Summary
Fast, accurate nucleotide incorporation by polymerases facilitates expression and maintenance of genomes. Many polymerases use conformational dynamics of a conserved α-helix to permit efficient nucleotide addition only when the correct nucleotide substrate is bound. This α-helix is missing in structures of RNA-dependent RNA polymerases (RdRps) and reverse transcriptases (RTs). Here we use solution-state NMR to demonstrate that the conformation of conserved structural motif D of an RdRp is linked to the nature (correct vs. incorrect) of the bound nucleotide and the protonation state of a conserved, motif-D lysine. Structural data also reveal the inability of motif D to achieve its optimal conformation after incorporation of an incorrect nucleotide. Functional data are consistent with the conformational change of motif D becoming rate limiting during and after nucleotide misincorporation. We conclude that motif D of RdRps and, by inference, RTs is the functional equivalent to the fidelity helix of other polymerases.
INTRODUCTION
Positive-strand RNA viruses cause a number of acute and chronic diseases, including the common cold, myocarditis, encephalitis, hepatitis and paralytic poliomyelitis (Knipe et al., 2007). Establishment of a population of distinct genetic variants contributes to the fitness of RNA viruses, including retroviruses, and is likely responsible for the ability of these viruses to escape bottlenecks and defenses of the host that would otherwise force the virus into extinction (Vignuzzi et al., 2006). It is now clear that virus population diversity is caused by nucleotide misincorporation of the viral RNA-dependent RNA polymerase (RdRp) (Arnold et al., 2005; Crotty et al., 2000; Pfeiffer and Kirkegaard, 2003) or reverse transcriptase (RT) (Anderson et al., 2004). Kinetic studies have suggested that the nucleotidyl transfer chemical step and a conformational change step preceding chemistry represent fidelity checkpoints for RdRps and RTs (Arnold and Cameron, 2004; Kati et al., 1992; Kellinger and Johnson, 2010). The structural basis for these checkpoints, however, remains unknown.
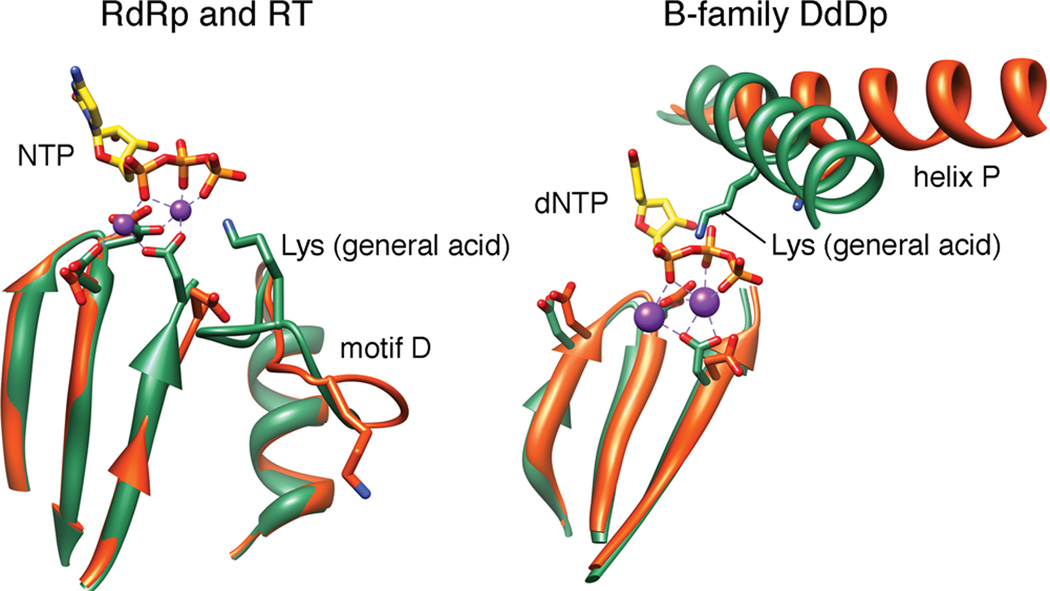
RdRps and RTs belong to a superfamily of template-directed nucleic acid polymerases, including DNA-dependent DNA polymerases (DdDp) and DNA-dependent RNA polymerases (DdRp). The polymerases share a ‘cupped right hand’ structure, including palm, fingers and thumb domains (Ng et al., 2008), catalyze phosphodiester bond formation through a conserved two metal ion mechanism and share a similar kinetic mechanism (Joyce and Steitz, 1995; Steitz, 1999) (Figure 1). Single subunit DdDps and DdRps use the movement of an α-helix (helix O in A-family and helix P in B-family polymerases) to complete the architecture of their active-sites (Franklin et al., 2001; Kaushik et al., 1996; Kiefer et al., 1998; Li et al., 1998; Tahirov et al., 2002; Temiakov et al., 2004; Yin and Steitz, 2002, 2004). The stability and equilibrium position of this helix is known to tune the efficiency of nucleotidyl transfer to the nature (correct vs. incorrect) of the incoming nucleotide (Franklin et al., 2001; Johnson and Beese, 2004; Kiefer et al., 1998; Wu and Beese, 2011). However, an equivalent helix is absent in viral RdRps and RTs (O'Reilly and Kao, 1998).
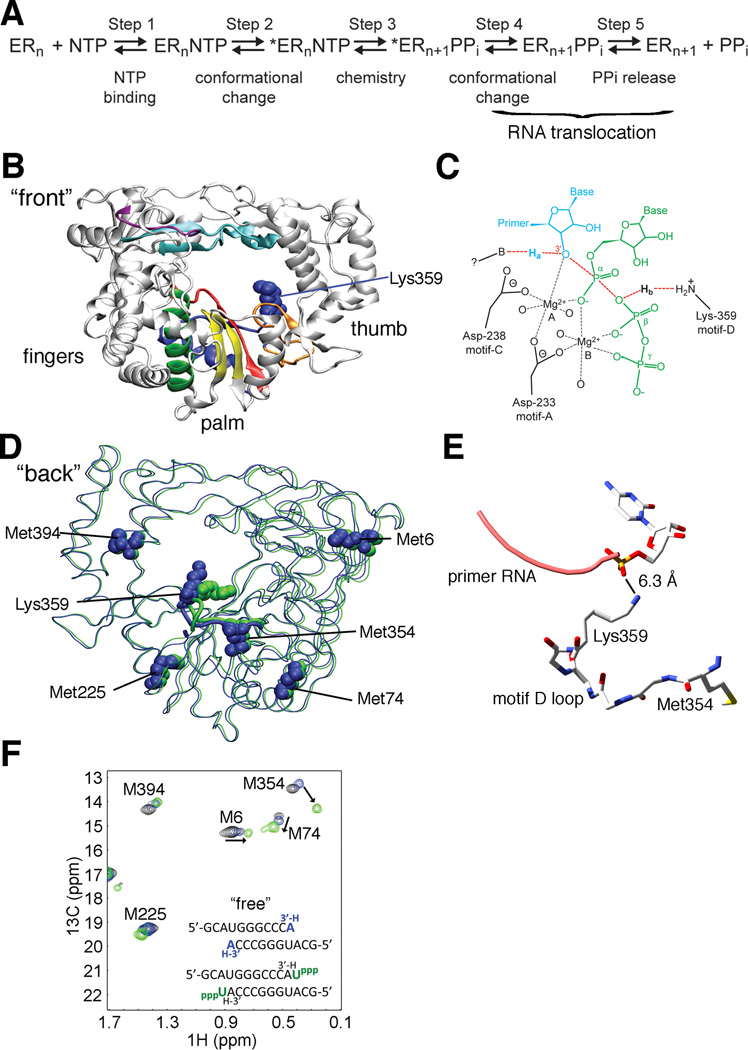
Figure 1.
