Abstract
Pancreatic ductal adenocarcinoma (PDAC) is a lethal malignancy, with most patients facing an adverse clinical outcome. Aberrant Notch pathway activation has been implicated in the initiation and progression of PDAC, specifically the aggressive phenotype of the disease. We used a panel of human PDAC cell lines, as well as patient-derived PDAC xenografts to determine whether pharmacological targeting of Notch pathway could inhibit PDAC growth and potentiate gemcitabine sensitivity. MRK-003, a potent and selective γ-secretase inhibitor, treatment is effective against PDAC as evidenced by the down-regulation of nuclear Notch1 intracellular domain (N1ICD), inhibition of anchorage independent growth, and reduction of tumor-initiating cells capable of extensive self-renewal. Pre-treatment of PDAC cells with MRK-003 in cell culture significantly inhibited the subsequent engraftment in immunocompromised mice. MRK-003 monotherapy significantly blocked tumor growth in 5 of 9 (56%) PDAC xenografts. A combination of MRK-003 and gemcitabine showed enhanced antitumor effects compared to gemcitabine in 4 of 9 (44%) PDAC xenografts, reduced tumor cell proliferation and induced both apoptosis and intra-tumoral necrosis. Gene expression analysis of untreated tumors indicated that up-regulation of nuclear factor kappa B (NFκB) pathway components were predictive of sensitivity to MRK-003, while up-regulation in B-cell receptor (BCR) signaling and nuclear factor erythroid-derived 2-like 2 (NRF2) pathway correlated with response to the combination of MRK-003 with gemcitabine. Our findings strengthen the rationale for small molecule inhibition of Notch signaling as a therapeutic strategy in PDAC.
Keywords: Notch, MRK-003, gamma-secretase inhibitor, pancreatic cancer, chemotherapy
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most devastating human malignancies characterized by extensive local invasion, early systemic dissemination, and pronounced resistance to chemotherapy and radiotherapy (1). Outcomes for patients with advanced PDAC remain poor with limited clinical benefits seen with currently available therapy. For example, gemcitabine (GEM) as a systemic agent in the treatment of advanced PDAC results in a median survival of less than 6 months. A recent Phase III trial of a combination regimen comprised of oxaliplatin, irinotecan, fluorouracil, and leucovorin (FOLFIRINOX) as first-line therapy has been shown to significantly improve median survival compared to GEM alone in patients with advanced PDAC (2). Nonetheless, the safety profile of FOLFIRINOX was less favorable than that of GEM, and many patients remain ineligible to receive FOLFIRINOX (3). Despite these ongoing advances, the continued poor survival in advanced PDAC underscores the need for new systemic therapies.
Notch signaling is an evolutionarily conserved pathway that plays an important role in multiple cellular and developmental processes (4). While the Notch pathway plays a pivotal role in normal cell development, aberrant Notch signaling pathway has been extensively linked to a range of human malignancies, rendering this pathway a compelling target for drug development (5, 6). Activation of canonical Notch signaling involves a series of proteolytic processing steps following ligand binding. The interaction of Notch ligands (Jagged and Delta-like) with their receptors promotes γ-secretase dependent cleavage of the Notch receptor and release of the Notch intracellular domain, which translocates to the nucleus, a requirement for activation of gene targets. Accumulating evidence supports the use of small molecule inhibitors of γ-secretase (GSIs) as a tractable avenue for Notch antagonism against a number of cancers linked with increased pathway activity (7, 8).
Several lines of preclinical evidence suggest that sustained activation of Notch signaling pathway contributes to the initiation and maintenance of PDAC (9). For example, in primary PDAC tissues, as well as in non-invasive precursor lesions of adenocarcinoma, multiple pathway components, including Notch ligands, receptors and target genes are over-expressed, relative to normal pancreas (10). In an autochthonous mouse model of PDAC, pharmacological Notch inhibition attenuates the development of intraductal precursor lesions and invasive cancer, implicating this pathway in tumor initiation (11). Further, down-regulation of Notch receptors by RNA interference or exposure to GSI in established human pancreatic cancer cells results in reduced proliferative rates, increased apoptosis, decreased anchorage independent growth and deceased invasive properties, suggesting a role for Notch signaling in PDAC maintenance as well (11, 12).
In this study, we assessed the effects of MRK-003, a potent and selective GSI (13, 14), in preclinical models of PDAC. MRK-003 is the preclinical analog of MK-0752, a GSI that is currently in clinical development (15, 16). We used a panel of human PDAC cell lines, as well as patient-derived PDAC xenografts, to determine whether pharmacological targeting of the Notch pathway using MRK-003 could curb tumor growth and potentiate GEM sensitivity. Herein, we also attempt to delineate a gene signature of responsiveness to Notch inhibition, which may aid in the selection of patient subsets more likely to benefit from this strategy in the clinic.
Materials and Methods
Drugs
MRK-003 was provided by Merck Research Laboratories. GEM (Eli Lilly) was purchased from the Johns Hopkins Hospital pharmacy. Structures of MRK-003 and GEM are shown in Figure 1 (17, 18).
Figure 1. Chemical Structures of MRK-003 and Gemcitabine.
Cell lines
Authenticated low-passage PDAC lines (Pa03C, Pa14C, Pa16C and Pa29C) established at Johns Hopkins was used for the study. Culture conditions and exome-wide somatic mutational status of these cell lines has been previously described (19). In addition, we used Capan-1 (ATCC), based on its Notch dependence (12). Cell line provider (ATCC) used cytochrome C oxidase I (COI) PCR assay, short tandem repeat (STR) profiling and ouchterlony diffusion method to characterize the cell line.
Quantitative real-time reverse transcription-PCR (qRT-PCR)
Cells were collected after incubation with MRK-003 (2 or 5μM) for 48 hours. Total RNA was extracted using RNeasy Mini kit (Qiagen). cDNA was synthesized with SuperScript First Strand System (Invitrogen). qRT-PCR was performed using FAST SYBR Green Master Mix (Applied Biosystems) on a Step One Plus Real-Time PCR System (Applied Biosystems). Human PGK1 and murine β-actin were used as housekeeping genes. Relative expression of the mRNA was estimated using the 2−ΔΔ CT method (20).
