Abstract
The hypothalamus is integral to the regulation of body homeostasis, including food intake, energy balance, and blood pressure. Dysfunction of the hypothalamus has been associated with a broad range of disorders; many of which are sex-dependent in prevalence. Small nucleolar (sno) RNAs are a group of small RNAs located in nucleoli that modulate chemical modifications and maturation of ribosomal or other RNAs. Recent data suggest that snoRNA Snord116 is important for the pathogenesis of Prader-Willi syndrome (PWS) characterized by hyperphagia and obesity. The current study was conducted to assess a potential cellular link between Snord116 and phenotypes of PWS. Data from mice revealed Snord116 expression in the medial hypothalamus, particularly within nuclei that are part of feeding circuitry. High expression of Snord116 was evident in the paraventricular (PVN) and ventromedial (VMH) nuclei, but particularly prevalent in the arcuate nucleus (ARC) according to in situ hybridization. Snord116 expression level in ventral hypothalamic dissections including ARC was significantly greater (by 2-fold) than that in cortex; and its expression level in dorsal hypothalamic dissections including PVN was double that in cortex. The enhanced expression pattern of Snord116 in hypothalamic nuclei was observed at weaning and young adult stages, but less obvious neonatally when expression was significantly more widespread. Therefore the expression of Snord116 likely is regulated developmentally. These results provide a new lead for understanding the mechanism(s) related to hyperphagia and obesity symptoms in PWS patients.
Keywords: snoRNA Snord116, hypothalamus, Prader-Willi syndrome (PWS), postnatal development, mouse
1. Introduction
The hypothalamus is located in the ventral part of the diencephalon. It functions as a master neuroendocrine control center by regulating the pituitary gland and controlling many functions throughout the body. The hypothalamus is implicated in a wide range of disorders. For instance, hypothalamic dysfunctions are associated with major depressive disorders (MDD) (Swaab et al., 2005) and metabolic disorders (e.g. hypothalamic obesity; Caqueret et al., 2005). Although diseases related to hypothalamic dysfunction often develop in later stages of life, the etiologic factors often are rooted in development (Tobet et al., 2009). Thus, studies of the hormonal and genetic factors that contribute to hypothalamic development are important in understanding disease mechanism, prevention, and treatment.
Much research on the molecular factors contributing to hypothalamic development is focused on signaling molecules (e.g., steroid hormones, transmitters, peptides, etc.,) and transcription factors (McClellan et al., 2006; Morton et al., 2006; Tobet et al., 2009, Shimogori et al., 2010). The current study focuses on a different class of molecules: small nucleic acid polymers in the brain. Non-coding RNAs account for about half of transcribed RNAs (Mattick, 2005). However, it has not been recognized until recent years that transcribed non-coding RNAs may have a number of important cellular functions (Mattick, 2005). Studies of neurodevelopment indicate that small RNAs play roles in cell fate determination during neuronal differentiation (Li et al., 2010). On the other hand, in mature neurons small RNAs may regulate gene expression and be involved in cognitive brain functions (Schratt, 2009). Considering that non-coding RNAs can be just as important as protein-coding RNAs in the nervous system, it is important to consider the possible regulatory roles of small RNAs on hypothalamic development and function and their potential involvement in disease.
Among non-coding RNAs, one particular small nucleic acid sequence has been related to the pathogenesis of Prader-Willi syndrome (PWS) and its symptoms of appetite disorder and obesity. PWS is a disease caused by defects in certain imprinted genes. About 1 in 10,000-15,000 newborns are affected, and without intervention, it usually leads to morbid obesity through adolescence and in adulthood. PWS is mainly caused by deletion or imprinting defect of paternal genetic material located at chromosome 15q11-13. The SNURF-SNRPN locus at chromosome 15q11-13 is associated with PWS, which contains multiple protein coding genes and several non-protein-coding snoRNA genes (Cavaille et al., 2000; de los Santos et al., 2000; Gunay-Aygun et al., 2001; Butler et al., 2006). SnoRNAs are a group of small RNAs ranging 80 to 300 nucleotides in length that are located primarily in nucleoli and are required for the chemical modification and maturation of rRNA.
Studies from human data and cases (Bürger et al., 2002; Gallagher et al., 2002; Runte et al., 2005; Sahoo et al., 2008; de Smith et al., 2009; Duker et al., 2010) indicate that genes in the SNURF-SNRPN locus are etiologic factors in PWS, which were supported by genetically engineered mouse models (Skryabin et al., 2007; Ding et al., 2008). In particular, it was reported (Gallagher et al., 2002) that SNORD116 potentially plays roles in the etiology of much or all of PWS symptoms. In recent years, this hypothesis was supported by the studies in several human cases showing that the deletion of the SNORD116 gene cluster leads to PWS phenotypes including hyperphagia and obesity (Sahoo et al., 2008; de Smith et al., 2009; Duker et al., 2010). Furthermore, recent progresses using knockout mice have suggested that the snoRNA Snord116 gene cluster, which has 27 copies in the SNURF-SNRPN locus, potentially was a critical element in PWS formation (Skryabin et al., 2007; Ding et al., 2008, Sahoo et al., 2008; de Smith et al., 2009; Duker et al., 2010). Other gene disruption studies in mice indicate that deletions of most protein-coding genes and snoRNA Snord115, another snoRNA found in this region, do not lead to PWS (Yang et al., 1998; Nicholls et al., 1999; Gerard et al., 1999; Tsai et al., 1999; Kozlov et al., 2007); while a region that mainly contains the Snord116 gene cluster termed PWS critical region (PWScr) was important in PWS formation (Ding et al., 2005; Schule et al., 2005). Knockout mice with deletion of only Snord116 genes showed the phenotype of hyperphagia (Ding et al., 2008). Interestingly, the obesity phenotype did not develop due to changes in energy expenditure (Ding et al., 2008).