Structural changes to PV RdRp motif D upon nucleotide binding as observed by solution-state NMR are not revealed by crystal structures. (A) RdRp kinetic mechanism includes a pre-chemistry conformational rearrangement (step 2) required for catalytic competence of the RdRp-RNA-NTP complex in the forward (nucleotide incorporation) direction. (B) The structure of viral RdRps is said to resemble a “cupped right hand” with fingers, palm and thumb subdomains. Sequence and structural alignments have identified seven conserved structural motifs in RdRps and RTs (A, red; B, green; C, yellow; D, blue; E, orange; F, cyan; G, purple). (C) Biochemical studies have implicated a conserved motif D lysine in RdRps (Lys359 in the PV RdRp) as a general-acid catalyst (Castro et al., 2009). As the transition state of nucleotidyl transfer (chemistry, step 3) is approached the primer 3′-OH proton, Ha, is abstracted by an unidentified base and the pyrophosphate leaving group is protonated (Hb) by the lysine general acid. Primer terminus, blue; conserved active site aspartates (233 and 328 in PV), black; conserved motif-D lysine general acid, orange; nucleoside triphosphate, green. (D) PV RdRp conformational rearrangements suggested by X-ray crystallography fail to demonstrate realignment of the motif-D general acid (Lys359) and show little to no changes in the position of five NMR probes (Met6, Met74, Met225, Met354 and Met394) or their surrounding environments. Overlapping crystal structures for the RNA-bound complex before (PDB 3OL6, blue) and after (PDB 3OL7, green) nucleotide addition, with Met NMR probes and general acid Lys359 shown as spheres. (E) Close-up view of the motif-D loop containing Met354 and Lys359 (PDB 3OL7). The terminal amine group on the side-chain of Lys359 appears to be too distant to perform general acid catalysis, although the structure presented is after CMP incorporation. Conformational changes in the motif-D loop would re-position Met354 and affect its associated chemical shift. (F) The open-complex to closed-complex structural transition upon correct nucleotide binding observed by solution-state NMR reveals repositioning of motif D. Met resonances in black are for free RdRp, in blue for the RdRp-RNA complex and in green are upon binding of correct UTP to RdRp-RNA. Arrows indicate chemical shift changes for three Met NMR probes that are particularly sensitive to nucleotide binding. The largest chemical shift change elicited by UTP binding is observed for Met354 which is located in motif D near the general acid catalyst Lys359. Spectra were collected at 293 K with 245 µM RdRp, 500 µM RNA and 4 mM UTP. See also Figure S1.
Studies of the chemical mechanisms of single-subunit polymerases have suggested the use of a general acid to protonate the pyrophosphate leaving group during nucleotidyl transfer (Castro et al., 2007; Castro et al., 2009). The proton transfer reaction, while not essential to catalysis, enhances the rate of nucleotidyl transfer 50 to 2000 fold. The general acid has been suggested to be a conserved lysine residue on helix O/P of A-/B-family members, or palm motif D of RdRps and RTs (Castro et al., 2009). Until recently, there was no function associated with conserved structural motif D in RdRps and RTs (Canard et al., 1999; O'Reilly and Kao, 1998).
Here, we use solution-state NMR to identify the structural determinants of catalytic efficiency and fidelity of a prototypical RdRp. These studies revealed that the structure of motif D is sensitive to formation of ternary complexes with both correct and incorrect nucleotide. Moreover, the protonation state of the motif-D lysine governs the open-closed conformational equilibrium, and mutation of the lysine leads to a dramatic increase in the fidelity of the polymerase reaction. We conclude that motif D of RdRps, and by inference RTs, is the structural and functional analogue of helix O/P of A-/B-family polymerases. Mutations to the motif-D lysine could serve as a starting framework for the design of live, attenuated vaccine viral strains as has been shown with other fidelity-governing mutations in RdRps (Vignuzzi et al., 2006; Vignuzzi et al., 2008).
RESULTS
Movement of motif D facilitates active-site closure
Poliovirus (PV) RdRp has been extensively studied both kinetically and structurally (Ng et al., 2008), and thus represents a good model system for the study of RdRps, and by extension, RTs. Previous structural and sequence alignments have identified seven conserved structural motifs (A-G) in RdRps (O'Reilly and Kao, 1998); five of the motifs (A-E) reside in the palm subdomain (Figure 1B). Most motifs have been assigned functions. For example, motifs A and C contain the absolutely conserved Asp residues important for chelating divalent metal ions (Figure 1C). Residues in motifs A-C have also been implicated in nucleotide and sugar binding and discrimination (Ng et al., 2008). However, until recently, structural motif D had no assigned function (Castro et al., 2009).
We previously assigned the [methyl-13C]methionine NMR spectrum of PV RdRp (Yang et al., 2010) and monitored the chemical shift changes in response to RNA binding, and formation of the correct RdRp-RNA-NTP ternary complex (Figure 1E; see Figure S1A for experimental outline) (Yang et al., 2010). Met354, located in structural motif D and near the proposed general acid Lys359, undergoes the largest chemical shift change upon formation of the RdRp-RNA-NTP ternary complex (Figure 1E). Other notable chemical shift changes include those to Met74 and Met225, which are located on structural elements that make direct interactions with motif D. The α-helix containing Met74 interacts with motif D through the H-bond interaction between Asp71 and Tyr350, and the short loop containing Met225 and the flanking β-strand (Lys228-Thr235) are closely packed against the motif-D loop (Figure S1C). Chemical shift changes to Met74, Met225 and Met354 thus can report on structural rearrangements in motif D. It should be noted that binding of NTP alone does not induce chemical shift changes to these Met resonances (Figure S1D). For simplification purposes, most of our discussion below focuses on the Met354 resonance as a reporter for conformational changes in motif D, but chemical shift changes for Met354 are generally accompanied by corresponding changes to the Met74 and Met225 resonances.
Comparison of PV RdRp crystal structures bound with RNA before (PDB 3OL6) and after (PDB 3OL7) nucleotide incorporation indicated only a small (1.4 Å) change in the position of the terminal methyl group of Met354 (Gong and Peersen, 2010). Other Met probes, including Met6, Met74 and Met225, that show chemical shift changes upon formation of the ternary complex also do not show appreciable changes to their conformations or surrounding protein environments when comparing these crystal structures (Gong and Peersen, 2010) (Figure 1D, E). These results suggest that there are additional conformational changes in PV RdRp observed in solution upon formation of the RdRp-RNA-NTP complex that may not reported by the X-ray crystal structures. Such changes may be required to form the catalytically-relevant “closed” state (step 2 of the kinetic mechanism, Figure 1A) that may involve the re-positioning of the general acid in motif D.