Anchorage independent growth
Anchorage independent growth of cells was determined by soft agar assays in 6-well plates. Briefly, cells were incubated in media containing 0.5% FBS with vehicle or MRK-003 (2 or 5μM). After incubation for 48 hours, the treated cells were recovered by media with 10% FBS for 24 hours. Thereafter, equal numbers of viable cells from each condition were quantified using a hemocytometer with trypan blue counterstain, and then plated for soft agar assays. A bottom layer of 1% agarose, a middle layer of 0.6% agarose including 10,000 cells and a top layer of medium only were applied into each well. After incubating the plates for 3 weeks, colonies were stained with crystal violet solution, visualized by trans-UV illumination and counted using the analysis software Quantity One (BioRad).
Stable over-expression of Notch 1 intracellular domain
Stable transfectants overexpressing the Notch 1 intracellular domain (N1ICD) was established in Pa03C cells, as previously described (12). The stable transfectants were maintained in media supplemented with 600 μg/mL of G418. Mock vector was transfected as a control. Overexpression of N1ICD compared with empty vector–transfected cells was confirmed by qRT-PCR (12).
Protein extraction and western blotting
Both N1ICD stable transfected as well as empty vector-transfected Pa03C cells were cultured separately in tissue culture flasks. Cells were trypsinized and cell pellets were lysed using lysis buffer. Western blots were performed as previously described (21). Membranes were incubated with primary antibodies against rabbit N1ICD (Val1744) and Hes-1 (Cell Signaling Technology, Inc. and Abcam respectively). Membranes were probed with secondary horseradish peroxidase-conjugated antibody (GE Healthcare) and bound antibodies were detected by SuperSignal West Pico/Femto chemiluminescent substrate (Thermo Scientific). Equal loading was verified with β-actin antibody.
Engraftment of ex vivo pre-treated PDAC cells in athymic mice
Male athymic nude mice (6-week-old, Harlan) were housed and maintained in accordance with the Institutional Animal Care and Use Committee and guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care International. PDAC cells were treated ex vivo with either vehicle or with MRK-003 (5μM) for 48 hours, followed by a recovery in full serum conditions for an additional 24 hours, prior to subcutaneous injection. Viable 5×106 cells in a total volume of 200μL of 1:1 (v/v) PBS/ Matrigel (BD Biosciences) were injected subcutaneously into bilateral flanks (right flank; cells pre-treated with vehicle, left flank; cells pre-treated with MRK-003) of mice (N=6). Tumor size was measured with digital calipers.
Fluorescence-activated cell sorter (FACS) analysis of tumor initiating cells (CD44+CD24+ and ALDH+ cancer cells)
PDAC cells were treated with MRK-003 (2 or 5μM) for 48 hours. The cells were harvested and stained with ALDEFLUOR. Briefly, one million cells were re-suspended in 1mL ALDEFLUOR buffer and 1μL ALDEFLUOR reagent in the presence or absence of the ALDH1 inhibitor, diethylamino-benzaldehyde (DEAB), for 30 minutes in a 37°C water bath. The cells were washed and incubated at 4°C for 15 minutes with monoclonal anti-CD44-allophycocyanin (APC) (1:20 dilution; clone G44-26, BD Biosciences) and anti-CD24-phycoerythrin (PE) (1:20 dilution; clone ML5, BD Biosciences) antibodies. The cells were washed and re-suspended in ALDEFLUOR buffer containing 2μg/mL propidium iodide (PI). A FACSCalibur flow cytometer (BD Biosciences) was used for flow cytometric analysis, as previously described (22). The cells were first gated based on side-scatter and forward-scatter properties, followed by exclusion of nonviable (PI-positive) cells. The ALDH+ gate was created based on DEAB-treated cells stained with ALDEFLUOR, anti-CD24-PE, and anti-CD44-APC. The CD44+CD24+ gates were created based on cells stained with ALDEFLUOR, mouse-specific IgG2b k-APC (1:100 dilution; BD Biosciences) and IgG2a k-PE (1:100 dilution; BD Biosciences) antibodies (22, 23). Gates were created based on cellular staining with isotype control antibodies. FACS plots for these controls are shown as Supplementary Figure 1.
Notch -1 gene expression
RNA isolated from baseline (untreated) tumors of 30 individual patient derived pancreatic cancer xenografts were profiled using Affymetrix U133 Plus 2.0 gene arrays. Sample preparation and processing procedure was conducted as described in the Affymetrix GeneChip Expression Analysis Manual (Affymetrix, Inc.). Gene expression levels were converted to a rank based matrix and standardized for each microarray (23).
Xenograft establishment and in vivo efficacy studies
Low-passage PDAC xenografts established at Johns Hopkins Hospital was used for the study (24). Nine independent patient-derived xenografts were selected for the in vivo efficacy study, based on elevated Notch-1 expression. Cohorts of athymic mice with tumor size of ~200 mm3 were randomized to four arms (5 mice; 10 tumors per group): (a) Vehicle control, (b) MRK-003 150mg/Kg p.o once weekly for 3 weeks (c) GEM 30mg/Kg i.p. once weekly, for 3 weeks (d) combination of MRK-003 and GEM in the above mentioned dose and schedule. Tumor size was evaluated twice weekly and tumor volume was calculated using the following formula: tumor volume = [length × width2]/2.
Histology, immunohistochemistry, and fluorescence microscopy
Excised tumors were fixed in 10% buffered formalin before paraffin embedding. Immunohistochemical staining of nuclear anti-Notch1 (Abcam, ab8925, 1: 400 dilutions) and nuclear anti-Hes1 (Abcam, ab49170, 1: 250 dilutions) were performed as per the manufacture’s protocol. Immunohistochemistry for proliferation (Ki-67 antigen) was performed using an anti-MIB-1 (Ki-67) antibody (clone K2, dilution 1:100, Ventana Medical Systems) as previously described (25). Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining was done by a commercial apoptosis detection kit (DeadEnd™ Fluorometric TUNEL System, Promega), according to the recommendations of the manufacturer (25). We used Vectashield® with DAPI to preserve fluorescence and to counterstain nuclei. Sections were examined microscopically and the average number of cells stained positive for Ki-67 and TUNEL were counted in 5 random fields each from the tumors of two separate animals under at 20x magnification.