Previous studies showed that Snord116 is primarily expressed in the embryonic brain, but its expression pattern in the brain remains unclear, especially at postnatal ages (Cavaillé et al., 2000; de los Santos et al. 2000; Lee et al., 2003). PWS symptoms, such as hyperphagia and obesity, are associated with the neuronal functions regulated by the hypothalamus (Holm et al., 1993; Gunay-Aygun et al., 2001). Based on the fact that a lack of Snord116 leads to PWS, it seemed likely that this gene would be expressed in the hypothalamus related to circuits responsible for feeding and energy balance. We examined the expression of Snord116 in the brain using in situ hybridization and quantitative reverse-transcription PCR. The result of these experiments provides a link between Snord116 expression and PWS phenotypes at a cellular level.
2. Materials and methods
2.1. Animals
All animal procedures were performed according to the NIH guide for the care and use of laboratory animal and were approved by the Colorado State University Animal Care and Use Committee (ACUC). Mice were housed in the animal facility of the Lab Animal Resource (LAR) at the Colorado State University. Mice used in the current study were on a C57BL/6J background.
2.2. Probe generation
The Snord116 probe was generated based on the sequence of MBII-85 (alias of Snord116) C/D box snoRNA (Lee et al., 2003): AAT GAT GAT TCC CAG TCA AAC ATT CCT TGG AAA AGC TGA ACA AAA TGA GTG AAA ACT CTG TAC TGC CAC TCT CAT CGG AAC TGA. A duplex oligonucleotide of sense and antisense MBII-85 was purchased from Integrated DNA Technologies (IDT, Inc.) and contained the above sequence and PstI overhang in the 3′-ends. The DNA generated from duplex oligonucleotide was then inserted into the PstI site of pBluescript plasmid. One insertion copy of Snord116 into pBluescript plasmid was confirmed by sequencing. The linearized pBluescript plasmids containing the Snord116 insertion were generated by restriction enzyme BamHI or EcoRI digestion. Antisense and sense riboprobes were then generated using T7 or T3 RNA polymerases and a Digoxygenin (DIG) RNA Labeling Kit (Roche, 11277073910).
2.3. In situ hybridization
Mice were examined at multiple ages including perinatal group (embryonic day (E) 17 and postnatal day (P)0), weaning age group (P19-P20), and adult group (6-10 weeks) for in situ hybridization (ISH). They were perfused intracardially with 4% paraformaldehyde/0.1M phosphate buffer (PB; pH= 7.4). The brains were post-fixed in 4% paraforamaldehyde/0.1M PB overnight. The brains were then removed and embedded in 5% agarose prepared in 1% diethyl pyrocarbonate (DEPC; Sigma, D5758) treated H2O. Coronal sections (100 μm) were cut though the hypothalamus using a vibrating microtome (Leica, VT1000S). The procedures for processing free-floating brain sections for ISH were carried out as described previously (Dellovade et al., 2000). Briefly, brain sections were treated with 6% hydrogen peroxide in PBT (0.05M PB, 1% Tween-20) for 1 h at RT, washed in PBT, then treated with 10μg/ml proteinase K (Sigma, P2308)/PBT for 15 min at RT, 2mg/ml glycine/PBT for 10 min at RT. Brain sections were post-fixed in 4% paraformaldehyde/0.2% glutaraldehyde for 20 min at RT. Before hybridization, sections were incubated in pre-hybridization solution (50% formamide in saline-sodium citrate (SSC; Sigma, S6639) buffer containing 8.8 units/ml heparin (Sigma, H9399), 0.05mg/ml yeast tRNA (Sigma, R5636), and 0.024mg/ml dextran sulfate (Sigma, D8906)) for 1h at 60°C. Riboprobes were denatured at 85°C for 5 min and added to pre-hybridization incubation solution overnight at 60°C. On the second day, sections were washed three times for 30 min in a 50% formamide/25% SSC solution, then three times for 30 min in 50% formamide/10% SSC. Sections were washed in TBST (Tris buffered saline (TBS) containing 1% Tween-20) and pre-blocked in 10% sheep serum in TBST. Sections then were incubated with 1:2000 dilution of alkaline phosphatase conjugated anti-DIG antibody (Roche, 11093274910) in 1% sheep serum/TBST overnight at 4°C. On the third day, sections were washed in TBST to clear out primary antibody and stored at 4°C overnight. On the last day, sections were washed in a NTMT solution (0.1M NaCl, 0.1M Tris/pH=9.5, 0.05M MgCl2, and 1% Tween-20) and incubated in a color detection mix solution (10% polyvinyl alcohol, 1 mM Levamisol (Vector Laboratories, SP5000), 2% NBT/BCIP (Roche, 11681451001), 0.1M Tris (pH=9.5), 0.1M NaCl, and 0.005M MgCl2) in the dark at RT. The color detecting reaction was stopped by washing the sections in NTMT in the dark.
2.4. Hypothalamic dissection
Fresh hypothalami were dissected from P0, P19 and 6-10 weeks old adult mice. For P0 mice, brains were removed and placed in 0.05M phosphate buffered saline (PBS)/1% DEPC in a petri dish with ventral side up. Coronal cuts were made caudal to the optic chiasm and rostral to the brainstem. For P0 pups, triangular cuts were then made from the top of the third ventricle medially to the hypothalamic slice laterally at the base of the brain to further dissect out a region that included the PVN, VMH, and ARC. A piece of cortex also was removed from the same coronal slice as a control for brain region. For P19 and adult mice, brains were removed and dissected similarly to create a single coronal slice. However, at these older ages, a ventral hypothalamic piece that included the ARC and a dorsal hypothalamic piece that included PVN were dissected further from the same coronal slice in addition to a piece of cortex. The dissected pieces were snap-frozen or subjected to RNA extraction immediately.
