affected cyclin D1 protein stability, UBP43 was co-transfected with HA-tagged cyclin D1 into BEAS-2B cells treated +/− CHX. UBP43 increased exogenous cyclin D1 protein stability +/− CHX treatments (Fig. 1B). Specific lysines in cyclin D1 affected its stability (26). Since lysines are targets of ISG15 conjugation (10), it was expected that lysine-less cyclin D1 would be resistant to UBP43 effects on its stability (Fig. 1C).
Figure 1.
Effects of UBP43 on cyclin D1 protein stability. (A) Effects of UBP43 on individual transiently-transfected D-type and cyclin E species. UBP43 transfection (+) or empty vector transfection (−) was accomplished in BEAS-2B HBE cells with immunoblot analyses subsequently performed. Actin expression served as a loading control. Quantification of each respective signal is displayed in the right panel. The percent change in expression of each of these cyclins relative to respective actin expression is presented. (B) Effects of UBP43 co-transfection on transfected HA-tagged cyclin D1 immunoblot expression in the presence (+) or absence (−) of cycloheximide (CHX) treatment of BEAS-2B cells. UBP43 stabilized (versus actin control) exogenous cyclin D1 protein, which was detected by an anti-HA antibody. Exogenous cyclin D1 expression was enhanced despite CHX treatment. Quantification of signals is provided in the panel below this immunoblot. (C) Consequences of UBP43 transfection on wild-type cyclin D1 and independently on lysine-less cyclin D1 (Mut-cyclin D1) species. These respective species were independently transfected into ED-1 lung cancer cells in the presence (+) or absence (−) of co-transfected UBP43. Compared with effects on wild-type cyclin D1, UBP43 did not appreciably augment lysine-less cyclin D1 expression. Quantification of signals is provided in right panels. (D) Effects of cyclin D1 protein stability on wild-type (wild-type UBP43) or an enzymatically-inactive UBP43 species (Mut-UBP43) after their respective co-transfections. Wild-type and mutant UBP43 species were independently co-transfected into ED-1 cells along with HA-tagged-cyclin D1 species. Compared to wild-type UBP43, this mutant UBP43 species did not stabilize cyclin D1 expression. Quantification of these respective signals is in the right panel.
UBP43 enzymatic activity is required for ISG15 deconjugation (28). Whether cyclin D1 stabilization by UBP43 depended on this activity was explored. Active site cysteine residues of UBP43 (C64 and C65) were transversed to serines to render UBP43 inactive (designated UBP43-S64.S65 or Mut-UBP43). Fig. 1D shows transfection of ED-1 lung cancer cells with this mutant UBP43 species did not appreciably affect cyclin D1 stability versus cells transfected with wild-type UBP43 that increased cyclin D1 expression.
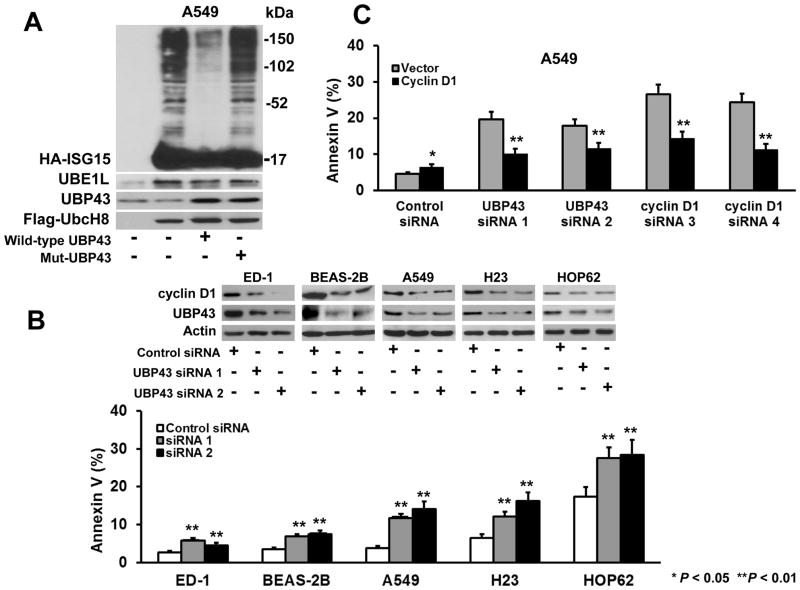
UBP43 is the protease specific for ISG15 (13,29). Complexes between ISG15 and UBP43 were detected in APL cells (11). Whether catalytically inactive UBP43-S64.S65 affected ISG15 conjugation or cyclin D1 stability was studied in lung cancer cells individually transfected without exogenous UBP43, with wild-type UBP43, or with this inactive UBP43 species. As expected, expression of the enzymatically-inactive UBP43 in A549 cells did not affect overall ISG15 conjugation while over-expression of wild-type UBP43 markedly reduced this conjugation (Fig. 2A).