Gene expression analysis
RNA isolated from the nine parental (untreated) PDAC xenografts were profiled using Affymetrix U133 Plus 2.0 gene arrays at least in duplicates. This gene array has ~54,000 probes comprising ~20,000 RefSeq genes. CEL files generated from the array were normalized using the robust multiarray average algorithm (26). Pearson correlation coefficients were calculated to identify genes that show significant correlation (P<0.01) with tumor growth inhibition upon drug treatment. Pathway and biological function analysis of gene sets was performed using Ingenuity Pathway Analysis (IPA, Ingenuity Pathway Analysis, Redwood City, CA), and overlap of gene sets with other publicly available gene signatures was examined using NextBio (NextBio, Cupertino, CA). Microarray raw data are available in GEO (Accession number GSE37645).
Statistical Analysis
All groups were studied in parallel and differences between groups were analyzed by ANOVA, as appropriate, and Bonferroni multiple comparison tests performed using GraphPad Prism. The difference between experimental groups were considered significant when the P-value was <0.05. Bar and line graphs show mean ± SEM, respectively if not otherwise indicated.
Results
MRK-003 treatment down-regulates the expression of Hes-1 in PDAC cells
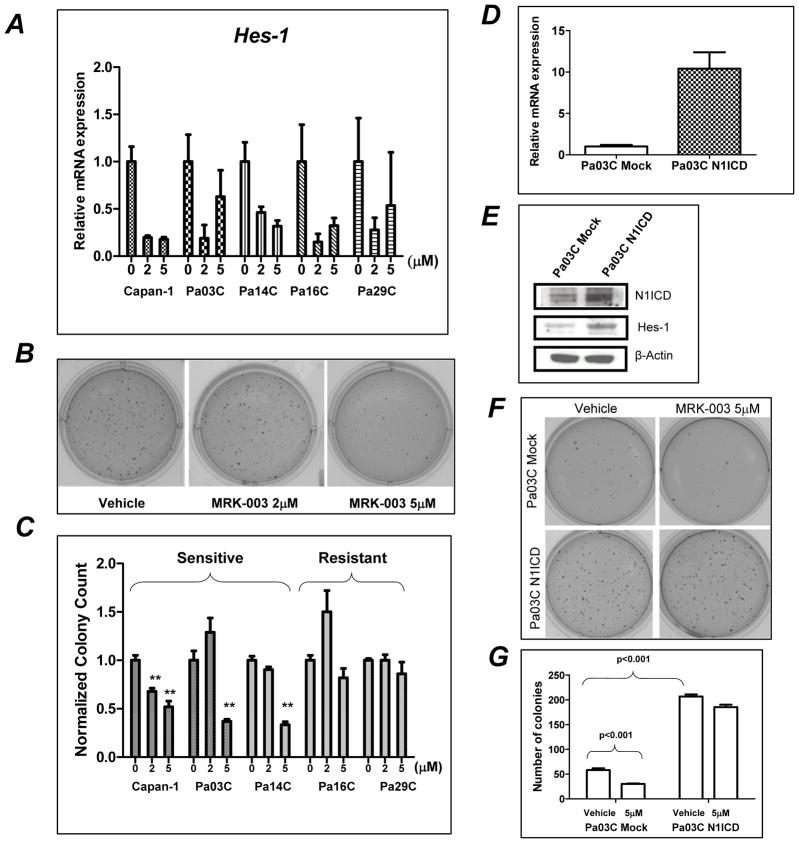
To determine whether MRK-003 could modulate Notch target genes, we examined the expression of Hes-1 transcripts in a panel of five PDAC cells lines (Capan-1, Pa03C, Pa14C, Pa16C, and Pa29C). Hes-1 transcripts were down-regulated in all pancreatic cancer cell lines treated with both 2μM and 5μM of MRK-003 (Figure 2A).
Figure 2. MRK-003 treatment inhibits anchorage-independent growth of PDAC cell lines, which is rescued by enforced expression of N1ICD.
A: Down-regulation of Hes-1 mRNA is seen in PDAC lines treated with MRK-003.
B: Colony formation in soft agar is inhibited in Pa03C cells with 5μM of MRK-003.
C: Normalized colony counts in PDAC cell lines, pre-treated with 2 and 5μM of MRK-003 (**, P<0.01). Three of the cells lines (Capan-1, Pa03C, and Pa14C) were “sensitive” based on significant inhibition of anchorage growth, while the other two (Pa16C and Pa29C) were “resistant”.
D and F: qRT-PCR and western blots of N1ICD stably transfected Pa03C showing fover-expression of transcripts corresponding to the ICD domain as compared to mock-trasfected cells.
F and G: Treatment of MRK-003 significantly reduced the number of colonies in mock Pa03C cells, while no significant effects are seen in N1ICD expressing Pa03C cells. Notably, expression of N1ICD per se results in a significant expansion of colonies in soft agar in untreated Pa03C compared to mock controls.
MRK-003 pre-treatment inhibits anchorage independent growth
Anchorage independent growth in soft agar was significantly reduced upon exposure to MRK-003 in Capan-1 at both 2 and 5μM dosages (P<0.01), while significant inhibition in the other two “sensitive” lines (Pa03C and Pa14C) was observed at 5μM dosing (Figure 2B). In contrast, the remaining two PDAC lines - Pa16C and Pa29C - were resistant to MRK-003 at both 2 and 5μM dosages (Figure 2C), despite the down-regulation of Hes-1 transcripts observed previously, suggesting other Notch effectors were involved in this phenotype.
Enforced expression of N1ICD rescues in vitro effects of MRK-003
To determine if enforced Notch expression could rescue the MRK-003 phenotype, stable transfectant of N1ICD and mock vectors were established in Pa03C cells (“Pa03C-N1ICD” and “Pa03C-mock”, respectively). Stable expression of N1ICD was confirmed by up-regulation of transcript corresponding to the ICD portion of NOTCH1, as well as the over-expression of N1ICD and Hes-1 protein compared to mock transfected Pa03C cells (Figure 2D and E). Enforced N1ICD expression per se augmented colony formation significantly compared with Pa03C-mock clones in vehicle treated cells (Figure 2F, P<0.001). In Pa03C-mock cells, treatment with MRK-003 at 5μM reduced colony numbers by ~50% (P<0.001), whereas in Pa03C- N1ICD cells, equimolar MRK-033 attenuated colony formation by approximately 10%, which was not statistically significant (Figure 2G).