2.5. Quantitative reverse-transcription PCR
Total RNA, including RNA smaller than 200bp, was purified using the mirVanaTM miRNA isolation kit (Applied Biosystems, AM1560) according to the manufacturer’s instructions. Total RNA was subjected to RNase-free DNAase (Qiangen; Cat#79254) digestion for 10 minutes at room temperature. After DNase digestion, total RNA was precipitated and eluted in RNase free water. One microgram of total RNA was reverse transcribed using the qScript cDNA synthesis kit (Quanta Biosciences, Cat# 95047). cDNA was diluted 1:4 and used immediately for PCR amplification in 10 microliter reactions containing SYBR Green qPCR Master Mix (2X, Roche applied sciences). Snord116 primers: forward 5′-TGATGATTCCCAGTCAAACATTC-3′; reverse 5′-ACCTCAGTTCCGATGAGAGTGG-3′: Gapdh primers: forward-5′GGGAAGCCCATCACCATCTT-3′; reverse-5′GCCTTCTCCATGGTGGTGAA-3′. Primers were tested using PCR to ensure a single amplicon was produced. Real-time PCR was performed using a LightCycler480 PCR system (Roche Applied Sciences) with the following cycle conditions: 95°C for 5 minutes and 45 cycles of 95°C for 10 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. Dissociation curve analysis was conducted to confirm amplification specificity.
2.6. Data analysis
To assess the expression pattern of Snord116, brain sections processed by ISH were examined under an Olympus BH2 microscope. Images of the hypothalamus were captured with 4x and 40x objectives and an Insight QE digital camera with Spot Advanced Software. For comparisons the images were taken with the same illumination intensity and capture time. To quantify the relative level of Snord116, qRT-PCR was performed using the LightCycler 480 PCR system and Cp values were determined by the LightCycler 480 SW 1.5 software. Each reaction was performed in triplicate and averaged to determine final Cp values. The relative level of Snord116 was obtained by normalizing the transcribed RNA signal of Snord116 to the mRNA signal of Gapdh internal control (Schmittgen and Livak, 2008). One-way ANOVA was used for the statistical analyses.
3. Results
3.1. Expression of Snord116 in newborn and adult brains
The brain expression pattern of Snord116 was visualized by ISH at 3 different ages: perinatal (E17and P0), weaning age (P19 and P20), and young adult (6-10 weeks) mice. The Snord116 was more widely and evenly distributed before and after birth with little evidence of selective expression at both long and short incubation times with the substrate (representative images in Figure 1A and 1B). In negative control sections using either sense Snord116 probe in ISH or hybridization buffer with no probe, no signals were observed (data not shown). This is consistent with a previous report that the expression of Snord116 was more uniformly distributed in the embryonic brain (Lee et al., 2003). In contrast, the expression of Snord116 in weaning and young adult mice was detectable more prominently in the hypothalamus, hippocampus, amygdala and pyriform cortex (Figures 2C, 2G, 2H, 2I); while most other brain areas, had a lower intensity signal (Figures 2D, 2E, 2F), with levels comparable to the area surrounding the hypothalamus (Figures 3A, 3B). These results suggest developmental regulation of Snord116 expression in the hypothalamus. Other than that, the expression of Snord116 in weaning and young adult mice appeared in cells throughout the brain if the substrate incubation time was relatively long (data not shown), suggesting its universal expression in the adult brain.
Figure 1.
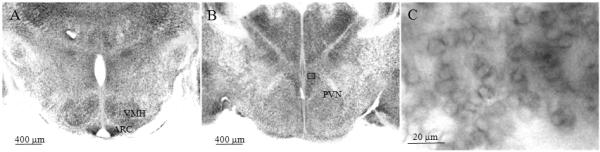
Digital images show digoxigenin-based in situ hybridization (ISH) illustrating the expression of Snord116 in the brain including hypothalamus and surrounding area in neonatal (P0) mice. Panels A and B show the representative images of Snord116 in mice at birth (P0). Panel C is the magnified image from the boxed area in panel B. Scale bars are located in the lower left of each panel.
Figure 2.
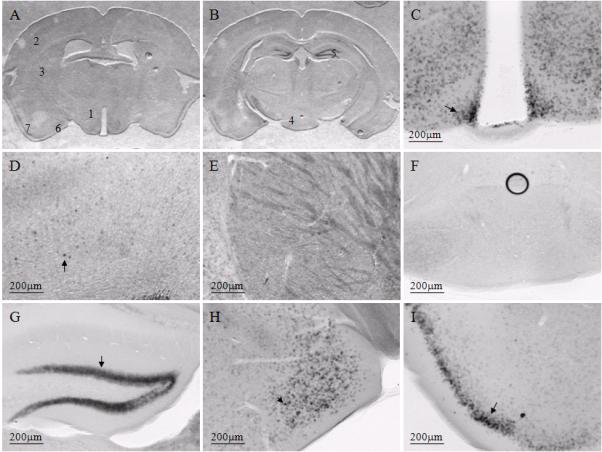
Representative images from a female mouse at weaning age to illustrate digoxigenin-based Snord116 in situ hybridization (ISH) signal in multiple brain regions. Panels A (caudal diencephalon) and B (rostral mesencephalon) illustrate coronal sections hundreds of microns apart containing Snord116 ISH labeling. The numbered brain regions correspond to images shown at higher magnifications in panels C through I (1=C, 2=D, 3=E, 4=F, 5=G, 6=H, 7=I). Representative positive cells are highlighted by arrows shown in panels C, D, G, H and I. The Snord116 ISH signal was most readily noticeable in hypothalamic nuclear groups (panel C). In most other brain regions, the Snord116 ISH signal was lower or not detectable compared to the hypothalamus, as shown in the representative regions of cortex (panel D), striatum (panel E) and brain stem (panel F). Other brain regions in the same coronal plane of hypothalamus that also have high Snord116 ISH signal include cells in the dentate gyrus of the hippocampus (panel G), medial amygdala (panel H) and pyriform cortex (panel I). Scale bars are located in the lower left of each panel.