Mechanism of active-site closure
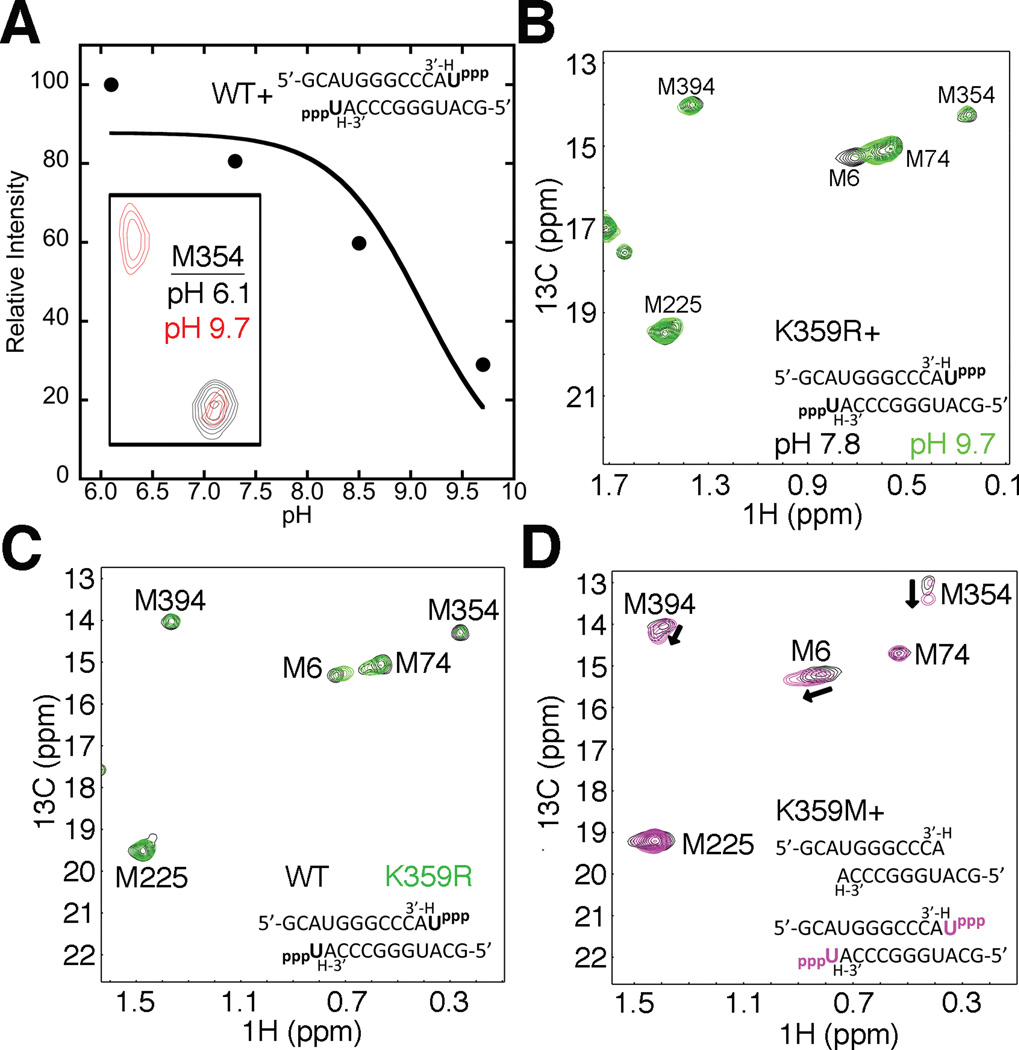
We previously suggested that the motif-D lysine in RdRps plays an analogous role to His1085 in the trigger loop of yeast RNA polymerase II (Castro et al., 2009). The interaction between His1085 and the β-phosphate of the incoming NTP is critical for the structural completion of the active-site (Wang et al., 2006). It has also been suggested that proton transfer from His1085 to the pyrophosphate leaving group links substrate recognition to catalysis, and could potentially trigger translocation (Wang et al., 2006). The general acid lysine on the O/P helix of A/B family DNA polymerases, or the motif-D lysine in RdRps and RTs could fulfill a similar function to that of His1085. Achieving the closed conformation in RdRps may thus depend on the protonation state of the motif-D lysine. For this reason, we investigated the influence of pH on the ε-13CH3-Met NMR spectra of both free PV RdRp (Figure S2A) and RdRp bound with RNA and UTP (Figure 2A, Figure S2B). It should be noted that unlike other nucleic acid polymerases, the PV RdRp-RNA complexes are very stable towards pH changes (Arnold and Cameron, 2000; Castro et al., 2009), although we were limited to pH < 10 since Mg2+ precipitates out of solution at higher pH values. For ligand-free PV RdRp, only Met6 changed chemical shift in response to differences in pH (Figure S2A). The RdRp-RNA-UTP complex showed additional changes to the NMR spectrum as pH was increased. At higher pH values (pH 8.5, 9.7), Met354 gave two resonances (Figure 2A inset, Figure S2B), suggesting that motif D is fluctuating on the slow NMR chemical shift timescale (i.e. < 180 s−1, which is on the same order of timescale as the pre-chemistry conformational change determined previously using stopped-flow (Arnold and Cameron, 2004)). We used the relative intensity of the Met354 resonance (using the only observed Met354 resonance at pH 6.1) at the different pH values to estimate a pKa (~9.2) for the motif-D conformational change in PV RdRp (Figure 2A).
Figure 2.
Formation of the closed ternary complex depends on the protonation state of the motif-D lysine. (A) A proton on Lys359 is required to form the closed complex. Inset- Met354 resonances for the RdRp-RNA-ternary complex at pH 6.1 (black) and 9.7 (red). At pH 9.7 the Met354 resonance shifts to the open complex position. The relative peak intensity of the Met354 resonance in the closed complex state (Intensity(pH n)/Intensity(pH 6.1)×100%) plotted versus pH was used to determine that the open to closed transition is associated with a pKa of 9.2. The data were fit to a model describing a single-ionizable group. (B) Formation of the closed complex at high pH is restored by substitution of Lys359 with Arg, consistent with the higher pKa of Arg. Met resonances for K359R RdRp-RNA-UTP ternary complexes at pH 7.8 are in black and at 9.7 are in green. The Met354 resonance remains unchanged between different pH regimes and is consistent with formation of the closed complex. (C) Changes in the rate constants for correct nucleotide incorporation are not due to structural differences between WT and K359R PV RdRp-RNA-NTP ternary complexes. Met resonances for WT (black) and K359R (green) RdRp-RNA-NTP ternary complexes at pH 8.0 are nearly identical. (D) Substitution of Lys359 with Met does not support the formation of a closed ternary complex. Met resonances for K359M RdRp-RNA binary complexes in the absence or presence of UTP are in black or magenta, respectively. There were small chemical shift changes to Met6, Met354 and Met394 consistent with UTP binding but these are not indicative of a closed complex. All spectra were collected at 293 K with 245 µM RdRp, 500 µM RNA and 4 mM UTP (where included). See also Figure S2.
The estimated pKa would be consistent with the involvement of the motif-D lysine in the pH dependent chemical shift changes in the RdRp-RNA-NTP complex. To further test this proposal, we carried out similar pH studies with the K359R mutant of PV RdRp (Figure 2B). In this case, the RdRp-RNA-NTP complex spectra at lower and higher pH were nearly identical, except for a chemical shift change to Met6 (Figure 2B). The lack of chemical shift changes at higher pH is consistent with the higher pKa value (~12) expected for Arg.
It should be noted that while the pH dependencies of wild-type (WT) and K359R PV RdRp are different, their spectra are nearly identical at pH 8.0 in their ligand-free form, bound with RNA (Figure S2C,D) and bound with RNA and NTP (Figure 2C). This finding is critical because it suggests that the previously determined differences in the catalytic function for the K359R mutant (e.g. K359R PV RdRp has a ~20 –fold decrease in kpol (Castro et al., 2009)) are due only to the change in the nature of the amino acid side chain at position 359, and not due to gross structural rearrangements of the enzyme.