MRK-003 pre-treatment delayed tumor engraftment
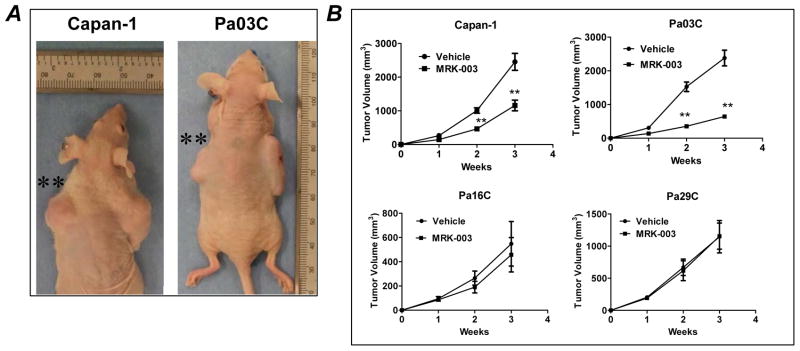
In order to determine whether pre-treatment with MRK-003 could modulate tumorigenicity in mice, we monitored the tumor growth of subcutaneously implanted Capan-1 and Pa03C cells, which had been pretreated ex vivo with either MRK-003 or vehicle. Engraftment was significantly delayed at day 14 and day 21 post-implantation in both cell lines (Figure 3A and B). In contrast, there was no difference in engraftment rates for either Pa16C or Pa29C cells, between the control and MRK-003 pre-treatment arms (Figure 3B). The differential sensitivity to MRK-003 was consistent with the prior data in anchorage independent growth obtained using this panel of cells.
Figure 3. Ex vivo pre-treatment with MRK-003 significantly inhibits engraftment in mice.
A: Representative photographs of mice injected with MRK-003 pretreated Capan-1 and Pa03C cells (tumor generated from vehicle and MRK-003 pretreatment treated cell lines are on the right and left side of mice, respectively).
B: Tumor growth curves demonstrate that MRK-003 pretreatment significantly delayed engraftment in Capan-1 and Pa03C cells (**P<0.01) compared to the tumor generated from vehicle treated cells. However, MRK-003 pretreatment did not impact the engraftment of Pa16C or Pa29C compared to the tumors generated from vehicle treated cells.
Effect of MRK-003 treatment on tumor initiating cells
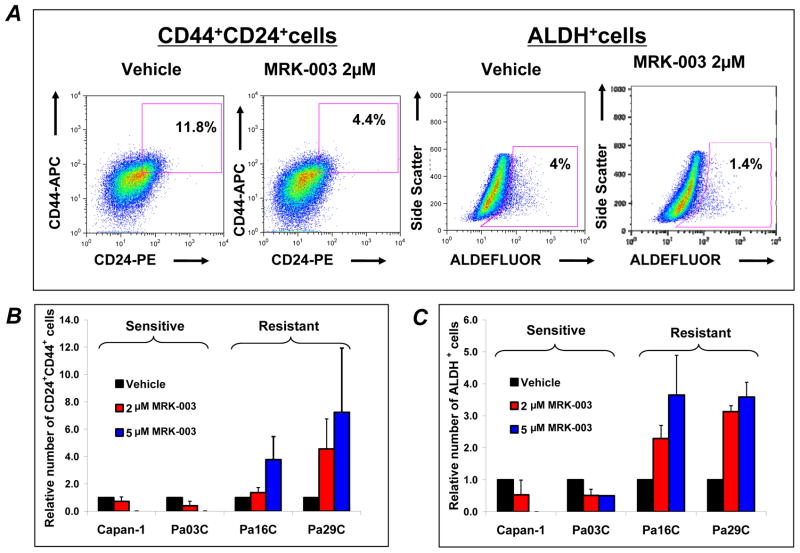
We examined the effect of MRK-003 treatment on two partially overlapping populations of cells - CD44+CD24+ and ALDH+, which have independently been associated with tumor-initiating properties in PDAC (22, 27). MRK-003 treatment in Capan-1 resulted in the reduction of CD44+CD24+ tumor cells at 2μM, and elimination of these cells at 5μM (Figure 4A and B). A comparable observation was noticed in the sensitive Pa03C cell line (Figure 4B). Similarly, MRK-003 treatment reduced the proportion of ALDH+ cells in Capan-1 and in Pa03C (Figure 4A and C). However MRK-003 treatment increased the relative proportion of CD44+CD24+ and ALDH+ cells in resistant lines (Pa16C and Pa29C), respectively (Figure 4B and C).
Figure 4. MRK-003 treatment modulates the population of tumor initiating cells in PDAC lines.
PDAC cells were treated with MRK-003 (2 μM or 5μM) for 48 hours. The cells were harvested and stained for FACS analysis.
A: Representative data for CD44+CD24+ and ALDH+ population in MRK-003 treated in Capan-1 cells showing reduction of CD44+CD24+ and ALDH+ cells compared to vehicle treated cells.
B and C: Percentage of CD44+CD24+ and ALDH+ population respectively in four pancreatic cancer cell lines treated with MRK-003 for 48 hours. Dose-dependent reduction in the proportion of CD44+CD24+ and ALDH+ cells were noticed in two sensitive cell lines (Capan-1 and Pa03C). How ever, MRK-003 increased the proportion of CD44+CD24+ and ALDH+ cells in Pa16C and Pa29C. Experiment were conducted in triplicate, N=3, error bars SD.