Figure 3.
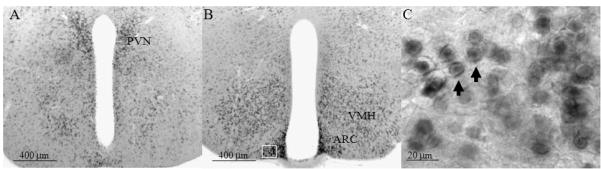
Representative images from a female mouse at weaning age show digoxigenin-based Snord116 in situ hybridization (ISH) signals in the hypothalamus. The ISH signal was detected in the paraventricular nucleus (PVN; panel A), ventromedial nucleus of the hypothalamus (VMH; panel B) and arcuate nucleus (ARC; panel B). Panel C shows a representative higher magnification image of Snord116 from the boxed area in the ARC in panel B. The arrows point to representative cells that show nucleoli labeling of Snord116. Scale bars are located in the lower left of each panel.
3.2. Snord116 localization in hypothalamus
In weaning and adult stages, the expression of Snord116 was prominent in the medial hypothalamic structures (Figures 3, 4), including the paraventricular nucleus (PVN), ventromedial nucleus of the hypothalamus (VMH) and particularly in the arcuate nucleus (ARC). Among those areas, the signal was greatest in the ARC (Figure 3B). At a higher magnification, Snord116 was found in nuclear compartments particularly in nucleoli and adjacent to nuclear membranes (Figure 3C) in weaning and adult mice. In neonatal mice, Snord116 was rarely detectable in nucleoli; in contrast, most Snord116 signal was adjacent to nuclear membranes (Figure 1C). Except for PVN, VMH and ARC, other regions in the hypothalamus with prominent Snord116 signal included the anteroventral periventricular nucleus (AvPV), rostral periventricular area of the third ventricle (RP3V) and posterior periventricular nucleus of the hypothalamus (PVp) (Figure 4).
Figure 4.
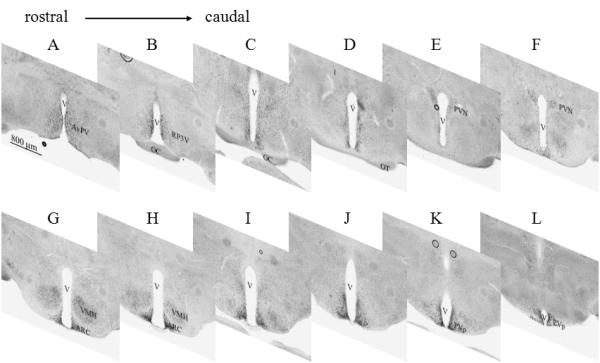
Representative digoxigenin-based in situ hybridization (ISH) images of Snord116 from the rostral to caudal hypothalamus in a female mouse at weaning age. The images from panels A to L are hypothalamic sections taken from rostral to caudal. The Snord116 ISH signals are prevalent in the nuclear groups of the anteroventral periventricular nucleus (AvPV; panel A), rostral periventricular area of the third ventricle (RP3V; panel B), paraventricular nucleus (PVN; panels E,F), ventromedial nucleus of the hypothalamus (VMH; panels G, H), arcuate nucleus (ARC; panels G, H), and posterior periventricular nucleus of the hypothalamus (PVp; Panels K, L). Letter V in the images indicates the third ventricle of the brain. Scale bar is located in the lower left of panel A and is applied for the rest of panels in this figure.
3.3. Snord116 expression according to qRT-PCR
To confirm the regional expression of Snord116, its relative levels were quantified by quantitative reverse-transcription PCR (qRT-PCR). The primers designed against Snord116 were validated using PCR, and a single amplicon was detected of the expected size in brain samples, but barely detectable in liver samples as predicted (Cavaillé et al., 2000) (Figure 5A).
Figure 5.
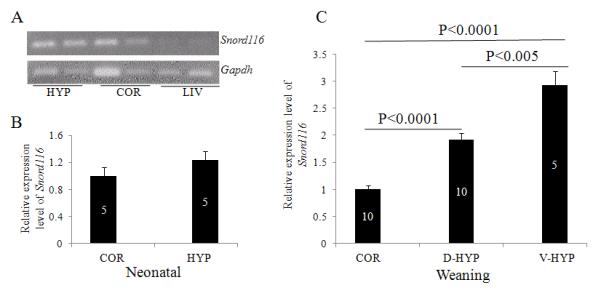
Relative levels of Snord116 determined by quantitative reverse-transcription PCR (qRT-PCR). Snord116 levels were detectable by RT-PCR in the hypothalamus (HYP; panel A) and cortex (COR; panel A) in the brain but not in the liver (LIV; panel A). qRT-PCR revealed the relative level of Snord116 in the HYP and COR in neonatal pups (panel B). Relative levels of Snord116 were determined at weaning age in the COR, dorsal hypothalami (D-HYP) including the paraventricular nucleus, and ventral hypothalami (V-HYP) including the arcuate nucleus (panel C). The mean value of Snord116 relative levels in the COR was normalized to 1. The values of other groups are the fold differences of the mean value in the COR. The number of animals per group is given within each of the bars, which represent mean values +/-SEM.