Figure 2.
UBP43 effects on ISG15-conjugation and cellular apoptosis. (A) A549 lung cancer cells were transiently transfected with vector-control or plasmids containing HA-ISG15, UBE1L, or Flag-tagged-UbcH8 species in the absence or the presence of wild-type or the described mutant UBP43 species. The level of overall ISG15 conjugation was assessed with an anti-HA antibody that recognized the HA-tagged ISG15. As compared with wild-type UBP43 transfection, transfected mutant UBP43 (Mut-UBP43) species did not reduce ISG15 conjugates, consistent with its enzymatic inactivity (B) UBP43 knock-down in lung cancer and HBE cells caused apoptosis. Independent transient transfection of two independent UBP43-targeting siRNAs versus a control siRNA for 24 hours reduced UBP43 and cyclin D1 immunoblot expression in ED-1, BEAS-2B, A549, H23 and HOP62 cells (upper panel). UBP43 knock-down by each siRNA-targeting UBP43 significantly induced an apoptosis marker versus transfected control siRNA (lower panel). (C) Cyclin D1 involvement in UBP43-dependent apoptosis in A549 lung cancer cells. Assays confirmed that siRNA-mediated knock-down of UBP43 led to an induced apoptosis marker (gray bars) versus control siRNA. Transient transfection of independent cyclin D1-targeting siRNAs increased apoptosis versus a control siRNA. Co-transfection of cyclin D1 with respective siRNAs-targeting UBP43 or cyclin D1 species significantly reduced apoptosis (black bars). The expected changes in expressed UBP43 and cyclin D1 mRNAs as well as findings from rescue experiments are shown in Supplemental Fig. 2. Standard deviation bars are shown. Symbols refer to *P < 0.05 and ** P < 0.01.
Gain of UBP43 expression increased cyclin D1 and PML/RARα expression (10,11). Whether UBP43 repression affected apoptosis in lung cancer cells via changes in cyclin D1 levels was studied. Two different siRNAs that targeted UBP43 and an inactive control siRNA were each independently transfected into murine ED-1 lung cancer and BEAS-2B HBE cells as well as A549, H23 and HOP62 human lung cancer cell lines (Fig. 2B). Knock-down of UBP43 by each of these UBP43-targeting siRNAs substantially decreased UBP43 and cyclin D1 immunoblot expression versus control cells (Fig. 2B). Actin expression was unaffected. Compared with controls, siRNA mediated knock-down of UBP43 significantly augmented apoptosis (Fig. 2B, lower panel). Supplemental Fig. 1A revealed UBP43 knock-down (versus controls) in ED-1 cells augmented an apoptosis marker at day 1. This marker increased over 3 days of study.
Forced cyclin D1 expression antagonized UBP43 knock-down effects on apoptosis in A549 (Fig. 2C) and ED-1 (data not shown) lung cancer cells. The respective increases or decreases in UBP43 and cyclin D1 expression profiles were confirmed (Supplemental Fig. 2A and 2B). As expected, engineered expression of wild-type, but not enzymatically inactive UBP43 antagonized the effects of siRNA-mediated UBP43 knock-down on cyclin D1 expression (Supplemental Fig. 2C). Representative FACS analyses are displayed in Supplemental Fig. 3.
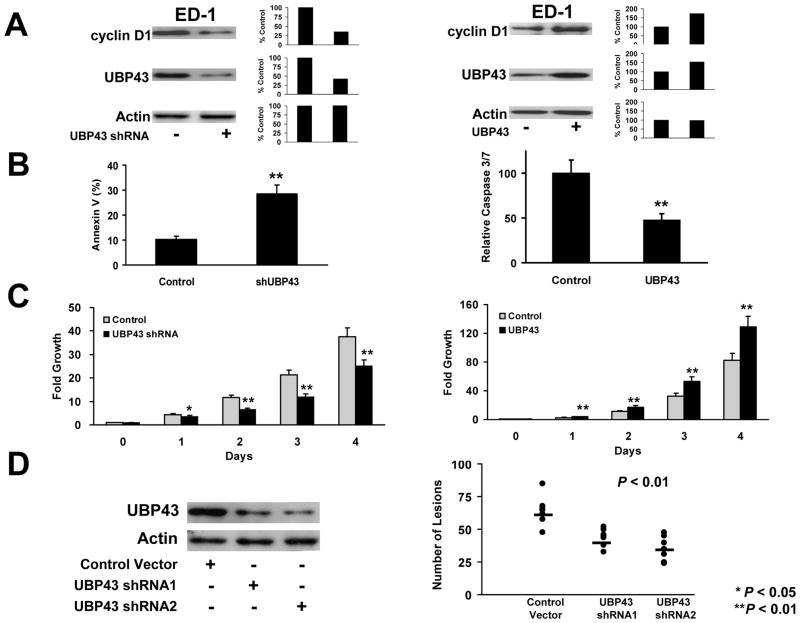
To confirm and extend these findings, stable shRNA-mediated repression of UBP43 was achieved in lung epithelial or cancer cells. Effects of loss of UBP43 expression on cyclin D1 and apoptosis were studied in ED-1 and BEAS-2B transductants. Stable UBP43 knock-down was engineered by lentiviral (for ED-1) or retroviral (for BEAS-2B) transductions followed by puromycin selection of desired shRNAs in these cells. Fig. 3A and Supplemental Fig. 4A (left panels) showed shRNA-mediated UBP43 knock-down reduced endogenous cyclin D1 protein expression in transductants versus controls. UBP43 knock-down significantly increased apoptosis in both examined cell lines (Fig. 3B, left panel and Supplemental Fig. 4B, left panel).
