SUMMARY
Nitric oxide (NO·) is an important mediator of innate immunity. The facultative intracellular pathogen Salmonella has evolved mechanisms to detoxify and evade the antimicrobial actions of host-derived NO· produced during infection. Expression of the NO·-detoxifying flavohemoglobin Hmp is controlled by the NO·-sensing transcriptional repressor NsrR and is required for Salmonella virulence. In this study we show that NsrR responds to very low NO· concentrations, suggesting that it plays a primary role in the nitrosative stress response. Additionally, we have defined the NsrR regulon in Salmonella enterica sv. Typhimurium 14028s using transcriptional microarray, qRT-PCR and in silico methods. A novel NsrR-regulated gene designated STM1808 has been identified, along with hmp, hcp-hcr, yeaR-yoaG, ygbA and ytfE. STM1808 and ygbA are important for S. Typhimurium growth during nitrosative stress, and the hcp-hcr locus plays a supportive role in NO· detoxification. ICP-MS analysis of purified STM1808 suggests that it is a zinc metalloprotein, with histidine residues H32 and H82 required for NO· resistance and zinc binding. Moreover, STM1808 and ytfE promote Salmonella growth during systemic infection of mice. Collectively, these findings demonstrate that NsrR-regulated genes in addition to hmp are important for NO· detoxification, nitrosative stress resistance and Salmonella virulence.
INTRODUCTION
Salmonella enterica sv. Typhimurium (S. Typhimurium) is a facultative intracellular pathogen that can invade and replicate within host immune cells, from which it can subsequently disseminate to infect new cells (Mastroeni and Grant, 2011). Innate host resistance is dependent in part on the generation of nitric oxide (NO·) by inducible nitric oxide synthase (iNOS) expressed by host phagocytes (Vazquez-Torres and Fang, 2001). Expression of iNOS is induced upon the recognition of Salmonella lipopolysaccharide by the TLR4 receptor (Vazquez-Torres et al., 2004), resulting in the production of NO·, which exerts direct antimicrobial effects (Fang, 2004). NO· can diffuse across cell membranes to interact with molecular targets within the bacterial cell that include protein metal centers and thiols as well as DNA bases (Fang, 2004). NO·-mediated cytotoxic effects on the bacterial cell are ameliorated by protective responses that detoxify NO· or bypass its antimicrobial actions (Fang, 2004; Spiro, 2006).
Many bacteria, including the enteric pathogens Salmonella enterica and Escherichia coli, harbor enzymes that detoxify NO· by converting it to non-toxic metabolites; these enzymes include flavohemoglobin (Hmp), flavorubredoxin and associated oxidoreductase (NorVW), and cytochrome c nitrite reductase (NrfA) (Crawford and Goldberg, 1998; Hausladen et al., 1998; Mills et al., 2005; Poock et al., 2002; van Wonderen et al., 2008). NorVW and NrfA play important roles in NO· detoxification under anaerobic conditions, in which NO· can be reduced to nitrous oxide (N2O) or ammonia (NH3) (Mills et al., 2008; Poock et al., 2002; van Wonderen et al., 2008). Hmp is particularly important for NO· detoxification under aerobic environments, in which nitrate (NO3−) is produced by the denitrosylase activity of Hmp (Hausladen et al., 2001), although Hmp can also reduce NO· to N2O in the absence of oxygen (Gardner et al., 2002; Kim et al., 1999; Mills et al., 2001; Mills et al., 2008).
In S. Typhimurium the principal regulator of hmp expression during nitrosative stress is the transcriptional repressor NsrR (Bang et al., 2006). Originally identified in Nitrosomonas europaea as a nitrite-sensitive repressor, NsrR is a member of the Rrf2 family of transcription factors (Tucker et al., 2010). Rrf2 family members are found prevalently in microorganisms and consist of small (12–18kDa) proteins that contain a helix-turn-helix DNA binding domain near the N-terminus (Tucker et al., 2010). Some Rrf2 family members, including NsrR, IscR, the regulator of iron-sulfur cluster biogenesis, and RirA, a regulator of iron metabolism in Rhizobium leguminosarum, incorporate iron-sulfur (Fe-S) clusters (Johnston et al., 2007; Schwartz et al., 2001; Tucker et al., 2008). The Fe-S clusters are thought to act as sensors that respond to the presence of iron (RirA) or NO· (NsrR) (Johnston et al., 2007; Tucker et al., 2008; Yukl et al., 2008). In vitro studies of purified NsrR suggest that NO· is sensed directly through the Fe-S cluster of NsrR, as nitrosylation of the cluster abrogates DNA binding by NsrR (Tucker et al., 2008).