In P0 mice, qRT-PCR data revealed Snord116 relative levels were similar between the cortex (n=5) and hypothalamus (n=5) (Figure 5B). The relative level of Snord116 in the cortex was normalized to 1 and the level of Snord116 in other regions was considered relative to the cortex. In contrast to the P0 mice (Figure 5B), at weaning Snord116 relative levels were different in the cortex, dorsal hypothalamus and ventral hypothalamus (Figure 5C, One-way ANOVA, F (2,22)=46.8, p<0.0001). The Snord116 expression level in the dorsal hypothalamic dissection that included PVN (n=10) was almost double that in the cortex (p<0.0001, n=10); and in the ventral hypothalamic dissection including the ARC (n=5) it was approximately two times greater than in the cortex (n=10; p<0.0001) (Figure 5C). Between the hypothalamic areas, Snord116 level in the ventral hypothalamic dissection that included ARC (n=5) was approximately twice as great as in the dorsal hypothalamic dissection that included PVN (n=10, p<0.005). Regional differences were similar between mice at weaning age and in adulthood. In adults, relative Snord116 level in the hypothalamus was higher than in other brain regions, including the cortex, striatum and brain stem (data not shown).
3.4. Relative level of Snord116 in female compared to male brains
Based on non-radioactive ISH, Snord116 immunoreactivity was greater in females (4 of 6 female and male brains run in pairs; data not shown) compared to males, however, the variability of the ISH signals rendered the results unreliable. Therefore, we compared relative levels of Snord116 between females and males by qRT-PCR. Data obtained from the experiment in figure 5C was broken down by sex. The relative level of Snord116 in male cortex was normalized to 1 and the level of Snord116 in other regions in males and in all regions in females was relative to the male cortex. The results revealed that Snord116 levels were similar between females and males in weaning age mice in the cortex, dorsal hypothalamus and ventral hypothalamus in weaning age of mice (Figure 6).
Figure 6.
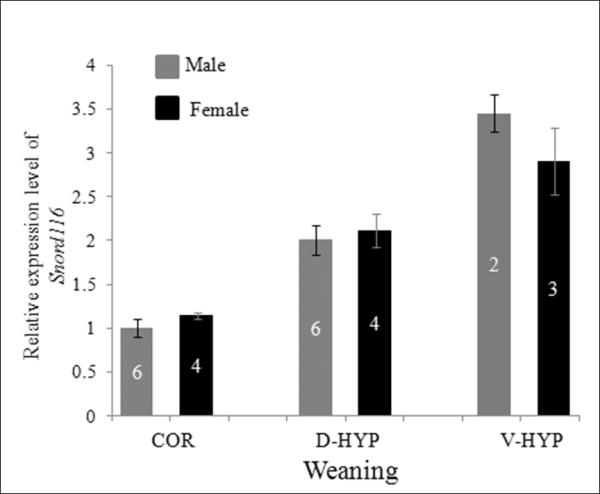
Relative level of Snord116 in female and male mouse brains determined by qRT-PCR. At weaning, the relative level of Snord116 was less in COR of males and females than in dorsal hypothalami (D-HYP) or ventral hypothalami (V-HYP), but similar by sex. The mean value of Snord116 relative levels in the male COR was normalized to 1. The values of other groups are the fold differences of the mean value in the male COR. The number of animals per group is given within each of the bars, which represent mean values +/-SEM.
4. Discussion
The current study examined the neuroanatomical distribution of Snord116, a factor in the pathogenesis of PWS related to overeating and obesity phenotypes (Sahoo et al., 2008; de Smith et al., 2009; Duker et al., 2010; Skryabin et al., 2007; Ding et al., 2008). Expression of Snord116 was examined qualitatively using ISH to determine the localization and quantitatively in dissected tissues using quantitative reverse-transcription PCR. Snord116 expression was prominent in hypothalamic regions that control food intake and regulate energy balance. Furthermore, expression of this gene became progressively more prominent in these hypothalamic areas after birth. The results support the view that Snord116 may be related to appetite disorders and obesity symptoms in PWS patients.
The current study links Snord116 expression to hypothalamic feeding circuitry at a cellular level. Snord116 ISH signals were detected throughout the brain, however, they were prominent in the hypothalamus in both weanling and adult mice. Specifically, Snord116 expression was high in the PVN and VMH, and particularly high in the ARC, all key components of hypothalamic feeding circuitry. The current data is consistent with prior evidence that Snord116 is involved in CNS regulation of food intake and energy balance. First, Snord116 is expressed in the brain (Cavaillé et al., 2000; de los Santos et al. 2000). Second, based on genetic studies in mice and human patients, Snord116 is believed to be a key etiologic factor leading to the PWS and its associated phenotypes related to obesity and hyperphagia (Sahoo et al., 2008; de Smith et al., 2009; Duker et al., 2010; Skryabin et al., 2007; Ding et al., 2008). Within the brain, the hypothalamus is considered one region that may be responsible for the development of PWS (Swaab, 1997). Data in the current study provides direct evidence for the neuroanatomical distribution of Snord116 in hypothalamic nuclei, including PVN, VMH and ARC that are associated with feeding.
In the current study, Snord116 levels were most prominent in the ARC, suggesting its involvement in the function(s) of cells in the ARC. Dysfunction or damage of the hypothalamus has been associated with obesity and hypothalamic hyperphagia (Hochberg et al., 2010). Among the nuclei in the hypothalamus, the ARC is one of the central structures controlling food intake and energy balance. In the ARC, two distinct cell groups related to the regulation of appetite and energy balance are the agouti-related peptide (AgRP) and the pro-opiomelanocortin (POMC)-expressing neurons. AgRP and POMC neurons are the receivers for peripheral hormone signals, such as leptin, insulin and ghrelin. AgRP neurons in the ARC project to the PVN, while POMC neurons project to additional nuclei including the PVN and VMH to further regulate feeding behaviour (Bouret, 2009). Determining that if AgRP or POMC neurons express Snord116 may help determine the role of Snord116 in feeding regulation. Based on the number of AgRP and POMC neurons compared to the number of Snord116 positive cells in the ARC, it is not likely that Snord116 is restricted to one population or the other.