Figure 3.
Engineered UBP43 knock-down versus UBP43 over-expression in ED-1 lung cancer cells. (A) Stable UBP43 knock-down by lentiviral-mediated shRNA transduction or engineered UBP43 over-expression via retroviral-mediated transduction of UBP43 in ED-1 cells. UBP43 shRNA transduction (+) destabilized endogenous cyclin D1 protein expression as compared with a transfected empty vector (−) (left panel). Retroviral transduction (+) of UBP43 increased UBP43 expression and augmented cyclin D1 expression in ED-1 cells (right panel). Actin expression confirmed similar protein loading in each line. Quantification of signals is provided. (B) Lentiviral-mediated knock-down of UBP43 triggered significant apoptosis (left panel). UBP43 retroviral-mediated transduction inhibited apoptosis (right panel) in ED-1 cells versus the empty control vector transduction. (C) Lentiviral-mediated knock-down of UBP43 significantly inhibited proliferation (left panel) and engineered gain of UBP43 expression independently augmented proliferation (right panel) of ED-1 cells versus these empty control transduced cells. (D) Engineered UBP43 knock-down in ED-1 cells significantly reduced (P< 0.01) tumorigenicity of these transduced lung cancer cells. The indicated transfected UBP43 shRNA knock-downs of ED-1 cells repressed in vivo lung tumorigenicity after tail vein injections into FVB mice. The horizontal bars represented the respective median numbers of lung cancers for UBP43 shRNA-mediated transduction of ED-1 cells relative to mice injected with control vector transduced ED-1 cells. Each circle in this figure represents an individual mouse. Standard deviation bars are shown. Symbols refer to * P < 0.05 and ** P < 0.01.
Gain of UBP43 expression was independently achieved in ED-1 and BEAS-2B cells in Fig. 3A (right panel) and Supplemental Fig. 4A (right panel). Unlike UBP43 knockdown, retroviral-mediated UBP43 expression augmented cyclin D1 in both of these cell lines (Fig. 3A, right panel and Supplemental Fig. 4A, right panel) and significantly reduced apoptosis (Fig. 3B, right panel and Supplemental Fig. 4B, right panel) versus empty vector controls.
UBP43 effects on growth of ED-1 and BEAS-2B cells were studied independently after loss and also after gain of UBP43 expression. CellTiter-Glo assays confirmed UBP43 knock-down markedly repressed growth of these cells (Fig. 3C and Supplemental Fig. 4C, left panels). In contrast, UBP43 over-expression significantly increased (P < 0.01) growth (versus empty vectors) of these cells (Fig. 3C, right panel and Supplemental Fig. 4C, right panel).
Tumorigenicity
UBP43 knock-down effects on tumorigenicity were next studied. FVB mice were injected via their tail veins with syngeneic ED-1 cells (24) engineered with different lentiviral-mediated shRNAs to achieve stable UBP43 knock-down. Results were compared to empty vector transductants. Four weeks after injections, lung lesions were scored. ED-1 cells independently engineered with reduced UBP43 expression using different UBP43-repressing shRNAs produced significantly fewer (P < 0.01) lung cancers versus controls (Fig. 3D). Representative photomicrographs of lung tumors that formed after tail-vein injections of these respective engineered cells are presented in Supplemental Fig. 5A. In vitro growth of these transductants (versus parental ED-1 cells) is displayed in Supplemental Fig. 5B.