In silico analysis has identified NsrR binding sites in various bacterial taxa including γ-and β-proteobacteria, Neisseria, Bacillus and Streptomyces spp. (Rodionov et al., 2005). It has been proposed the genes regulated by NsrR play distinct roles in denitrifying and non-denitrifying organisms such as Neisseria meningitidis and Escherichia coli, respectively (Tucker et al., 2010), controlling NO· production and consumption during denitrification in the former and mediating nitrosative stress resistance in the latter (Tucker et al., 2010). In S. Typhimurium, previous studies have determined that NsrR negatively regulates the expression of the hmp, hcp, ygbA and ytfE genes (Bang et al., 2006; Gilberthorpe et al., 2007). Hcp belongs to the family of hybrid cluster proteins that are found in a wide range of microorganisms including archaea, strict anaerobes and facultatively anaerobic bacteria (Rodionov et al., 2005). Hybrid cluster proteins (HCP) contain two Fe-S clusters, either 4Fe-4S or 2Fe-2S, along with a unique 4Fe-2S-2O cluster that enables four oxidation states (Arendsen, 1998; Cooper et al., 2000). HCPs are differentiated into 3 classes based on their iron-sulfur cluster-binding motifs. (Overeijnder et al., 2009). Purified hybrid cluster proteins from Class I (Desulfovibrio desulfuricans), Class II (Escherichia coli and Rhodobacter capsulatus) and Class III (Pyrococcus furiosus) have been shown to reduce hydroxylamine in vitro (Aragao et al., 2003; Cabello et al., 2004; Overeijnder et al., 2009; Wolfe et al., 2002). YgbA is a small cytoplasmic basic protein (MW 13.5 kDa, pI = 9.74) of unknown function. A Pfam motif search revealed that YgbA contains a motif (pf:AFOR_C) from aldehyde ferredoxin oxidoreductase domains 2 and 3 (Finn et al., 2010). The amino acid sequence of YgbA shows the presence of a CXXCXXXC motif that is also found in ferredoxin oxidoreductases, suggesting that YgbA may bind an Fe-S cluster but lacks the DXXGLC/AX domains critical for molybdopterin ligand binding (Chan et al., 1995; Kletzin et al., 1995). Previous studies in E. coli have shown that YtfE is a di-iron protein important for iron-sulfur cluster assembly (Justino et al., 2006; Vine et al., 2010). In addition, in silico analysis has predicted an NsrR consensus binding site upstream of a tehB homolog, encoding a putative tellurite resistance determinant (Rodionov et al., 2005).
Earlier studies in Salmonella have also shown that Hmp is required for survival in murine macrophages and virulence in mice, demonstrating that NO· detoxification by Hmp plays an important role in Salmonella pathogenesis (Bang et al., 2006; Gilberthorpe et al., 2007). In this study we performed microarray, RT-PCR and in silico analysis to define the NsrR regulon in S. Typhimurium, and examined the roles of NsrR-regulated genes other than Hmp in the Salmonella nitrosative stress response. Our findings include the identification of a novel NsrR-regulated gene (STM1808) that is important for nitrosative stress resistance and virulence in mice. Furthermore we have identified key histidine residues in STM1808 that are important for NO· resistance and metal binding, and provide evidence for a role of the hcp-hcr locus in NO· detoxification.