At the cellular level Snord116 is located inside cell nuclei, mostly in nucleoli and adjacent to the nuclear membrane in the older mice. Previous reports show that the accumulation of Snord116 in nucleoli increased from perinatal to adult, correlating with maturation of nucleoli (Leung et al, 2009). During early postnatal development, Snord116 was primarily located outside the nucleoli adjacent to the nuclear membrane. Snord116 belongs to the C/D box family of snoRNA, which guide the 2′-O-methylation of ribosomal RNA (rRNA) by formation of RNA duplexes at target RNA modification site. However, increasing evidence suggests that snoRNAs may direct the chemical modification of or edit other types of RNA, such as snRNA, tRNA and even mRNA (Bachellerie et al., 2002). For example, snoRNA Snord115, another PWS related snoRNA, regulates the alternative splicing and RNA editing of the serotonin 2C receptor (5HT2CR) (Flomen et al., 2004; Vitali et al., 2005; Kishore et al., 2006). Subsequent research showed that lack of Snord115 was related to unusual behavior mediated by 5HT2CR (Doe et al., 2009). Other than chemical modification and RNA editing, snoRNAs also likely play roles in gene regulation, such as gene silencing (Brameier et al., 2010). Collectively, snoRNAs potentially have complex roles and engage in multiple cellular functions. The extra-nucleolar localization of Snord116 seen in the current study indicates the possibility that other than rRNA maturation, Snord116 may have other cellular functions, especially during developmental stages.
Results of the current study show that, in contrast to adulthood, Snord116 expression was not prominent in the hypothalamus at birth. This observation is consistent with a previous report (Lee et al., 2003). In that study Snord116 expression was tested in the developing brains from embryo day 9.5 to 18.5 and Snord116 was expressed at a low level throughout the brain. The current study extends these observations and reveals that the predominance of expression of Snord116 in the hypothalamus occurs after birth, including at weaning and adult ages. The later development of this restricted expression may relate to the observation that over-eating in PWS patients is not present immediate after birth, but rather develops gradually during early childhood (McAllister et al., 2010). In addition, regulation of Snord116 specific expression in the hypothalamus falls within the time window of postnatal programming of the hypothalamic feeding circuitry, which develops during the first few weeks after birth (Caqueret et al., 2005). During that time, ARC neuronal projections to other hypothalamic nuclei are maturing (Bouret et al., 2004; Bouret et al., 2006). Snord116 may contribute to the development of hypothalamic feeding circuitry due to its timing and pattern of expression in development. Leptin is known to regulate food intake and energy expenditure in adults. However, leptin signals also play an important role in guiding postnatal development of hypothalamic feeding circuits (Bouret et al., 2004; Pinto et al., 2004). Without leptin signals, the number of axonal projections from ARC to PVN is reduced (Bouret et al., 2004; Pinto et al., 2004). It remains to be determined whether Snord116 expression affects, or is affected by, leptin signaling during development.
Our results showed prominent expression of Snord116 in hypothalamic nuclei associated with feeding regulation, indicating a linkage of Snord116 neuroanatomical distribution and its involvement in the PWS formation. It will be important to further examine if and how these hypothalamic nuclei are functional molecular targets of Snord116. For feeding circuits, two potential mechanisms exist: 1) Snord116 potentially affects feeding circuitry formation. It would be interesting to examine if axonal projections are defective in feeding circuitry when Snord116 is deleted. 2) Snord116 potentially indicates maturation of feeding circuitry. Thereafter, Snord116 could play roles in feeding control by regulating downstream effectors, possibly through its chemical modification, editing or gene silencing functions (Bachellerie et al., 2002; Flomen et al., 2004; Vitali et al., 2005; Kishore et al., 2006; Brameier et al., 2010). Interestingly, a previous study showed similar gene expression profiling in the hypothalamus between wild type and Snord116 deletion mice in the neonatal period including day 5 and day 13 (Ding et al., 2010). These results suggested Snord116 does not play direct roles in feeding regulation in neonatal period. However, it might be more important to test the gene profiling at a later stage, such as after weaning or in adulthood. In the current study the expression of Snord116 in hypothalamus became restricted at weaning age and into adulthood, while previously the hyperphagic symptoms appeared in Snord116 deletion mice after 3 months of age (Ding et al., 2008). Other than simply looking at different time points, it might also be important to test other changes in mRNA, since Snord116 plays roles in RNA modification and editing that may not be detected by standard gene profiling method.
The hypothalamus is a highly sexually dimorphic brain region that is influenced by steroid hormones in development (Tobet & Fox, 1992; McCarthy, 2008; Tobet et al., 2009). The downstream effectors of steroid hormones include γ-aminobutyric acid (GABA), nitric oxide (NO), and brain-derived neurotrophic factor (BDNF) among many others (Tobet et al., 2009). As snoRNA editing may influence neurotransmitter receptor function there may be interesting sex dependent interactions in conditions where transmitter signaling is altered. For example, disruption of GABAB receptor signaling by deleting the R1 subunit of the GABAB receptor affected the organization of cell populations in the PVN and surrounding area in a sex dependent manner when no sex difference was evident in wild-type mice (McClellan et al., 2010). In the current study, results from ISH experiments comparing female and male hypothalami were suggestive of greater levels of Snord116 in females, but inconclusive. Quantitative analyses revealed that Snord116 levels were similar levels in tissue between males and females. It remains to be determined whether changes in transmitter function (e.g., GABAB receptor disruption; McClellan et al., 2010) might reveal changes in Snord116 function.
Highlights.
The expression of snoRNA Snord116 was examined in perinatal, weaning age and young adult mouse brain by in situ hybridization and qPCR.
Snord116 expression in weaning and adult mice is prominent in the medial hypothalamus, particularly within nuclei that are part of feeding circuitry.
This prominent expression pattern of Snord116 was observed in adult but not in newborn, indicating it was developmentally regulated.
The result provides a link between Snord116 expression and PWS phenotypes at a cellular level.