Antineoplastic Drug Responses
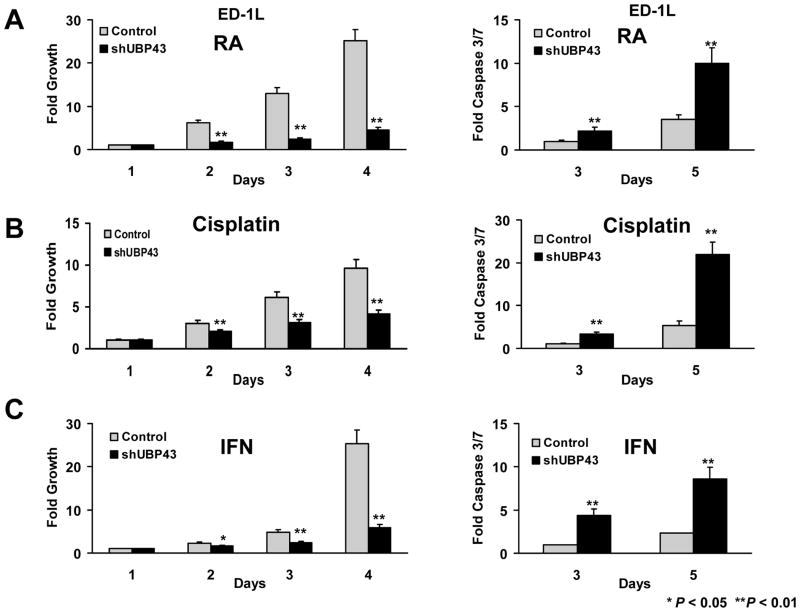
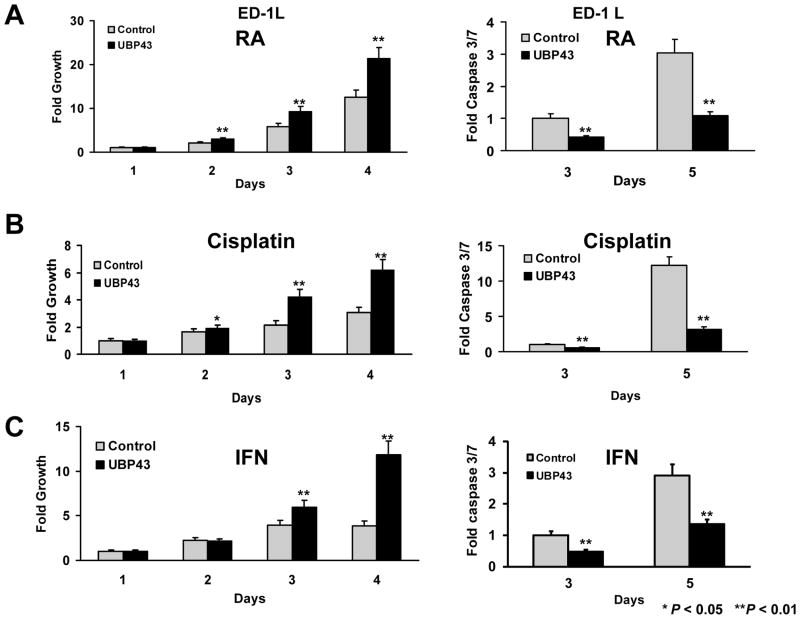
UBP43 is an IFN and retinoid-regulated species (10,11,13,30,31). This implied that regulating UBP43 levels would affect response to these and other anti-neoplastic agents. A single cell subclone of ED-1 cells (ED-1L) was engineered with gain and also independently with loss of UBP43 expression (see Supplemental Fig. 1). Individual treatment effects of RA, cisplatin, or IFN on growth and apoptosis assays were studied. UBP43 knock-down followed by RA, cisplatin, or IFN treatments increased growth-inhibitory and pro-apoptotic effects of each agent (Fig. 4). In contrast, forced UBP43 expression antagonized these ED-1L cellular effects (Fig. 5).
Figure 4.
Growth and apoptotic effects of drug treatments in lung cancer cells engineered with UBP43 knock-down (designated as shUBP43) versus control vector (Control). (A) Treatment with RA (1μM) inhibited cell growth (left panel) and triggered a significant increase in apoptosis (right panel) in these ED-1L cells having lentiviral-mediated UBP43 knock-down. (B) Cisplatin (2.5μM) treatments of the same UBP43-mediated knock-down cells inhibited proliferation (left panel) and promoted apoptosis (right panel). (C) IFN (1000U) treatments decreased cell growth and increased apoptosis in the same UBP43 knock-down cells. Standard deviation bars are shown. Symbols refer to * P < 0.05 and ** P < 0.01.
Figure 5.