Effect of MRK-003 treatment on tumor growth, Notch targets, proliferation and apoptosis
Thirty patient-derived PDAC xenografts were mined in silico for relative expression of NOTCH1 transcripts, using microarray data (Figure 5A). Based on the elevated NOTCH1 expression in the baseline xenografts (relative expression is illustrated as red to green in order of decreasing values), we selected nine individual patient derived xenografts for further expansion in mice, and randomization to MRK-003 or GEM monotherapy, or the combination of MRK-003 and GEM. As shown in Figure 5B and C, MRK-003 monotherapy significantly reduced tumor volume in five of nine xenografts (56%) as compared to control, which were Panc374 (P<0.05), Panc219 (P<0.05), Panc265 (P<0.001), Panc420 (P<0.01) and JH033 (P<0.01). Moreover, there was a significant decrease in tumor growth in four of nine (44%) xenografts in the combination therapy group as compared to single-agent GEM, and this subset of tumors was comprised of Panc198 (P<0.05), Panc219 (P<0.01), Panc291 (P<0.001) and JH033 (P<0.05) (Figure 5C). Baseline Notch-1 expression (Figure 5A) was not predictive of γ-secretase inhibitor (MRK-003) sensitivity in this tumor model.
Figure 5. In vivo efficacy of MRK-003, GEM and combination of MRK-003 and GEM in a panel of patient-derived PDAC xenografts.
A: Nine individual cases for this study were selected from a pool of 30 PDAC xenografts based on expression of NOTCH1 transcripts at baseline. In the illustrated heatmap, red to green indicates decreasing transcript expression. The numbers on the top panel indicate each individual patient-derived PDAC xenograft.
B: Growth curves of two representative PDAC xenografts, Panc374 and Panc198 demonstrates that the combination of GEM and MRK-003 can significantly inhibit tumor growth compared to the vehicle and/or GEM treated animals. Each treatment arm was comprised of ten tumors, implanted as bilateral subcutaneous xenografts in five mice.
C: Summary of tumor growth inhibition data. MRK-003 monotherapy significantly reduced tumor volume in 5 of 9 xenografts compared to vehicle treated xenografts, which were Panc420, Panc374, Panc219, Panc265 and JH033. There was a significant reduction in tumor growth of 4 of 9 xenografts in the combination therapy group compared to GEM alone, which were Panc198, Panc219, Panc291 and JH033. (*P<0.05, **P<0.01, *** P <0.001 versus vehicle treated mice; # P <0.05, # # P <0.01 compared to GEM treated mice).
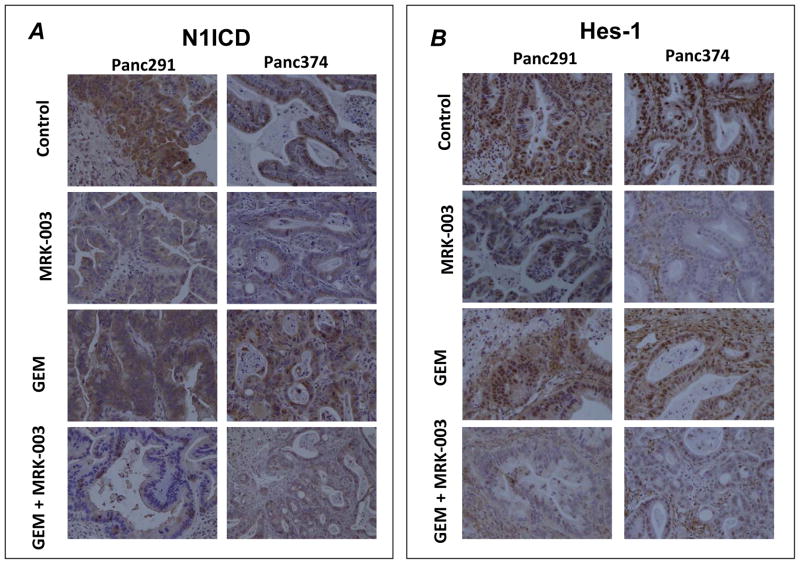
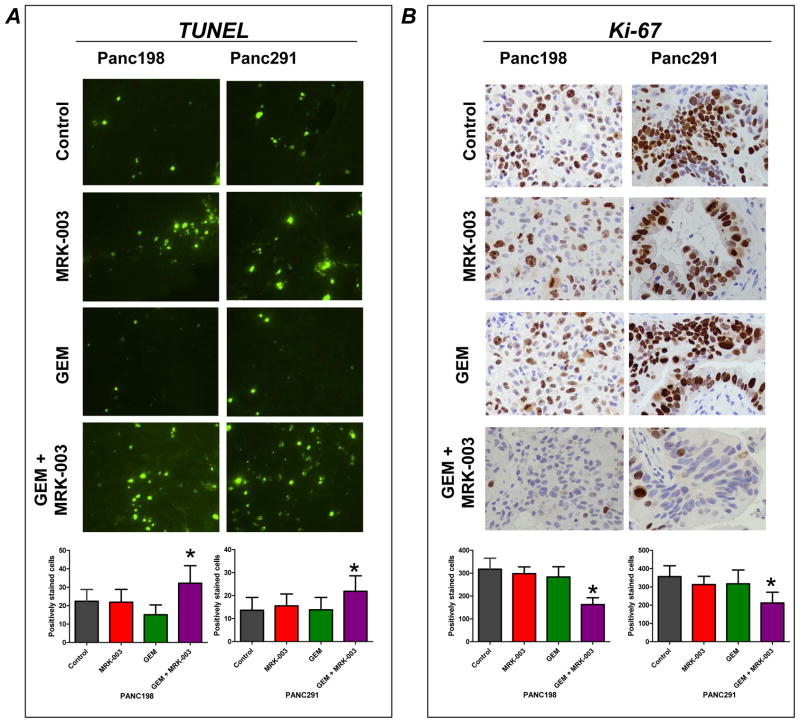
MRK-003 as well as combination of GEM with MRK-003 treatments reduced the expression of both nuclear Notch1 (N1ICD) and Hes-1 as compared to control and GEM treated tumors (Figure 6A and B). We then sought to determine the effects on proliferation and apoptosis in the post-treatment samples, using two representative xenografts (Panc198 and Panc291). Compared to single agent therapy, xenografts receiving MRK-003 and GEM demonstrated significantly increased apoptosis (as assessed by TUNEL staining, Figure 7A), as well as significantly reduced tumor cell proliferation (as assessed by Ki-67, Figure 7B).