Acknowledgements
We thank members of the Tobet laboratory for their help and Juliano da Silveira for his help with the qRT-PCR experiment. We thank Drs. Jill Goldstein and Bob Handa for helpful comments throughout the project. This work was supported by ORWH P50-MH082679 (Goldstein, Tobet and Handa).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- •.Bachellerie JP, Cavaillé J, Hüttenhofer A. The expanding snoRNA world. Biochimie. 2002;84(8):775–90. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- •.Bouret SG, Draper SJ, Simerly RB. Trophic action of leptin on hypothalamic neurons that regulate feeding. Science. 2004;304(5667):108–10. doi: 10.1126/science.1095004. [DOI] [PubMed] [Google Scholar]
- •.Bouret SG. Early life origins of obesity: role of hypothalamic programming. J Pediatr Gastroenterol Nutr. 2009;48(Suppl 1):S31–8. doi: 10.1097/MPG.0b013e3181977375. [DOI] [PubMed] [Google Scholar]
- •.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39(2):675–86. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Bürger J, Horn D, Tönnies H, Neitzel H, Reis A. Familial interstitial 570 kbp deletion of the UBE3A gene region causing Angelman syndrome but not Prader-Willi syndrome. Am J Med Genet. 2002;111(3):233–7. doi: 10.1002/ajmg.10498. [DOI] [PubMed] [Google Scholar]
- •.Butler MG, Hanchett JM, Thompson TE. Clinical Findings and Natural History of PWS. In: Butler MG, Lee PDK, Whitman BY, editors. Managment of Prader-Willi Syndrome. Springer; 2006. pp. 3–48. [Google Scholar]
- •.Caqueret A, Yang C, Duplan S, Boucher F, Michaud JL. Looking for trouble: a search for developmental defects of the hypothalamus. Horm Res. 2005;64(5):222–30. doi: 10.1159/000088977. [DOI] [PubMed] [Google Scholar]
- •.Cavaillé J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Hüttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci U S A. 2000;97(26):14311–6. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.de los Santos T, Schweizer J, Rees CA, Francke U. Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader-Willi deletion region, which is highly expressed in brain. Am J Hum Genet. 2000;67(5):1067–82. doi: 10.1086/303106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.de Smith AJ, Purmann C, Walters RG, Ellis RJ, Holder SE, Van Haelst MM, Brady AF, Fairbrother UL, Dattani M, Keogh JM, Henning E, Yeo GS, O’Rahilly S, Froguel P, Farooqi IS, Blakemore AI. A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum Mol Genet. 2009;18(17):3257–65. doi: 10.1093/hmg/ddp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Dellovade TL, Young M, Ross EP, Henderson R, Caron K, Parker K, Tobet 4 SA. Disruption of the gene encoding SF-1 alters the distribution of 5 hypothalamic neuronal phenotypes. J Comp Neurol. 2000;423:579–89. doi: 10.1002/1096-9861(20000807)423:4<579::aid-cne4>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- •.Ding F, Prints Y, Dhar MS, Johnson DK, Garnacho-Montero C, et al. Lack of Pwcr1/MBII-85 snoRNA is critical for neonatal lethality in Prader-Willi syndrome mouse models. Mamm Genome. 2005;16:424–431. doi: 10.1007/s00335-005-2460-2. [DOI] [PubMed] [Google Scholar]
- •.Ding F, Li HH, Zhang S, Solomon NM, Camper SA, Cohen P, Francke U. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PLoS One. 2008;3(3):e1709. doi: 10.1371/journal.pone.0001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Ding F, Li HH, Li J, Myers RM, Francke U. Neonatal maternal deprivation response and developmental changes in gene expression revealed by hypothalamic gene expression profiling in mice. PLoS One. 2010 Feb 24;5(2):e9402. doi: 10.1371/journal.pone.0009402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Doe CM, Relkovic D, Garfield AS, Dalley JW, Theobald DE, Humby T, Wilkinson LS, Isles AR. Loss of the imprinted snoRNA mbii-52 leads to increased 5htr2c pre-RNA editing and altered 5HT2CR-mediated behaviour. Hum Mol Genet. 2009;18(12):2140–8. doi: 10.1093/hmg/ddp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Duker AL, Ballif BC, Bawle EV, Person RE, Mahadevan S, Alliman S, Thompson R, Traylor R, Bejjani BA, Shaffer LG, Rosenfeld JA, Lamb AN, Sahoo T. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur J Hum Genet. 2010;18(11):1196–201. doi: 10.1038/ejhg.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Flomen R, Knight J, Sham P, Kerwin R, Makoff A. Evidence that RNA editing modulates splice site selection in the 5-HT2C receptor gene. Nucleic Acids Res. 2004;32(7):2113–22. doi: 10.1093/nar/gkh536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Gallagher RC, Pils B, Albalwi M, Francke U. Evidence for the role of PWCR1/HBII-85 C/D box small nucleolar RNAs in Prader-Willi syndrome. Am J Hum Genet. 2002;71(3):669–78. doi: 10.1086/342408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Gerard M, Hernandez L, Wevrick R, Stewart CL. Disruption of the mouse necdin gene results in early post-natal lethality. Nat Genet. 1999;23:199–202. doi: 10.1038/13828. [DOI] [PubMed] [Google Scholar]
- •.Gunay-Aygun M, Schwartz S, Heeger S, O’Riordan MA, Cassidy SB. The changing purpose of Prader-Willi syndrome clinical diagnostic criteria and proposed revised criteria. Pediatrics. 2001;108(5):E92. doi: 10.1542/peds.108.5.e92. [DOI] [PubMed] [Google Scholar]
- •.Hochberg I, Hochberg Z. Expanding the definition of hypothalamic obesity. Obes Rev. 2010;11(10):709–21. doi: 10.1111/j.1467-789X.2010.00727.x. [DOI] [PubMed] [Google Scholar]