While Arg at position 359 would be able to hydrogen bond with the β-phosphate of the incoming NTP, the side chain of an amino acid such as Met would not. A substitution to Met at position 359 would also potentially provide another NMR spectral probe to monitor structural changes in motif D. The K359M substitution results in an additional, very intense resonance with similar chemical shifts as the free Met amino acid, indicating that Met359 is very solvent exposed (Figure S2E). However, titration of PV K359M RdRp with RNA and/or nucleotide did not result in any major chemical shift changes to the Met359 resonance. Moreover, the K359M RdRp failed to achieve the closed state of the enzyme as monitored by the lack of a large chemical shift change in Met354 (and smaller chemical shift changes to Met6, Met74 and Met225) upon formation of the RdRp-RNA-UTP complex (Figure 2D). However, K359M RdRp still binds UTP, as evidenced by the smaller Met chemical shift changes from RNA binary complex, and we would expect a similar Kd,app for NTP binding as the K359L RdRp (Kd,app ~ 700 µM; [UTP] > 4 mM; (Castro et al., 2009)). These findings further support the notion that the structural rearrangements reported by the chemical shift changes in WT RdRp required to form the RNA-UTP complex are dependent on the nature of the side-chain at residue 359.
We also tested the importance of the nucleotide β-phosphate to the formation of the closed conformation (Figure S2F,H). Intriguingly, formation of the RdRp-RNA-UDP complex resulted in chemical shift changes similar to that observed for the RdRp-RNA-UTP, but formation of the RdRp-RNA-UMP complex did not (UMP is bound as evidenced by the chemical shift change to Met6, Figure S2G). These results strongly suggest that an interaction between the β-phosphate and the enzyme (likely through Lys359) is critical for the achievement of the closed state.
Motif D structure is sensitive to incorrect NTP binding
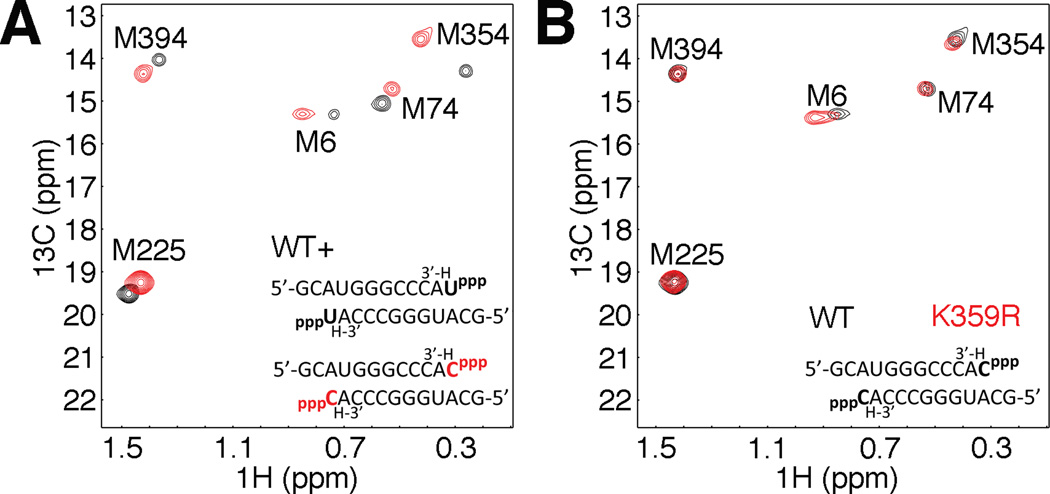
Achievement of the closed, active-state in RdRps, especially the position of motif D, should be dependent on the nature of the incoming nucleotide (correct vs. incorrect) as previously observed in DNA polymerases and the O/P helix (Franklin et al., 2001; Johnson and Beese, 2004; Kiefer et al., 1998; Wu and Beese, 2011). As such, we extended our NMR titration studies to ternary complexes bound with incorrect nucleotides (i.e. GTP, ATP and CTP templated by AMP). Binding of incorrect nucleotide(s) resulted in chemical shift changes from the RdRp-RNA binary complex, but these changes were distinct from the RdRp-RNA-NTP ternary complex with correct UTP bound (Figure 3, Figure S3). In particular, binding of correct nucleotide elicits a large chemical shift change to Met354 consistent with a conformational change in motif D in preparation for catalysis, whereas incorrect nucleotide binding fails to stimulate the large Met354 chemical shift change, suggesting that complex closure is defective with incorrect nucleotide bound.
Figure 3.
The equilibrium position of motif D is altered in response to binding of an incorrect nucleotide and correlates with reduced catalytic efficiency. (A) Comparison of Met resonances between WT RdRp-RNA complexes in the presence of correct nucleotide, UTP, (black) or incorrect nucleotide, CTP (red). (B) The difference in polymerase fidelity between WT and K359R PV RdRp is not due to gross structural differences between the enzymes. Met resonances for WT (black) and K359R (red) RdRp-RNA binary complexes in the presence of incorrect CTP. Spectra were collected at 293 K with 245 µM RdRp, 500 µM RNA, and either 4 mM UTP, or 8 mM GTP, ATP, CTP or 12 mM CTP (K359R only, panel d). See also Figure S3.
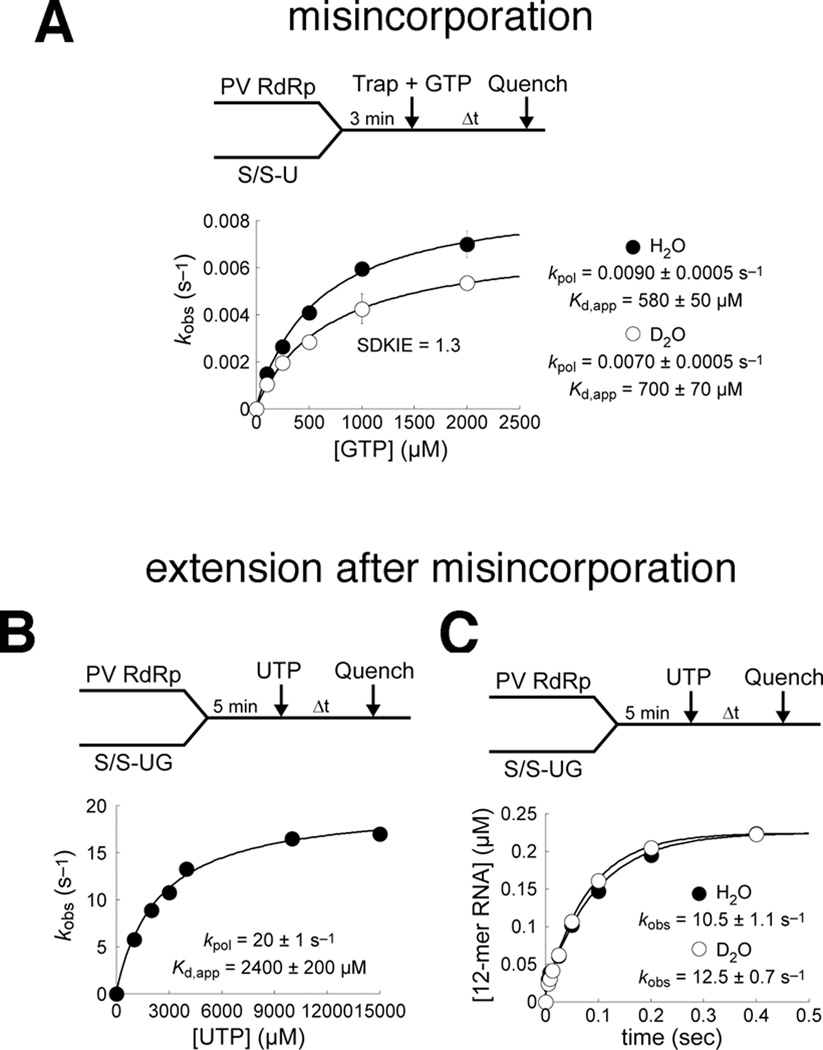
Binding of incorrect nucleotide appears to perturb the position of motif D and shift the conformational equilibrium away from the closed, active-state (i.e. shift the equilibrium to the left in step 2 of the kinetic mechanism, Figure 1A). The chemical shift differences in the RdRp-RNA-NTP ternary complexes bound with correct and incorrect nucleotide may further suggest that incorrect nucleotide incorporation proceeds through a distinct RdRp conformation compared to correct nucleotide incorporation, as previously suggested for T7 DNA polymerase (Tsai and Johnson, 2006). Closure of the active-site may also be slower and contribute more to the overall rate for incorrect nucleotide incorporation, or stated in another way, chemistry would make less of a contribution to the overall rate. To assess the overall rate contribution of chemistry (step 3 in Figure 1A), we measured the solvent deuterium kinetic isotope effect (SDKIE defined as kpol(H2O)/kpol(D2O)) for GMP misincorporation templated by UMP (Figure 4A, Figure S4). The SDKIE reflects in part the donation of a proton from the active-site acid Lys359 to the pyrophosphate group (Castro et al., 2009), and thus, a lower SDKIE is expected if the chemistry step makes less of a contribution to the overall rate. Indeed, the SDKIE for incorrect GMP incorporation (~1.3) was lower than for correct AMP incorporation (~3) (Castro et al., 2007) (Figure 4A). The SDKIE approaching unity for misincorporation suggests defects in the formation of a closed complex prior to chemistry, consistent with the NMR data.