Growth and apoptotic effects of drug treatments in lung cancer cells engineered with UBP43 over-expression (designated UBP43) versus control vector (Control). (A) Treatment with RA (1μM) reduced growth inhibition (left panel) and significantly decreased apoptosis (right panel) in engineered ED-1L cells having UBP43 over-expression. (B) Cisplatin (2.5μM) treatments inhibited growth suppression (left panel) and apoptosis (right panel) in ED-1L cells over-expressing UBP43 relative to controls. (C) IFN (1000U) treatments increased cell growth and decreased apoptosis in UBP43 over-expressing ED-1L cells versus controls. Standard deviation bars are shown. Symbols refer to * P < 0.05 and ** P < 0.01.
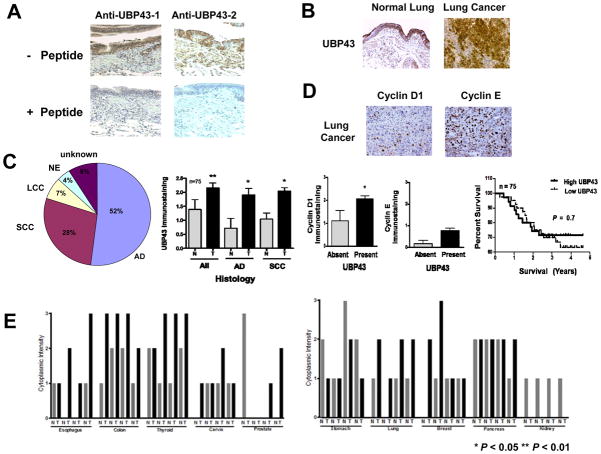
Differential UBP43 expression profiles were examined in normal versus malignant lung tissues. Two different polyclonal anti-UBP43 antibodies detected UBP43 in immunoblot assays (11). These antibodies were used in immunohistochemical assays. Both the anti-UBP43 antibody recognizing a domain nearer to the amino terminus (anti-UBP43-1) than the second anti-UBP43 antibody (anti-UBP43-2) detected UBP43 immunohistochemical expression in the normal lung with specificity confirmed using blocking peptides (Fig 6A). A representative lung cancer and a representative adjacent normal lung tissue are displayed in Fig. 6B. To extend this analysis, a paired normal-malignant human lung tissue array from the New Hampshire and Dartmouth lung cancer registry (25) was examined individually for UBP43, cyclin D1 and cyclin E immunohistochemical expression. UBP43 levels from 75 NSCLC cases (see Venn diagram) versus normal lung are shown in Fig. 6C. UBP43 expression was significantly increased in the malignant versus adjacent normal lung tissues (P = 0.04 adenocarcinoma and P = 0.02 for squamous cell carcinoma). Significant associations were found between UBP43 and cyclin D1, but not for cyclin E (Fig. 6C). There was not a significant difference in survival in lung cancer cases with high versus low UBP43 levels. Representative individual immunohistochemical assays for cyclin D1 and cyclin E are shown in lung cancers Fig. 6D. To determine whether UBP43 was differentially expressed in other cancers, a normal-malignant tissue array was examined from many cancer subtypes. Imunohistochemical findings in Fig. 6E and Supplemental Fig. 6 revealed increased UBP43 expression in diverse cancers versus corresponding normal tissues. Reduced or absent UBP43 expression was found in several kidney cancers or normal as well as malignant prostate tissues.
Figure 6.
UBP43 expression profiles in paired normal-malignant tissue arrays. (A) Immunohistochemical assays for UBP43 expression levels were performed using two different anti-UBP43 antibodies. Blocking peptides for each respective antibody were used. UBP43 protein expression was detected in the normal human lung. These peptides blocked UBP43 staining by each respective antibody. (B) UBP43 immunohistochemical expression was enhanced in the malignant versus normal human lung tissues. (C) Differential expression profiles for UBP43, cyclin D1 and cyclin E in a paired normal-malignant lung tissue array. Different histopathologic lung cancers are shown (All = NSCLCs, AD = adenocarcinoma, and SCC = squamous cell carcinoma). Significant associations were found between UBP43 and cyclin D1, but not with cyclin E. The Venn diagram displayed lung cancer histopathologies (NE = neuroendocrine, LCC = large cell carcinoma, AD = adenocarcinoma, and SCC = squamous cell carcinoma). There was no significant survival difference in lung cancer cases with high versus low UBP43 expression (right panel). (D) Representative immunohistochemical expression of cyclin D1 and cyclin E in human lung cancers. (E) Differential UBP43 immunohistochemical expression profiles in a tissue array with diverse normal-malignant human tissues. Symbols refer to * P < 0.05 and ** P < 0.01.