- •.Holm VA, Cassidy SB, Butler MG, Hanchett JM, Greenswag LR, Whitman BY, Greenberg F. Prader-Willi syndrome: consensus diagnostic criteria. Pediatrics. 1993;91(2):398–402. [PMC free article] [PubMed] [Google Scholar]
- •.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311(5758):230–2. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- •.Kozlov SV, Bogenpohl JW, Howell MP, Wevrick R, Panda S, et al. The imprinted gene Magel2 regulates normal circadian output. Nat Genet. 2007;39:1266–1272. doi: 10.1038/ng2114. [DOI] [PubMed] [Google Scholar]
- •.Lee S, Walker CL, Wevrick R. Prader–Willi syndrome transcripts are expressed in phenotypically significant regions of the developing mouse brain. Gene Expr Patterns. 2003;3(5):599–609. doi: 10.1016/s1567-133x(03)00113-3. [DOI] [PubMed] [Google Scholar]
- •.Leung KN, Vallero RO, DuBose AJ, Resnick JL, LaSalle JM. Imprinting regulates mammalian snoRNA-encoding chromatin decondensation and neuronal nucleolar size. Hum Mol Genet. 2009;18(22):4227–38. doi: 10.1093/hmg/ddp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Li X, Jin P. Roles of small regulatory RNAs in determining neuronal identity. Nat Rev Neurosci. 2010;11(5):329–38. doi: 10.1038/nrn2739. [DOI] [PubMed] [Google Scholar]
- •.Mattick JS. The functional genomics of noncoding RNA. Science. 2005;309(5740):1527–8. doi: 10.1126/science.1117806. [DOI] [PubMed] [Google Scholar]
- •.McAllister CJ, Whittington JE, Holland AJ. Development of the eating behaviour in Prader-Willi Syndrome: advances in our understanding. Int J Obes (Lond) 2010:1–10. doi: 10.1038/ijo.2010.139. [DOI] [PubMed] [Google Scholar]
- •.McClellan KM, Parker KL, Tobet S. Development of the ventromedial nucleus of the hypothalamus. Front Neuroendocrinol. 2006;27(2):193–209. doi: 10.1016/j.yfrne.2006.02.002. [DOI] [PubMed] [Google Scholar]
- •.McClellan KM, Stratton MS, Tobet SA. Roles for gamma-aminobutyric acid in the development of the paraventricular nucleus of the hypothalamus. J Comp Neurol. 2010;518(14):2710–28. doi: 10.1002/cne.22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central nervous system control of food intake and body weight. Nature. 2006;443(7109):289–95. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- •.Nicholls RD. Incriminating gene suspects, Prader-Willi style. Nat Genet. 1999;23:132–134. doi: 10.1038/13758. [DOI] [PubMed] [Google Scholar]
- •.Olney JW. Brain lesions, obesity, and other disturbances in mice treated with monosodium glutamate. Science. 1969;164(880):719–21. doi: 10.1126/science.164.3880.719. [DOI] [PubMed] [Google Scholar]
- •.Pinto S, Roseberry AG, Liu H, Diano S, Shanabrough M, Cai X, Friedman JM, Horvath TL. Rapid rewiring of arcuate nucleus feeding circuits by leptin. Science. 2004;304(5667):110–5. doi: 10.1126/science.1089459. [DOI] [PubMed] [Google Scholar]
- •.Runte M, Varon R, Horn D, Horsthemke B, Buiting K. Exclusion of the C/D box snoRNA gene cluster HBII-52 from a major role in Prader-Willi syndrome. Hum Genet. 2005 Feb;116(3):228–30. doi: 10.1007/s00439-004-1219-2. [DOI] [PubMed] [Google Scholar]
- •.Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40(6):719–21. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- •.Schratt G. Fine-tuning neural gene expression with microRNAs. Curr Opin Neurobiol. 2009;19(2):213–9. doi: 10.1016/j.conb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- •.Schule B, Albalwi M, Northrop E, Francis DI, Rowell M, et al. Molecular breakpoint cloning and gene expression studies of a novel translocation t(4;15)(q27;q11.2) associated with Prader-Willi syndrome. BMC Med Genet. 2005;6:18. doi: 10.1186/1471-2350-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, Qi L, Qian J, Blackshaw S. A genomic atlas of mouse hypothalamic development. Nat Neurosci. 2010;13(6):767–75. doi: 10.1038/nn.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Skryabin BV, Gubar LV, Seeger B, Pfeiffer J, Handel S, Robeck T, Karpova E, Rozhdestvensky TS, Brosius J. Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet. 2007;3(12):e235. doi: 10.1371/journal.pgen.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Swaab DF, Bao AM, Lucassen PJ. The stress system in the human brain in depression and neurodegeneration. Ageing Res Rev. 2005;4(2):141–94. doi: 10.1016/j.arr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- •.Swaab DF. Prader-Willi syndrome and the hypothalamus. Acta Paediatr Suppl. 1997;423:50–4. doi: 10.1111/j.1651-2227.1997.tb18369.x. [DOI] [PubMed] [Google Scholar]
- •.Tobet S, Knoll JG, Hartshorn C, Aurand E, Stratton M, Kumar P, Searcy B, McClellan K. Brain sex differences and hormone influences: a moving experience? J Neuroendocrinol. 2009;21(4):387–92. doi: 10.1111/j.1365-2826.2009.01834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Tsai TF, Armstrong D, Beaudet AL. Necdin-deficient mice do not show lethality or the obesity and infertility of Prader-Willi syndrome. Nat Genet. 1999;22:15–16. doi: 10.1038/8722. [DOI] [PubMed] [Google Scholar]
- •.Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaillé J, Huttenhofer A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol. 2005;169(5):745–53. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Yang T, Adamson TE, Resnick JL, Leff S, Wevrick R, et al. A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet. 1998;19:25–31. doi: 10.1038/ng0598-25. [DOI] [PubMed] [Google Scholar]