Figure 4.
The kinetics of nucleotide incorporation are perturbed both before and after incorrect nucleotide misincorporation. (A) The solvent deuterium kinetic isotope effect (SDKIE) for nucleotide misincorporation is much smaller than for correct incorporation. Values of kpol for GMP misincorporation templated by U were 0.009 s−1 or 0.007 s−1 in H2O or D2O, respectively, revealing a SDKIE of ~1.3. By comparison, the SDKIE for corrrect AMP incorporation was ~3(Castro et al., 2009). The SDKIE approaching unity for misincorporation suggests defects in formation of a closed complex prior to chemistry, consistent with suppression of NMR chemical shift changes upon binding of an incorrect nucleotide shown. (B) The kinetics of correct nucleotide incorporation are severely perturbed by the presence of a mispaired primer terminus. The value for kpol was depressed by ~16-fold and that for Kd,app was elevated by ~6-fold, resulting in ~100-fold decrease in catalytic efficiency, kpol/Kd,app, relative to the kinetic performance in the presence of a correct RNA primer terminus(Arnold and Cameron, 2004). (C) Presence of a mispaired primer terminus results in the absence of a solvent deuterium kinetic isotope effect (SDKIE) for correct nucleotide incorporation. Indistinguishable values for kobs in H2O and D2O suggest kinetic masking of the chemistry step by a slow, perturbed pre-chemistry conformational rearrangement. Absence of a SDKIE for correct incorporation is again consistent with defective complex closure revealed by NMR. See also Figure S4.
Motif D is sensitive to mismatches at the RNA terminus
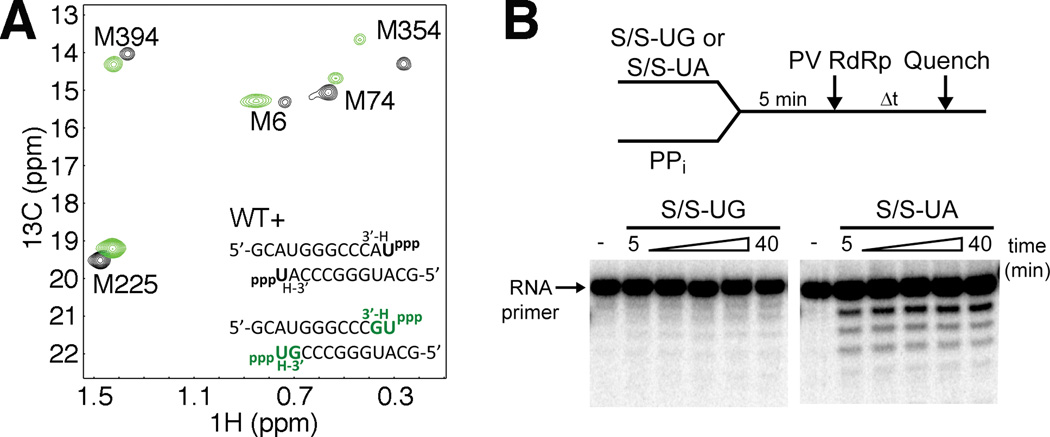
For DNA polymerases, it is known that misinsertion of nucleotides can have a profound effect on subsequent DNA extension (Johnson and Beese, 2004; Kunkel and Bebenek, 2000) by perturbing the closure of the active-site (Johnson and Beese, 2004). We expected that PV RdRp would behave in a similar manner, and that specifically, the conformation of motif D would be impacted. In this case, we pre-incubated PV RdRp with RNA and 3’-dGTP to generate a terminal G:U mispair and 3’block of the RNA (Figure S1B). Following a desalting column to remove excess 3’-dGTP, UTP (the next correct nucleotide) was then added to generate the ternary complex (Figure 5, Figure S5). The ternary complex with the G:U terminal mismatch in the RNA substrate did not result in the same chemical shift changes to Met354 and other methionine resonances as the ternary complex with the correct A:U Watson-Crick base pair. Rather, the Met resonance chemical shifts were similar to that of PV RdRp bound with RNA and incorrect incoming nucleotide (Figure S5F). Thus, similar to incorrect nucleotide binding, motif D is likely not achieving a conformation conducive to catalysis.
Figure 5.
A mispaired primer terminus in the RdRp-RNA binary complex results in perturbed equilibrium positioning of motif D upon correct nucleotide binding. (A) The Met resonance positions for complexes having mispaired RNA termini (G:U) and next correct UTP binding are shown (green) along with Met resonance positions for complexes having the G:U mispaired terminus but without UTP binding (black). This comparison reveals the perturbing influence of the mispaired RNA terminus. Spectra were collected at 293 K with RdRp (245 µM), RNA (500 µM) and UTP where included (4 mM with correct primer terminus or 13 mM with incorrect primer terminus). (B) The PV RdRp reverse reaction (pyrophosphorolysis) is inhibited by the presence of an incorrect base at the RNA primer 3′ terminus. WT PV RdRp (1 µM) was added to 5′-32P-labeled RNA (0.5 µM) with incorrect (S/S-UG, Fig. S8) or correct (S/S-UA) 3′ - terminating base and pyrophosphate, PPi (500 µM). After varying amounts of time reactions were quenched by addition of EDTA (50 mM). S/S-UA is the same as S/S-UG except with A instead of G at RNA 3′ terminus (opposite templating base U). RNA reaction products were resolved by denaturing PAGE and visualized by phosphorimaging. The 11-mer RNA primer is indicated by an arrow. WT PV RdRp was unable to utilize the PPi substrate to accomplish pyrophosphorolysis when incorrect G resided at the primer terminus, but was when correct A was present in the primer terminal position. See also Figure S5.