Figure 6. MRK-003 treatment down-regulates nuclear N1ICD and Hes-1 protein expressions in PDAC xenografts as shown by immunohistochemical staining.
A and B: Representative photomicrographs of N1ICD and Hes-1 from Panc291 and Panc374 xenografts showing down-regulation of nuclear N1ICD and Hes-1 expression in MRK-003 and combination of GEM and MRK-003 treatment compared to vehicle treated and GEM treated tumor samples.
Figure 7. Combination of GEM and MRK-003 induces apoptosis and reduces cell proliferation in PDAC xenografts.
A: Representative photomicrograph of TUNEL staining from Panc198 and Panc291. Histogram of TUNEL-positive nuclei per high power field (lower panel), showing that combination of GEM and MRK-003 significantly enhanced apoptotic cells per field as compared to GEM treatment.
B: Representative photomicrograph of Ki-67 staining from Panc198 and Panc291. Histogram of Ki-67-positive nuclei per high power field (lower panel), showing that combination of GEM and MRK-003 significantly reduced proliferating cells as compared to GEM treatment. Histograms for TUNEL and Ki-67 were generated by evaluating five high power fields per xenograft section from two independent tumors per treatment arms; mean ± SEM (*P<0.001 compared to GEM).
Gene expression analysis for predictive signatures of response to MRK-003
Probe sets that show significant correlation with response to MRK-003, or the combination of MRK-003 plus GEM (692 and 967 probes, respectively, Supplementary Tables 1 and 2) were identified with Pearson P value <0.01, and subjected to Ingenuity Pathway Analysis (IPA), as well as overlap with publicly available expression datasets using NextBio. IPA demonstrated that up-regulation of nuclear factor kappa B (NFκB) signaling function in the parental xenografts correlated with response to MRK-003 in vivo (Supplementary Figure 2A). NextBio analysis of MRK-003 sensitivity associated probes showed that 75% of the single agent response signature in PDAC xenografts overlapped with that of trimethylated histone H3 (H3K4me3) bound genes (28) with high significance (overlap P value = 8.2E-53). We then assessed for potential signatures of response to the combination of MRK-003 and GEM using the IPA platform. Up-regulation of B-cell receptor (BCR) signaling (Supplementary Figure 2B), as well as the nuclear factor erythroid-derived 2-like 2 (NRF2)-mediated oxidative stress response pathways (data not shown) were significantly associated with the sensitivity of MRK-003 plus GEM combination. NextBio analysis of the combination response signature demonstrated that, in addition to the previously observed overlap with the H3K4me bound genes, 9% of the response signature also overlapped with NRF1 binding site gene set from the Broad MSigDB – Regulatory Motifs (overlap P value = 2.6E-17), in agreement with IPA’s association with NRF2-mediated canonical signaling pathway. Of interest, there was a negative correlation between the combination response signature with the signature attributable to Rituximab (anti-CD20 antibody) response, which is in agreement with the positive association with BCR signaling detected by IPA, as Rituximab specifically inhibits this signaling axis (29).
Discussion
Given its well-defined role in PDAC growth, Notch signaling has been an area of intense investigation (30, 31). A recent exomic profiling of PDAC has identified Notch as one of the twelve “core” signaling pathways that are abnormal in this neoplasm (19). Notch pathway components are enriched in PDAC patients resistant to GEM treatment and other molecular targeted agents (24, 32). The overwhelming majority of PDACs harbor activating mutations of KRAS (19), and sustained Notch-1 signaling was shown to be essential in maintaining the transformed phenotype of Ras-mutant cells (33). These multiple lines of evidence strongly support the pharmacological targeting of Notch signaling in PDAC, particularly in combination with GEM.
MRK-003 is a potent and selective GSI (13, 14), which is the preclinical analog of MK-0752, currently in clinical development (15, 16). As a “pan” Notch inhibitor, MRK-003 is expected to inhibit the enzymatic cleavage of all four Notch receptors, which bypasses the need for receptor specific targeting based on tumor context. For example, our studies have shown that Notch1 is the primary oncogenic influence in pancreatic cancer (10), while lung cancers harbor greater dependence on Notch3 signaling (13), and medulloblastomas on Notch2 (34). In the context of breast cancer, activation of Notch-1 is observed in tumors exposed to trastuzumab, contributing to therapeutic resistance (35). On the contrary, combining MRK-003 with trastuzumab completely prevented tumor recurrence in trastuzumab-sensitive tumor, while the combination of lapatinib with MRK-003 significantly reduced tumor growth in trastuzumab-resistant cancers (36).
Our in vitro results confirm that MRK-003 exposure leads to differential effects in PDAC cells, with a subset of lines unequivocally responding to the agent by significant reduction in anchorage independent growth and delay in tumor engraftment in athymic mice; in contrast, other lines remain refractory to the drug. While MRK-003 treatment could diminish the proportion of CD24+CD44+or ALDH+ subpopulation in Capan-1 and Pa03C cells, the treatment leads to the enrichment of CD24+CD44+ and ALDH+ cells in Pa16C and Pa29C cells. While our current study was entirely geared towards the use of MRK-003 as a therapeutic strategy in established PDAC, Plentz and colleagues have previously shown the significant efficacy of MRK-003 in chemoprevention using a genetically engineered mouse model (11), further underscoring the importance of Notch signaling to tumor initiation in the pancreas. It is important to note that the cellular phenotypes implicated in tumor initiation are also highly enriched for in pancreatic cancer metastases (22, 37), and thus pharmacological Notch inhibition would conceivably be beneficial in advanced disease settings as well.
A recent report indicated that a combination of MRK-003 and GEM could induce intra-tumoral necrosis in KPC mouse model (38). In light of this, we examined the histological sections from treated xenografts and find that stroma-rich xenografts (for example Panc374, Supplementary Figure 3) do, in fact, show increased necrosis in the combination therapy arm. While central necrosis to some degree is observed in all of the cohorts, the MRK-003 plus gemcitabine treated tumors demonstrate sheets of confluent necrosis. However, this phenomenon was not uniformly observed, especially in stroma-poor xenografts (data not shown). We believe that the relative hypovascularity of stroma-rich xenografts (a feature also reflected in human pancreatic cancers and the KPC model) might explain why these tumors demonstrate increased necrosis in the setting of combination therapy.