Discussion
This study advances prior work (10) by showing the ubiquitin protease UBP43 can regulate cyclin D1 stability. UBP43 is important in IFN and immune responses, leukemogenesis, among other biological effects (11,12,14,17,18). UBP43 acts by removing ISG15 from conjugated proteins (12,13). Our prior work implicated UBP43 in regulating stability of the oncogenic protein, PML/RARα (11). This study identified cyclin D1 as a growth regulatory protein whose stability was also affected by UBP43. UBE1L-ISG15-UBP43 pathway effects on protein stability, apoptosis, antineoplastic drug responses, and tumorigenicity are summarized in Supplemental Fig. 7.
Aberrant cyclin expression occurs in lung carcinogenesis (5–7). This study found that UBP43 stabilized cyclin D1 (but not other D-type cyclins or cyclin E) (Fig. 1A). This established UBP43 as a critical regulator of cyclin D1 protein stability. Cyclin D1 stabilization by UBP43 required UBP43 enzymatic activity (Fig. 1D, Fig. 2B and Supplemental Fig. 2C), but not de novo protein synthesis (Fig. 1). Cyclin D1 destabilization was engaged in diverse cells (Fig. 2B). Reduction of UBP43 repressed cyclin D1 expression. Specific cyclin D1 residues affected proteasomal degradation (26). The current work showed cyclin D1 stabilization by UBP43 also depended on lysines within cyclin D1 (Fig. 1C).
Cyclin D1 can affect apoptosis, as reviewed (32). UBE1L also regulated cyclin D1 stability and apoptosis (10,15). Results presented here build on these findings by showing UBP43 knock-down decreased cyclin D1 expression and significantly increased apoptosis in lung cancer cells (Fig. 2B). Forced cyclin D1 expression antagonized pro-apoptotic effects of UBP43 knock-down (Fig. 2C), indicating cyclin D1 played a key role in this process.
Increased UBE1L expression augmented apoptosis (15). Loss of UBP43 expression caused a similar effect. UBP43 was shown here to regulate cyclin D1 expression, cell growth, apoptosis and tumorigenicity. UBP43 knock-down also inhibited lung cancer formation in vivo (Fig. 3D). As expected, gain of UBP43 expression antagonized these growth and apoptotic effects.
Combination therapy is a tenet of cancer therapy (2,33). A desired regimen is one where a target like UBP43 can modulate responses to antineoplastic agents. UBP43 was found in this study to regulate response to RA, cisplatin, and IFN. Loss of UBP43 expression increased growth inhibitory and pro-apoptotic effects of these agents, but gain of UBP43 expression antagonized these activities (Fig. 4 and Fig. 5). These findings have therapeutic implications. An inhibitor of UBP43 should exert antineoplastic effects. Its effects would likely cooperate with RA, cisplatin, IFN and other agents.
Translational research was extended by studies of paired normal and malignant tissue arrays (Fig. 6). UBP43 immunohistochemical expression was significantly increased in the malignant versus normal lung. The same paired normal-malignant lung tissue array was probed for cyclin D1 and cyclin E expression. A significant association was found between UBP43 and cyclin D1, but not cyclin E (Fig. 6C). Yet, UBP43 levels alone did not predict prognosis in lung cancer (Fig. 6D). Prior work implicating UBE1L as a tumor suppressor in the lung (34) is extended here by showing UBP43 regulates cyclin D1 expression as well as apoptosis, proliferation and tumorigenicity of cancer cells.
Interactions between UBE1L-ISG15-UBP43 pathway members may prove important. Intriguingly, these species and their E2 (UbcH8) and E3 (Herc5a) undergo ISG15yaltion (35), indicating an autoregulatory UBP43 loop might exist. Therapeutic implications are highlighted by finding that UBP43 is over-expressed in diverse human cancers (Fig. 6).
In summary, this study uncovered UBP43 as an anti-neoplastic target. Enzymatically-active UBP43 directly affected cyclin D1 stability. UBP43 knock-down repressed cyclin D1 expression, promoted apoptosis, and cooperated with effects of antineoplastic agents. Loss of UBP43 also repressed cyclin D1 expression and reduced tumorigenicity in a murine syngeneic lung cancer model. UBP43 expression was augmented in human lung cancers (and other cancers) relative to the corresponding normal tissues. A therapeutic window likely exists in lung and other cancers for an inhibitor of UBP43 to exert anti-tumorigenic effects. Future work should explore this possibility given the need for improved treatments for lung cancers.
Supplementary Material
Acknowledgments
The authors thank all of the members of the Dmitrovsky Laboratory for their helpful consultations in the completion of this work.