The nature of the terminal base pair of the RNA primer/template substrate also has a significant influence on the kinetics of nucleotide incorporation. Chemical-quench flow assays for correct UMP incorporation in the presence of a G:U mispaired RNA terminus revealed the kpol value, the maximal rate constant for nucleotide incorporation, and the Kd,app value, the apparent dissociation constant for UTP binding to the RdRp-RNA binary complex, to be 20 ± 1s−1 and 2400 ± 200 µM, respectively (Figure 4B). Comparison to kinetic parameters for correct nucleotide incorporation in the presence of correct, Watson-Crick base-paired RNA termini (Arnold and Cameron, 2004) suggests that a terminal mispair leads to depression of kpol and elevation of Kd,app by several- to approximately 10-fold, resulting in decreased catalytic efficiency, kpol/Kd,app, by 1–2 orders of magnitude. The finding that a SDKIE for correct nucleotide incorporation is absent when the RNA terminus is mispaired (Figure 4C), whereas the SDKIE is ~ 3 when the RNA terminus consists of a correct base pair (Castro et al., 2009), indicates that chemistry is no longer rate limiting and suggests a rate-limiting conformational change. An incorrect nucleotide at the primer terminus also destabilizes the RdRp-RNA complex (Figure S4). Taken together, these kinetic data suggest that an incorrect nucleotide at the RNA primer terminus perturbs PV RdRp interactions with its substrates, in agreement with the NMR chemical shift findings described above.
An alternative interpretation of the NMR results would be that PV RdRp becomes “locked” in a post-translocated state following nucleotide incorporation into RNA primer with a mismatch. One way to rule out this interpretation is to monitor the rate of the reverse reaction i.e. pyrophosphorolysis. We would expect active-site closure to be perturbed in both forward and reverse directions and thus, decrease pyrophosphorolysis. Indeed, PV RdRp was unable to utilize the pyrophosphate (PPi) substrate to accomplish pyrophosphorolysis when incorrect G resided at the primer terminus (Figure 5B).
Mutation of the motif-D lysine leads to higher RdRp fidelity
The protonation state of the motif-D lysine appears to govern the pre-chemistry conformational changes necessary for the closure of the active-site in preparation for catalysis, and active-site closure is highly dependent on the nature of the RNA primer terminus and the incoming nucleotide. Changes to the motif-D lysine may thus also result in changes in nucleotide discrimination and polymerase fidelity. Our previous studies indicated that mutations to PV RdRp Lys359 can lower kpol (up to ~10-fold) and/or increase Kd,app (up to ~4-fold) for correct nucleotide insertion (in this case, ATP) compared to WT RdRp (Castro et al., 2009). Here, we have expanded these studies to compare the kinetics for incorrect nucleotide incorporation, either incorrect sugar (2’-dATP) or incorrect nucleobase (GTP) (Table 1).
Table 1.
The motif-D lysine is a molecular determinant of RdRp fidelity.
| Enzyme | NTP1 | kobs (s−1) | kobs,cor/ kobs,incor |
|---|---|---|---|
| WT | ATP | 50 ± 5 | − |
| GTP | 0.010 ± 0.001 | 5,000 | |
| 2’-dATP | 0.50 ± 0.05 | 100 | |
| K359R | ATP | 5.0 ± 0.5 | − |
| GTP | 0.0002 | 25,000 | |
| 2’-dATP | 0.010 ± 0.001 | 500 |
These assays all used S/S-U RNA (Figure S4), in which case ATP is the “correct” incoming nucleotide
The K359R substitution of RdRp discriminated much more against incorrect nucleotide compared to WT RdRp (Table 1). The fidelity of nucleotide incorporation was substantially increased for the K359R (kobs,correct/kobs,incorrect for GTP=25,000) mutant compared to WT RdRp (kobs,correct/kobs,incorrect for GTP=5,000). However, similar to WT RdRp (Arnold and Cameron, 2004), K359R was less able to discriminate against the incorrect sugar configuration (kobs,correct/kobs,incorrect for 2’-dATP = 500 for K359R) compared to the incorrect nucleobase. Changes in function and fidelity induced by the K359R mutation are not due to any gross structural rearrangements in the active-site, considering that the NMR spectra for the ternary complexes of WT and K359R RdRp bound with correct (Figure 2C) and incorrect (Figure 3B, Figure S3C,D) nucleotide essentially overlap. These results place the motif-D lysine in a central role for both catalytic efficiency and nucleotide incorporation fidelity in RdRp and by inference, RT enzymes.
DISCUSSION
Processive, faithful addition of nucleotides by DNA and RNA polymerases is essential for maintenance and expression of the genomes of all organisms. Studies of A-and B-family polymerases have shown that distinct conformational states of a single helix (O and P, respectively) are associated with each step of the nucleotide-addition cycle (Beard and Wilson, 2003; Franklin et al., 2001; Johnson and Beese, 2004; Kiefer et al., 1998; Li et al., 1998; Wu and Beese, 2011; Yin and Steitz, 2004). Binding of incorrect or non-natural nucleotides prevents helix O/P from achieving its closed state, leading to reduced incorporation efficiency (Johnson and Beese, 2004; Wu and Beese, 2011). Once a nucleotide is incorporated, a conformational change of helix O/P facilitates pyrophosphate release, which may also be linked to translocation (Johnson et al., 2003; Kiefer et al., 1998; Yin and Steitz, 2004). RdRps and RTs lack a corresponding helix O/P (O'Reilly and Kao, 1998). The motivation for this study was to identify the helix O/P equivalent in RdRps and, by inference, RTs.
The catalytic site, in the so-called palm subdomain, of RdRps and RTs superimposes quite well with A- and B-family polymerases (Hansen et al., 1997; Kohlstaedt et al., 1992; Singh and Modak, 1998). Unique to RdRps and RTs is a flexible loop referred to as conserved structural motif D. Since the first structures of RdRps and RTs appeared, motif D has been considered “scaffolding” for the palm (Hansen et al., 1997; Kohlstaedt et al., 1992; O'Reilly and Kao, 1998). Recently, we showed that motif D of RdRps and RTs contain a lysine that protonates the pyrophosphate leaving group, thus accelerating catalysis (Castro et al., 2009). That study revealed an analogous lysine on helix O/P of A- and B-family polymerases (Castro et al., 2009). Therefore, motif D may be the functional equivalent of helix O/P.
In general, X-ray crystal structural studies of RdRps and RTs have failed to reveal a conformational change of motif D of sufficient magnitude to place the conserved lysine within reach of the β-phosphate of the nucleotide substrate (Ferrer-Orta et al., 2007; Gong and Peersen, 2010; Zamyatkin et al., 2009); one exception exists (Gillis et al., 2008). A similar circumstance existed when some of the highest resolution structures appeared for the DNA polymerases. For example, in one case, helix O occluded the nucleotide-binding pocket (Kiefer et al., 1998). Removal of crystal packing restraints was necessary to permit the full range of motion of helix O to be observed crystallographically (Johnson et al., 2003). The use of solution NMR to study conformational changes during the nucleotide-addition cycle should not suffer these complications.