As an attempt to identify predictive signatures in response to MRK-003, we examined the baseline gene expression profiles of the nine patient-derived xenografts as it related to variable in vivo responses to either MRK-003 alone, or the combination regimen. Up-regulation of NFκB signaling components was correlated with response to single agent MRK-003, a finding that attains significance in light of the recent observation of crosstalk between Notch and NFκB signaling in murine PDAC models (30). In that study, NFκB activation collaborated with basal Notch signals to enhance Notch target gene expression, suggesting that a signature consistent with NFκB activation might indicate a greater degree of Notch dependence in PDAC cells. The existence of crosstalk between Notch and NFκB pathways has also been reported by other groups using in vitro models of pancreatic cancer (39, 40), underscoring the validity of our findings. In terms of the combination regimen, we identified two activation signatures that correlated with response in xenografts - BCR signaling and Nrf2, respectively. While an interaction between BCR and Notch signaling has not been reported previously in solid tumors, not unexpectedly a potential synergy does exist in normal and neoplastic hematopoeitic cells (41, 42). Thus, in both normal B-cells, as well as in B-cell lymphoma lines, Notch promotes BCR-mediated proliferation, which is blocked by administration of GSIs (41, 42). In these aforementioned studies, putative downstream effectors that have been implicated in the observed synergy include the MAP kinase signaling pathway and Myc, both of which retain considerable significance in PDAC pathogenesis. The correlation between sensitivity to GSI plus GEM and NRF1/2 signaling (which was identified by two disparate strategies - IPA and NextBio analyses, respectively) is, to the best of our knowledge, previously unreported. As recently demonstrated, NRF2 expression is crucial to most oncogene driven cancers (including PDAC), in light of its ability to attenuate deleterious reactive oxygen species (ROS) that accumulate from oncogene-induced cellular stress (43). The precise cellular mechanisms by which an NRF2-activation signature leads to sensitivity to MRK-033/GEM regimen remains to be determined; nonetheless, the in silico correlations generated with regards to various activation signatures (NFκB, BCR, and Nrf2) in PDAC and response to either single agent GSI or combination regimens should be testable hypotheses, as these agents are increasingly being evaluated in the clinic.
Substantial preclinical evidence supports the role of Notch signaling in the initiation, progression and maintenance of PDAC. Pharmacological targeting of Notch signaling in PDAC patients using GSIs such as MK-0752 are warranted based on this evidence. A combination regimen of GEM and MK-0752 is currently being evaluated in advanced PDAC patients (ClinicalTrials.gov, NCT ID: NCT01098344) (44). Our results strengthen the rationale for small molecule inhibition of γ-secretase inhibition as a therapeutic strategy in PDAC and identification of a putative sensitivity signature to the combination of GEM and MRK-003 may provide insights into clinical response in the ongoing and future trials.
Supplementary Material
Acknowledgments
The authors are thankful to Shweta G Pai (Johns Hopkins) for help in tissue culture experiments.
Grant Support: N.V. Rajeshkumar, A. Maitra and M. Hidalgo are supported by a Stand Up To Cancer Dream Team Translational Cancer Research Grant, a Program of the Entertainment Industry Foundation (SU2C-AACR-DT0509). A. Maitra is supported by Merck Research Laboratories and NIH CA113669.
Footnotes
Disclosure of Potential Conflicts of Interest: Q. Zhang and O. Puig are employees of Merck Research Laboratories.
References
- 1.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–20. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–25. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 3.Saif MW, Chabot J. Chemotherapy: Metastatic pancreatic cancer--is FOLFIRINOX the new standard? Nat Rev Clin Oncol. 2011;8:452–3. doi: 10.1038/nrclinonc.2011.107. [DOI] [PubMed] [Google Scholar]
- 4.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miele L, Golde T, Osborne B. Notch signaling in cancer. Curr Mol Med. 2006;6:905–18. doi: 10.2174/156652406779010830. [DOI] [PubMed] [Google Scholar]
- 6.Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–63. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 8.Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27:5124–31. doi: 10.1038/onc.2008.226. [DOI] [PubMed] [Google Scholar]
- 9.Leach SD. Epithelial differentiation in pancreatic development and neoplasia: new niches for nestin and Notch. J Clin Gastroenterol. 2005;39:S78–82. doi: 10.1097/01.mcg.0000155547.83901.a3. [DOI] [PubMed] [Google Scholar]
- 10.Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–76. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 11.Plentz R, Park JS, Rhim AD, Abravanel D, Hezel AF, Sharma SV, et al. Inhibition of gamma-secretase activity inhibits tumor progression in a mouse model of pancreatic ductal adenocarcinoma. Gastroenterology. 2009;136:1741–9. e6. doi: 10.1053/j.gastro.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullendore ME, Koorstra JB, Li YM, Offerhaus GJ, Fan X, Henderson CM, et al. Ligand-dependent Notch signaling is involved in tumor initiation and tumor maintenance in pancreatic cancer. Clin Cancer Res. 2009;15:2291–301. doi: 10.1158/1078-0432.CCR-08-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konishi J, Kawaguchi KS, Vo H, Haruki N, Gonzalez A, Carbone DP, et al. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer research. 2007;67:8051–7. doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Kesari S, Rooney C, Strack PR, Shen H, Wu L, et al. Inhibition of Notch Signaling Blocks Growth of Glioblastoma Cell Lines and Tumor Neurospheres. Genes Cancer. 2011;1:822–35. doi: 10.1177/1947601910383564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deangelo DJ, Stone RM, Silverman LB, Stock W, Attar EC, Fearen I, et al. A phase I clinical trial of the notch inhibitor MK-0752 in patients with T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) and other leukemias. J Clin Oncol. 2006;24;18S:6585. [Google Scholar]