Grant Support
This work was supported by National Institutes of Health (NIH) and National Cancer Institute (NCI) grants R01-CA087546 (E.D.), R01-CA062275 (E.D.), and R01-CA111422 (E.D.), by a Samuel Waxman Cancer Research Foundation (SWCRF) grant (E.D.), and an American Cancer Society Institutional Grant (S.J.F). E. Dmitrovsky is an American Cancer Society Professor supported by a generous gift from the F.M. Kirby Foundation.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparity on premature deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Dragnev KH, Stover D, Dmitrovsky E. Lung cancer prevention: the guidelines. Chest. 2003;123:60S–71S. doi: 10.1378/chest.123.1_suppl.60s. [DOI] [PubMed] [Google Scholar]
- 3.Freemantle SJ, Guo Y, Dmitrovsky E. Retinoid chemoprevention trials: cyclin D1 in the crosshairs. Cancer Prev Res. 2009;2:3–6. doi: 10.1158/1940-6207.CAPR-08-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petty WJ, Dragnev KH, Dmitrovsky E. Cyclin D1 as a target for chemoprevention. Lung Cancer. 2003;41:S155–61. doi: 10.1016/s0169-5002(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 5.Lonardo F, Rusch V, Langenfeld J, Dmitrovsky E, Klimstra DS. Overexpression of cyclins D1 and E is frequent in bronchial preneoplasia and precedes squamous cell carcinoma development. Cancer Res. 1999;59:2470–6. [PubMed] [Google Scholar]
- 6.Ratschiller D, Heighway J, Gugger M, Kappeler A, Pirnia F, Schmid RA, et al. Cyclin D1 overexpression in bronchial epithelia of patients with lung cancer is associated with smoking and predicts survival. J Clin Oncol. 2003;21:2085–93. doi: 10.1200/JCO.2003.03.103. [DOI] [PubMed] [Google Scholar]
- 7.Betticher DC, Heighway J, Hasleton PS, Altermatt HJ, Ryder WD, Cerny T, et al. Prognostic significance of CCND1 (cyclin D1) overexpression in primary resected non-small-cell lung cancer. Br J Cancer. 1996;73:294–300. doi: 10.1038/bjc.1996.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyle JO, Langenfeld J, Lonardo F, Sekula D, Reczek P, Rusch V, et al. Cyclin D1 proteolysis: a retinoid chemoprevention signal in normal, immortalized, and transformed human bronchial epithelial cells. J Natl Cancer Inst. 1999;91:373–9. doi: 10.1093/jnci/91.4.373. [DOI] [PubMed] [Google Scholar]
- 9.Langenfeld J, Kiyokawa H, Sekula D, Boyle J, Dmitrovsky E. Posttranslational regulation of cyclin D1 by retinoic acid: a chemoprevention mechanism. Proc Natl Acad Sci USA. 1997;94:12070–4. doi: 10.1073/pnas.94.22.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Q, Sekula D, Guo Y, Liu X, Black CC, Galimberti F, et al. UBE1L causes lung cancer growth suppression by targeting cyclin D1. Mol Cancer Ther. 2008;7:3780–8. doi: 10.1158/1535-7163.MCT-08-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Y, Dolinko AV, Chinyengetere F, Stanton B, Bomberger JM, Demidenko E, et al. Blockade of the ubiquitin protease UBP43 destabilizes transcription factor PML/RARα and inhibits growth of acute promyelocytic leukemia. Cancer Res. 2010;70:9875–85. doi: 10.1158/0008-5472.CAN-10-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baek KH. Conjugation and deconjugation of ubiquitin regulating the destiny of proteins. Exp Mol Med. 2003;35:1–7. doi: 10.1038/emm.2003.1. [DOI] [PubMed] [Google Scholar]
- 13.Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang DE. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277:9976–81. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 14.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6:776–88. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 15.Kitareewan S, Pitha-Rowe I, Sekula D, Lowrey CH, Nemeth MJ, Golub TR, et al. UBE1L is a retinoid target that triggers PML/RARα degradation and apoptosis in acute promyelocytic leukemia. Proc Natl Acad Sci USA. 2002;99:3806–11. doi: 10.1073/pnas.052011299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pitha-Rowe I, Petty WJ, Feng Q, Koza-Taylor PH, Dimattia DA, Pinder L, et al. Microarray analyses uncover UBE1L as a candidate target gene for lung cancer chemoprevention. Cancer Res. 2004;64:8109–15. doi: 10.1158/0008-5472.CAN-03-3938. [DOI] [PubMed] [Google Scholar]