RESULTS
NsrR senses very low concentrations of NO·
The iron-containing transcriptional regulators NsrR, NorR, SoxR and Fur have been shown to respond to NO· in E. coli (Crack et al., 2012; Fleischhacker and Kiley, 2011; Spiro, 2006; Zheng and Storz, 2000). The transcriptional repressor NsrR is inactivated by exposure to NO·, resulting in the derepression of genes including hmp, encoding a flavohemoglobin responsible for NO· detoxification (Fleischhacker and Kiley, 2011; Tucker et al., 2010). NorR is activated by NO· and positively regulates the expression of genes that include norVW, encoding proteins responsible for NO· reductase activity (Bush et al., 2011; Fleischhacker and Kiley, 2011; Tucker et al., 2006). The ferric uptake regulator Fur is also inactivated by NO·, and the derepression of Fur-regulated is observed in cells subjected to nitrosative stress (D’Autreaux et al., 2002; Spiro, 2006). SoxR, a transcription factor that is responsive to redox-cycling agents, has been shown to be activated by NO·, although the functional importance of SoxR activation by NO· is unclear (Ding and Demple, 2000).
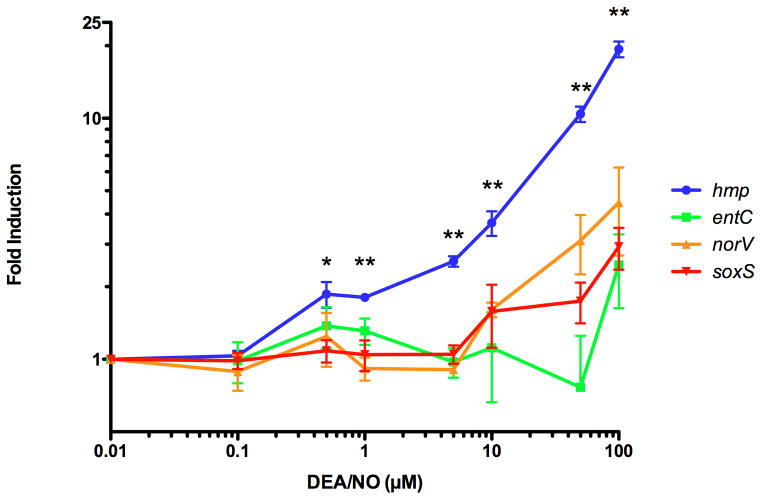
To determine the relative responsiveness of the NsrR, NorR, Fur and SoxR transcriptional regulators, the expression of individual genes that are specifically dependent on each regulator was measured following exposure to NO·. S. Typhimurium was treated with varying concentrations of the NO·-releasing compound diethylamine NONOate (DEA/NO), and hmp, norV, entC and soxS expression was determined by quantitative RT-PCR as a measure of NsrR, NorR, Fur and SoxR expression, respectively (Experimental Procedures). NsrR-dependent expression of hmp was significantly greater at DEA/NO concentrations as low as 1 μM (corresponds to approximately 1.5 μM of released NO·) in comparison to genes regulated by NorR, Fur or SoxR (Fig. 1). Expression of hmp, norV, entC and soxS following treatment with 100 μM DEA/NO was 10.24%, 0.03%, 1.23% and 0.83% of maximal levels of expression (see Experimental Procedures), respectively. These observations indicate that NsrR has a low threshold for sensing NO· in vivo relative to NorR, SoxR and Fur, suggesting that NsrR plays a primary role in responding to nitrosative stress.
Figure 1. Measurement of the NO· responsiveness of iron-containing transcription factors by quantitative RT-PCR.
Quantitative RT-PCR was performed on RNA samples isolated from S. Typhimurium 14028s cultures grown to early log-phase in LB then treated with increasing concentrations of diethylamine NONOate (DEA/NO) for 15 m. (Experimental Procedures). A representative gene was measured as an indicator of activation of the following transcription factors: NsrR (hmp, blue circles), Fur (entC, green squares), NorR (norV, orange up triangles) and SoxR (soxS, red down triangles). P values were calculated using the Wilcoxon Rank Sum Test. *P = 0.05 hmp vs. soxS fold-induction, **P = 0.05 hmp vs. norV, entC or soxS fold-induction.