We have used methyl-13C-labeled methionines as NMR probes (Yang et al., 2010) to identify conformational changes of the PV RdRp that occur in response to nucleotide binding on the path to formation of a catalytically competent state. Met354 is located in motif D in the vicinity of the general acid (Lys359) and has been a very informative probe for the conformation/equilibrium position of motif D (Figure 1D). Formation of the closed, catalytically competent RdRp-RNA-NTP complex, as monitored by the Met354 resonance, requires hydrogen bonding between the β-phosphate of incoming NTP and the lysine at position 359 (Figure S2). The protonation state of Lys359 is also critical for achievement of the closed conformation (Figure 2). The ability of motif D to achieve the closed conformation is antagonized by binding of an incorrect nucleotide (Figure 3). Indeed, closure of motif D may be rate limiting for nucleotide misincorporation (Figure 4). Unexpectedly, the ability of motif D to form the closed state continues to be affected after misincorporation (Figure 5), perhaps again contributing to the rate-limiting step for incorporation (Figure 4). These studies link the conformation of motif D to the efficiency and fidelity of nucleotide addition. Consistent with these NMR observations is the recent findings that MD simulations predict extraordinarily high flexibility and mobility for motif D (Moustafa et al., 2011). Moreover, NMR studies on the cystoviral RdRp have demonstrated that Ile488 in motif D undergoes conformational exchange on the same timescale as catalysis (Ren et al.). The responses of motif D to the nature of the bound nucleotide and complementarity of the basepairing at the primer terminus are remarkably similar to the responses of helix O/P to these same circumstances (Franklin et al., 2001; Johnson and Beese, 2004; Johnson et al., 2003; Kaushik et al., 1996; Kiefer et al., 1998; Tahirov et al., 2002; Temiakov et al., 2004; Wu and Beese, 2011; Yin and Steitz, 2002). Mutations on motif D (i.e. to Lys359) also impact polymerase fidelity similar to mutations on helix O/P in DNA polymerases (Bell et al., 1997; Suzuki et al., 1997; Suzuki et al., 2000). We conclude that motif D is the functional equivalent of helix O/P (Figure 6).
Figure 6.
Conformational changes associated with the general acid of nucleic acid polymerases. Our NMR data are consistent with movement of Lys359 of PV RdRp from the open (orange) to closed (green) conformation shown in the panel on the left. We propose that all RdRps and RTs undergo a similar conformational change. The conformations shown derive from structural data for Norovirus RdRp (apo: 1SH0; liganded: 3BSO) (Zamyatkin et al., 2008). Most replicative DNA polymerases of animal cells and their viruses belong to the B family. The general acid of this family undergoes a similar open-to-closed transition. The conformations shown derive from structural data for RB69 DdDp (apo: 1IH7; liganded: 1IG9) (Franklin et al., 2001). Ligands include (d)NTP and divalent metal ions (purple spheres).
It is unclear why X-ray crystal structures of PV RdRp and related enzymes have failed to reveal a conformation of motif D that positions Lys359 to participate in catalysis. None of the methyl-13C-labeled methionines whose resonances change in response to nucleotide binding show a change in the “open-to-closed” transition of PV RdRp observed crystallographically (Figure 1C). Superposition of the spectra for wild type and the K359R derivative of PV RdRp show that the general architecture of their ternary complexes is identical (Figure 2C). This observation is consistent with the more than 10-fold reduction in the rate constant for nucleotide incorporation measured for the K359R derivative of PV RdRp resulting solely from the elevated pKa of this residue and the corresponding reduction in the ability of Arg to protonate the pyrophosphate leaving group (Castro et al., 2009). Such a function could only occur if position 359 interacts directly with NTP.
Recent molecular dynamics simulations of the RT from human immunodeficiency virus (HIV) revealed movement of the motif-D lysine (Lys220) into the active site (Michielssens et al., 2011). This substantial conformational change of Lys220 required simulation on the microsecond timescale for observation (Michielssens et al., 2011). Hydrogen/deuterium-exchange mass spectrometry experiments (Seckler et al., 2009) also suggest that motif D in HIV RT is conformationally dynamic across longer timescales. Quantum and molecular mechanics calculations provide additional evidence for the use of Lys220 as a general acid in RT-catalzyed nucleotidyl transfer (Michielssens et al., 2011). Collectively, these data provide support for movement of motif D of HIV RT into the active site as suggested here for PV RdRp.
Studies of PV have shown that increasing the fidelity of its RdRp leads to restricted population diversity causing attenuation of the virus (Pfeiffer and Kirkegaard, 2005; Vignuzzi et al., 2006). These high-fidelity variants of PV have been shown to serve as vaccine strains (Vignuzzi et al., 2008). In our studies of the high fidelity Gly64Ser derivative of PV RdRp, we observed that binding of an incorrect nucleotide caused conformational differences in motif D relative to the wild-type enzyme, consistent with the position of motif D contributing to the fidelity of nucleotide incorporation (Yang et al., 2010). How Gly64 and motif D communicate is not completely clear, but both the NMR data (Yang et al., 2010) and MD simulations (Moustafa et al., 2011) suggest the existence of a network of interacting residues that connect residues of related function, even residues as far as 20 Å apart. In this regard, it is worth noting that the Sabin type 1 PV vaccine strain includes RdRp mutations Tyr73His (i.e. next to Met74) and Thr362Ile (near motif D) (Nomoto et al., 1982); both Met74 and Met354 are responsive to RNA and nucleotide binding, and MD simulations show that the motions of motif D and the α-helix containing Tyr73/Met74 are strongly anti-correlated (Moustafa et al., 2011). The mechanism of Sabin type 1 remains unclear (Racaniello, 2006). It is tempting to speculate that mutations to Tyr73 and/or Thr362 alter the conformational dynamics of motif D, and consequentially RdRp fidelity, thereby contributing to the attenuated phenotype of Sabin type 1. Likewise, mutations of the motif-D lysine of RdRps and RTs that result in changes to polymerase fidelity may underpin a general strategy for the rational design of live, attenuated vaccine strains. These targeted strategies can complement more genome-wide approaches (e.g. (Coleman et al., 2008)).
EXPERIMENTAL PROCEDURES
Expression and purification of PV RdRp
The PV RdRp constructs in this work contain the L446D and R455D mutations on the thumb subdomain and were expressed and purified using previously published procedures (Arnold et al., 2006; Arnold and Cameron, 2000; Gohara et al., 1999; Yang et al.).
NMR sample preparation
NMR sample preparation followed previously published procedures (Yang et al., 2010) with any modifications indicated in the Supplementary Methods. All NMR experiments were performed at 293 K on a Bruker Avance III 600MHz spectrometer equipped with a 5 mm “inverse detection” triple-resonance (1H/13C/15N) single axis gradient TCI cryoprobe similar to previous procedures (Yang et al., 2010).
Kinetic analysis of WT PV RdRp and Lys359 derivatives
Nucleotide-incorporation experiments were performed essentially as described previously (Arnold and Cameron, 2004; Castro et al., 2009). All details, including modifications, are provided in the Supplemental Information.
Supplementary Material
Highlights.