- 17.Rempel LA, Austin KJ, Ritchie KJ, Yan M, Shen M, Zhang DE, et al. UBP43 gene expression is required for normal Isg15 expression and fetal development. Reprod Biol Endocrinol. 2007;5:13. doi: 10.1186/1477-7827-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritchie KJ, Hahn CS, Kim KI, Yan M, Rosario D, Li L, et al. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat Med. 2004;10:1374–8. doi: 10.1038/nm1133. [DOI] [PubMed] [Google Scholar]
- 19.Dragnev KH, Pitha-Rowe I, Ma Y, Petty WJ, Sekula D, Murphy B, et al. Specific chemopreventive agents trigger proteasomal degradation of G1 cyclins: implications for combination therapy. Clin Cancer Res. 2004;10:2570–7. doi: 10.1158/1078-0432.ccr-03-0271. [DOI] [PubMed] [Google Scholar]
- 20.Dragnev KH, Petty WJ, Shah S, Biddle A, Desai NB, Memoli V, et al. Bexarotene and erlotinib for aerodigestive tract cancer. J Clin Oncol. 2005;23:8757–64. doi: 10.1200/JCO.2005.01.9521. [DOI] [PubMed] [Google Scholar]
- 21.Papadimitrakopoulou VA, Izzo J, Mao L, Keck J, Hamilton D, Shin DM, et al. Cyclin D1 and p16 alterations in advanced premalignant lesions of the upper aerodigestive tract: role in response to chemoprevention and cancer development. Clin Cancer Res. 2001;7:3127–34. [PubMed] [Google Scholar]
- 22.Petty WJ, Dragnev KH, Memoli VA, Ma Y, Desai NB, Biddle A, et al. Epidermal growth factor receptor tyrosine kinase inhibition represses cyclin D1 in aerodigestive tract cancers. Clin Cancer Res. 2004;10:7547–54. doi: 10.1158/1078-0432.CCR-04-1169. [DOI] [PubMed] [Google Scholar]
- 23.Witschi H, Espiritu I, Suffia M, Pinkerton KE. Expression of cyclin D1/2 in the lungs of strain A/J mice fed chemopreventive agents. Carcinogenesis. 2002;23:289–94. doi: 10.1093/carcin/23.2.289. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Sempere LF, Galimberti F, Freemantle SJ, Black C, Dragnev KH, et al. Uncovering growth-suppressive microRNAs in lung cancer. Clin Cancer Res. 2009;15:1177–83. doi: 10.1158/1078-0432.CCR-08-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120:1298–309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng Q, Sekula D, Müller R, Freemantle SJ, Dmitrovsky E. Uncovering residues that regulate cyclin D1 proteasomal degradation. Oncogene. 2007;26:5098–106. doi: 10.1038/sj.onc.1210309. [DOI] [PubMed] [Google Scholar]
- 27.Ma Y, Fiering S, Black C, Liu X, Yuan Z, Memoli VA, et al. Transgenic cyclin E triggers dysplasia and multiple pulmonary adenocarcinomas. Proc Natl Acad Sci USA. 2007;104:4089–94. doi: 10.1073/pnas.0606537104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potu H, Sgorbissa A, Brancolini C. Identification of USP18 as an important regulator of the susceptibility to IFN-alpha and drug-induced apoptosis. Cancer Res. 2010;70:655–65. doi: 10.1158/0008-5472.CAN-09-1942. [DOI] [PubMed] [Google Scholar]
- 29.Catic A, Fiebiger E, Korbel GA, Blom D, Galardy PJ, Ploegh HL. Screen for ISG15-crossreactive deubiquitinases. PLoS One. 2007;2:679. doi: 10.1371/journal.pone.0000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim KI, Yan M, Malakhova O, Luo JK, Shen MF, Zou W, et al. Ube1L and protein ISGylation are not essential for alpha/beta interferon signaling. Mol Cell Biol. 2006;26:472–9. doi: 10.1128/MCB.26.2.472-479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KG, Fuchs SY, et al. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25:2358–67. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han EK, Ng SC, Arber N, Begeman M, Weinstein IB. Roles of cyclin D1 and related genes in growth inhibition, senescence and apoptosis. Apoptosis. 1999;4:213–9. doi: 10.1023/a:1009618824145. [DOI] [PubMed] [Google Scholar]
- 33.Dmitrovsky E, Sporn MB. Pharmacology of cancer chemoprevention. In: Bertino J, editor. Encyclopedia of Cancer. 2. St. Louis: Academic Press; 2002. pp. 444–55. [Google Scholar]
- 34.Kok K, Hofstra R, Pilz A, van den Berg A, Terpstra P, Buys CH, et al. A gene in the chromosomal region 3p21 with greatly reduced expression in lung cancer is similar to the gene for ubiquitin-activating enzyme. Proc Natl Acad Sci USA. 1993;90:6071–5. doi: 10.1073/pnas.90.13.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao C, Denison C, Huibregtse JM, Gygi S, Krug RM. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc Natl Acad Sci USA. 2005;102:10200–5. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.