Microarray analysis of the S. Typhimurium NsrR regulon
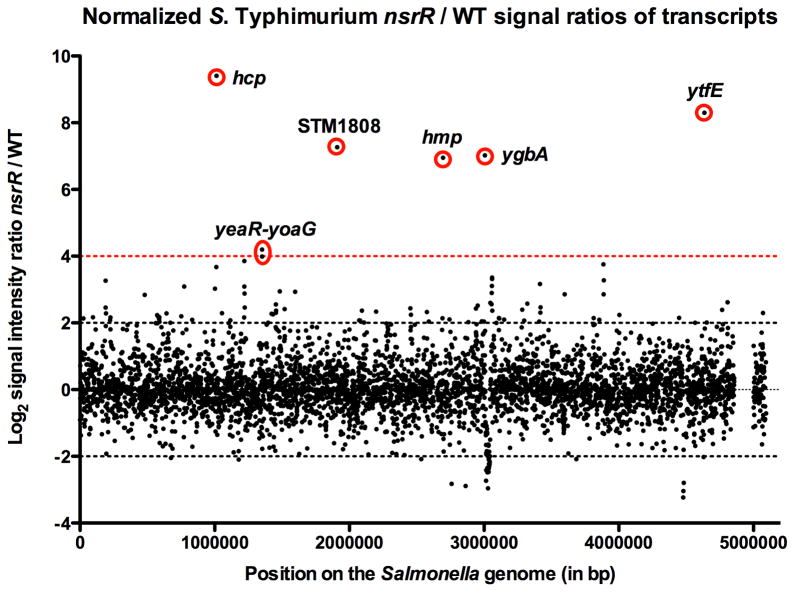
In previous studies, NsrR was identified as the major regulator of hmp transcription in S. Typhimurium (Bang et al., 2006). Comparative genomic studies of NsrR binding sites in γ-proteobacteria have suggested that operons containing hcp, hmp, ytfE, and a homolog of tehB are also regulated by NsrR in S. Typhimurium (Rodionov et al., 2005). Regulation of the ytfE, hcp and ygbA operons by NsrR was validated by RT-PCR (Gilberthorpe et al., 2007). To comprehensively define the NsrR regulon in Salmonella, we performed microarray analysis using cDNA from S. Typhimurium 14028s and an isogenic nsrR mutant. For each microarray analysis, RNA was isolated from three independent cultures of nsrR mutant and wild-type Salmonella grown aerobically in rich medium to mid-log phase (Experimental Procedures). A number of genes exhibited differential expression in nsrR mutant and wild-type cells (Fig. 2 and Supplemental Table 1). The steady state mRNA concentrations of 90 genes comprising 40 operons were increased more than 4-fold in an nsrR mutant compared to wild-type (Supplemental Table 1). Moreover, the operons of hcp-hcr, ytfE, ygbA, hmp, and yeaR-yoaG, previously shown to be regulated by NsrR in Escherichia coli and Salmonella (Bang et al., 2006; Filenko et al., 2007; Gilberthorpe et al., 2007; Lin et al., 2007), were found to be induced 678.1-, 314.5-, 130.2-, 123.7- and 15.9-fold, respectively, by the absence of NsrR (Fig. 2 and Supplemental Table 1). In addition, our microarray analysis revealed that a novel gene designated STM1808 was highly induced (153.7-fold) in an nsrR mutant relative to wild-type (Fig. 2 and Supplemental Table 1). Conversely, expression levels of 26 genes were found to be decreased by more than 4-fold in nsrR mutant Salmonella compared to wild-type (Supplemental Table 1). The majority of these genes were contained within Salmonella pathogenicity islands 1 and 4 (SPI1 and SPI4), important for eukaryotic cell adherence, invasion and intestinal translocation (Altier, 2005; Gerlach et al., 2007; Lostroh and Lee, 2001; Morgan et al., 2007).
Figure 2. Normalized S. Typhimurium nsrR/WT signal ratios of mRNA from microarray analysis of S. Typhimurium 14028s.
The x-axis represents position on the Salmonella genome in base pairs (bp) (pSLT genes at bp 5,000,000), and the y-axis represents the Log2 signal intensity ratio nsrR/wild-type (WT) mRNA from microarray data (Supplemental Table 1). Log2 signal intensity ratio nsrR/wild-type -2 and 2 of nsrR/WT mRNA transcripts are indicated with dotted black lines, whereas the log2 signal intensity ratio of 4 is indicated by a dotted red line. Genes negatively regulated by NsrR are indicated with red circles.