motif D changes conformation upon binding “correct”, but not “incorrect”, NTP
structural change in motif D depends on the protonation state of the motif D lysine
the motif D lysine is also an important determinant of polymerase fidelity
motif D has an analogous function to helix O/P in A/B family DNA polymerases
ACKNOWLEDGEMENTS
We thank Dr. Ibrahim Moustafa for constructing Figure 5, and for his critical insights on this manuscript. This study was supported by a grant (AI45818) from the US National Institutes of Health to C.E.C., and start-up funds from Pennsylvania State University to D.D.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderson JP, Daifuku R, Loeb LA. Viral error catastrophe by mutagenic nucleosides. Annu Rev Microbiol. 2004;58:183–205. doi: 10.1146/annurev.micro.58.030603.123649. [DOI] [PubMed] [Google Scholar]
- Arnold JJ, Bernal A, Uche U, Sterner DE, Butt TR, Cameron CE, Mattern MR. Small ubiquitin-like modifying protein isopeptidase assay based on poliovirus RNA polymerase activity. Anal Biochem. 2006;350:214–221. doi: 10.1016/j.ab.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub) J Biol Chem. 2000;275:5329–5336. doi: 10.1074/jbc.275.8.5329. [DOI] [PubMed] [Google Scholar]
- Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+ Biochemistry. 2004;43:5126–5137. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JJ, Vignuzzi M, Stone JK, Andino R, Cameron CE. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J Biol Chem. 2005;280:25706–25716. doi: 10.1074/jbc.M503444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard WA, Wilson SH. Structural insights into the origins of DNA polymerase fidelity. Structure. 2003;11:489–496. doi: 10.1016/s0969-2126(03)00051-0. [DOI] [PubMed] [Google Scholar]
- Bell JB, Eckert KA, Joyce CM, Kunkel TA. Base miscoding and strand misalignment errors by mutator Klenow polymerases with amino acid substitutions at tyrosine 766 in the O helix of the fingers subdomain. J Biol Chem. 1997;272:7345–7351. doi: 10.1074/jbc.272.11.7345. [DOI] [PubMed] [Google Scholar]
- Canard B, Chowdhury K, Sarfati R, Doublie S, Richardson CC. The motif D loop of human immunodeficiency virus type 1 reverse transcriptase is critical for nucleoside 5'-triphosphate selectivity. J Biol Chem. 1999;274:35768–35776. doi: 10.1074/jbc.274.50.35768. [DOI] [PubMed] [Google Scholar]
- Castro C, Smidansky E, Maksimchuk KR, Arnold JJ, Korneeva VS, Gotte M, Konigsberg W, Cameron CE. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc Natl Acad Sci U S A. 2007;104:4267–4272. doi: 10.1073/pnas.0608952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro C, Smidansky ED, Arnold JJ, Maksimchuk KR, Moustafa I, Uchida A, Gotte M, Konigsberg W, Cameron CE. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, Andino R, Cameron CE. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. Sequential structures provide insights into the fidelity of RNA replication. Proc Natl Acad Sci U S A. 2007;104:9463–9468. doi: 10.1073/pnas.0700518104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- Gohara DW, Ha CS, Kumar S, Ghosh B, Arnold JJ, Wisniewski TJ, Cameron CE. Production of "authentic" poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr Purif. 1999;17:128–138. doi: 10.1006/prep.1999.1100. [DOI] [PubMed] [Google Scholar]
- Gong P, Peersen OB. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A. 2010;107:22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JL, Long AM, Schultz SC. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc Natl Acad Sci U S A. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce CM, Steitz TA. Polymerase structures and function: variations on a theme? J Bacteriol. 1995;177:6321–6329. doi: 10.1128/jb.177.22.6321-6329.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kati WM, Johnson KA, Jerva LF, Anderson KS. Mechanism and fidelity of HIV reverse transcriptase. J Biol Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- Kaushik N, Pandey VN, Modak MJ. Significance of the O-helix residues of Escherichia coli DNA polymerase I in DNA synthesis: dynamics of the dNTP binding pocket. Biochemistry. 1996;35:7256–7266. doi: 10.1021/bi960537i. [DOI] [PubMed] [Google Scholar]
- Kellinger MW, Johnson KA. Nucleotide-dependent conformational change governs specificity and analog discrimination by HIV reverse transcriptase. Proc Natl Acad Sci U S A. 2010;107:7734–7739. doi: 10.1073/pnas.0913946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer JR, Mao C, Braman JC, Beese LS. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. Field's Virology. 5th Edition. Philadelphia, PA: Lippincott, Williams and Wilkins; 2007. [Google Scholar]
- Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielssens S, Moors SL, Froeyen M, Herdewijn P, Ceulemans A. Structural basis for the role of LYS220 as proton donor for nucleotidyl transfer in HIV-1 reverse transcriptase. Biophys Chem. 2011 doi: 10.1016/j.bpc.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Moustafa IM, Shen H, Morton B, Colina CM, Cameron CE. Molecular dynamics simulations of viral RNA-dependent RNA polymerases link conserved and correlated motions of functional elements to fidelity. J. Mol. Biol. 2011 doi: 10.1016/j.jmb.2011.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KK, Arnold JJ, Cameron CE. Structure-function relationships among RNA-dependent RNA polymerases. Curr Top Microbiol Immunol. 2008;320:137–156. doi: 10.1007/978-3-540-75157-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto A, Omata T, Toyoda H, Kuge S, Horie H, Kataoka Y, Genba Y, Nakano Y, Imura N. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc Natl Acad Sci U S A. 1982;79:5793–5797. doi: 10.1073/pnas.79.19.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly EK, Kao CC. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- Pfeiffer JK, Kirkegaard K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc Natl Acad Sci U S A. 2003;100:7289–7294. doi: 10.1073/pnas.1232294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer JK, Kirkegaard K. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 2005;1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello VR. One hundred years of poliovirus pathogenesis. Virology. 2006;344:9–16. doi: 10.1016/j.virol.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Ren Z, Wang H, Ghose R. Dynamics on multiple timescales in the RNA-directed RNA polymerase from the cystovirus phi6. Nucleic Acids Res. 2010;38:5105–5118. doi: 10.1093/nar/gkq210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seckler JM, Howard KJ, Barkley MD, Wintrode PL. Solution structural dynamics of HIV-1 reverse transcriptase heterodimer. Biochemistry. 2009;48:7646–7655. doi: 10.1021/bi900790x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Modak MJ. A unified DNA- and dNTP-binding mode for DNA polymerases. Trends Biochem Sci. 1998;23:277–281. doi: 10.1016/s0968-0004(98)01250-x. [DOI] [PubMed] [Google Scholar]
- Steitz TA. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Avicola AK, Hood L, Loeb LA. Low fidelity mutants in the O-helix of Thermus aquaticus DNA polymerase I. J Biol Chem. 1997;272:11228–11235. doi: 10.1074/jbc.272.17.11228. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Yoshida S, Adman ET, Blank A, Loeb LA. Thermus aquaticus DNA polymerase I mutants with altered fidelity. Interacting mutations in the O-helix. J Biol Chem. 2000;275:32728–32735. doi: 10.1074/jbc.M000097200. [DOI] [PubMed] [Google Scholar]
- Tahirov TH, Temiakov D, Anikin M, Patlan V, McAllister WT, Vassylyev DG, Yokoyama S. Structure of a T7 RNA polymerase elongation complex at 2.9 A resolution. Nature. 2002;420:43–50. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- Temiakov D, Patlan V, Anikin M, McAllister WT, Yokoyama S, Vassylyev DG. Structural basis for substrate selection by t7 RNA polymerase. Cell. 2004;116:381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignuzzi M, Wendt E, Andino R. Engineering attenuated virus vaccines by controlling replication fidelity. Nat Med. 2008;14:154–161. doi: 10.1038/nm1726. [DOI] [PubMed] [Google Scholar]
- Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu EY, Beese LS. The structure of a high fidelity DNA polymerase bound to a mismatched nucleotide reveals an "ajar" intermediate conformation in the nucleotide selection mechanism. J Biol Chem. 2011;286:19758–19767. doi: 10.1074/jbc.M110.191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Welch JL, Arnold JJ, Boehr DD. Long-range interaction networks in the function and fidelity of poliovirus RNA-dependent RNA polymerase studied by nuclear magnetic resonance. Biochemistry. 2010;49:9361–9371. doi: 10.1021/bi100833r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- Zamyatkin DF, Parra F, Alonso JM, Harki DA, Peterson BR, Grochulski P, Ng KK. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J Biol Chem. 2008;283:7705–7712. doi: 10.1074/jbc.M709563200. [DOI] [PubMed] [Google Scholar]
- Zamyatkin DF, Parra F, Machin A, Grochulski P, Ng KK. Binding of 2'-amino-2'-deoxycytidine-5'-triphosphate to norovirus polymerase induces rearrangement of the active site. J Mol Biol. 2009;390:10–16. doi: 10.1016/j.jmb.2009.04.069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.