- 16.Fouladi M, Stewart CF, Olson J, Wagner LM, Onar-Thomas A, Kocak M, et al. Phase I Trial of MK-0752 in Children With Refractory CNS Malignancies: A Pediatric Brain Tumor Consortium Study. J Clin Oncol. 2011;29:3529–34. doi: 10.1200/JCO.2011.35.7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis HD, Leveridge M, Strack PR, Haldon CD, O’Neil J, Kim H, et al. Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of notch signaling. Chem Biol. 2007;14:209–19. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Pourquier P, Gioffre C, Kohlhagen G, Urasaki Y, Goldwasser F, Hertel LW, et al. Gemcitabine (2′,2′-difluoro-2′-deoxycytidine), an antimetabolite that poisons topoisomerase I. Clin Cancer Res. 2002;8:2499–504. [PubMed] [Google Scholar]
- 19.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Rajeshkumar NV, De Oliveira E, Ottenhof N, Watters J, Brooks D, Demuth T, et al. MK-1775, a Potent Wee1 Inhibitor, Synergizes with Gemcitabine to Achieve Tumor Regressions, Selectively in p53-Deficient Pancreatic Cancer Xenografts. Clin Cancer Res. 2011;17:2799–806. doi: 10.1158/1078-0432.CCR-10-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasheed ZA, Yang J, Wang Q, Kowalski J, Freed I, Murter C, et al. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J Natl Cancer Inst. 2010;102:340–51. doi: 10.1093/jnci/djp535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajeshkumar NV, Rasheed ZA, Garcia-Garcia E, Lopez-Rios F, Fujiwara K, Matsui WH, et al. A combination of DR5 agonistic monoclonal antibody with gemcitabine targets pancreatic cancer stem cells and results in long-term disease control in human pancreatic cancer model. Mol Cancer Ther. 2011;9:2582–92. doi: 10.1158/1535-7163.MCT-10-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrido-Laguna I, Uson M, Rajeshkumar NV, Tan AC, de Oliveira E, Karikari C, et al. Tumor engraftment in nude mice and enrichment in stroma-related gene pathways predict poor survival and resistance to gemcitabine in patients with pancreatic cancer. Clin Cancer Res. 2011;17:5793–800. doi: 10.1158/1078-0432.CCR-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pramanik D, Campbell NR, Karikari C, Chivukula R, Kent OA, Mendell JT, et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10:1470–80. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 27.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer research. 2007;67:1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 28.He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–7. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kheirallah S, Caron P, Gross E, Quillet-Mary A, Bertrand-Michel J, Fournie JJ, et al. Rituximab inhibits B-cell receptor signaling. Blood. 2010;115:985–94. doi: 10.1182/blood-2009-08-237537. [DOI] [PubMed] [Google Scholar]
- 30.Maniati E, Bossard M, Cook N, Candido JB, Emami-Shahri N, Nedospasov SA, et al. Crosstalk between the canonical NF-kappaB and Notch signaling pathways inhibits Ppargamma expression and promotes pancreatic cancer progression in mice. J Clin Invest. 2011;121:4685–99. doi: 10.1172/JCI45797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Banerjee S, Ahmad A, Li Y, Azmi AS, Gunn JR, et al. Activated K-ras and INK4a/Arf deficiency cooperate during the development of pancreatic cancer by activation of Notch and NF-kappaB signaling pathways. PLoS One. 2011;6:e20537. doi: 10.1371/journal.pone.0020537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Feldmann G, Mishra A, Bisht S, Karikari C, Garrido-Laguna I, Rasheed Z, et al. Cyclin-dependent kinase inhibitor Dinaciclib (SCH727965) inhibits pancreatic cancer growth and progression in murine xenograft models. Cancer Biol Ther. 2011;12:598–609. doi: 10.4161/cbt.12.7.16475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat Med. 2002;8:979–86. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 34.Fan X, Mikolaenko I, Elhassan I, Ni X, Wang Y, Ball D, et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer research. 2004;64:7787–93. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- 35.Osipo C, Patel P, Rizzo P, Clementz AG, Hao L, Golde TE, et al. ErbB-2 inhibition activates Notch-1 and sensitizes breast cancer cells to a gamma-secretase inhibitor. Oncogene. 2008;27:5019–32. doi: 10.1038/onc.2008.149. [DOI] [PubMed] [Google Scholar]
- 36.Pandya K, Meeke K, Clementz AG, Rogowski A, Roberts J, Miele L, et al. Targeting both Notch and ErbB-2 signalling pathways is required for prevention of ErbB-2-positive breast tumour recurrence. Br J Cancer. 2011;105:796–806. doi: 10.1038/bjc.2011.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, Mullendore M, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–96. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cook N, Frese KK, Bapiro TE, Jacobetz MA, Gopinathan A, Miller JL, et al. Gamma secretase inhibition promotes hypoxic necrosis in mouse pancreatic ductal adenocarcinoma. J Exp Med. 2012;209:437–44. doi: 10.1084/jem.20111923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Zhang Y, Banerjee S, Li Y, Sarkar FH. Inhibition of nuclear factor kappab activity by genistein is mediated via Notch-1 signaling pathway in pancreatic cancer cells. Int J Cancer. 2006;118:1930–6. doi: 10.1002/ijc.21589. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Zhang Y, Li Y, Banerjee S, Liao J, Sarkar FH. Down-regulation of Notch-1 contributes to cell growth inhibition and apoptosis in pancreatic cancer cells. Mol Cancer Ther. 2006;5:483–93. doi: 10.1158/1535-7163.MCT-05-0299. [DOI] [PubMed] [Google Scholar]
- 41.He F, Wang L, Hu XB, Yin DD, Zhang P, Li GH, et al. Notch and BCR signaling synergistically promote the proliferation of Raji B-lymphoma cells. Leuk Res. 2009;33:798–802. doi: 10.1016/j.leukres.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 42.Thomas M, Calamito M, Srivastava B, Maillard I, Pear WS, Allman D. Notch activity synergizes with B-cell-receptor and CD40 signaling to enhance B-cell activation. Blood. 2007;109:3342–50. doi: 10.1182/blood-2006-09-046698. [DOI] [PubMed] [Google Scholar]
- 43.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–9. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.ClinicalTrials.gov [homepage on the internet] A service of the US Natioanal Institutes of Health. Bethesda: [updated 2012 February 27]. Identifier NCT01098344. Available from: http://clinicaltrials.gov. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.