Our microarray analysis also suggested that NsrR might play a role in the positive regulation of Salmonella Pathogenicity Island 1 and 4 (SPI1 and SPI4) genes (See above). Quantitative RT-PCR confirmed that representative genes from SPI1 (invA) and SPI4 (siiC) display reduced expression in the absence of NsrR (Supplemental Fig. S1A). The SPI1 locus was previously shown to be important for invasion of epithelial cells (Altier, 2005; Lostroh and Lee, 2001), and Salmonella strains lacking the SPI1 gene invA are impaired for invasion into HeLa epithelial cells in vitro (Supplemental Fig. S1B). We observed that nsrR mutant S. Typhimurium is comparably defective to an invA mutant strain for HeLa cell invasion, suggesting a possible role for NsrR in the SPI1 regulation (Supplemental Fig. S1B). However, in silico analysis failed to identify an NsrR consensus binding site upstream of SPI1 or SPI4 operons or their known regulators. Therefore, the positive regulatory effect of NsrR on Salmonella pathogenicity islands 1 and 4 appears to be indirect.
In silico determination of the NsrR Consensus Binding Site in S. Typhimurium
To identify the NsrR consensus DNA binding site in Salmonella, upstream DNA sequences of 6 operons that were observed to be increased more than 16-fold in the nsrR mutant compared to wild-type (Fig. 2 and Supplemental Table 1) were analyzed bioinformatically using the motif-based sequence analysis tool MEME (Bailey and Elkan, 1994). A 27-bp consensus DNA binding site was identified upstream of the hcp-hcr, yeaR-yoaG, STM1808, hmp, ygbA, and ytfE operons (Table 1). The putative Salmonella NsrR consensus DNA binding site is similar to the 19-bp γ-proteobacteria NsrR consensus site determined in comparative genomic studies by Rodionov et al. (Rodionov et al., 2005) and also contains the 11-bp NsrR half-site binding motif (AANATGCATTT) identified by ChIP-on-chip (chromatin immunoprecipitation on microarray) analysis in E. coli (Partridge et al., 2009). The MEME generated NsrR consensus binding site from Salmonella (Table 1) was compared to DNA sequences upstream of open reading frames in the entire S. Typhimurium LT2 genome, as well as to DNA sequences upstream in the operons induced 4–16 fold as well as operons with 4-fold reduced expression in an nsrR mutant compared to wild-type (Supplemental Table 1), using the motif-based sequence analysis tool MAST (Bailey and Gribskov, 1998). In addition to hcp-hcr, yeaR-yoaG, STM1808, hmp, ygbA, and ytfE, the MAST analysis identified putative NsrR consensus binding sites upstream of the dsdXA, tehAB and STM1267 operons (Table 1).
Table 1.
MEME analysis of NsrR binding sites.
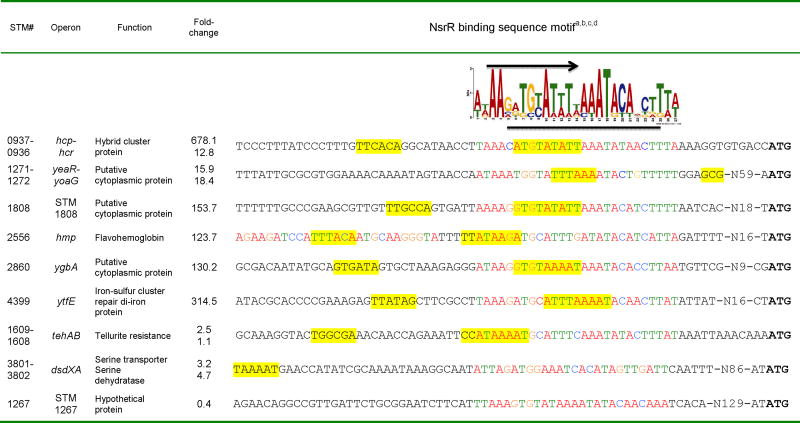
|
Logo of NsrR binding site (in color) was determined by analysis tool MEME (Bailey and Elkan, 1994).
Underline indicates the 19bp γ-proteobacteria NsrR consensus site determined by comparative genomic studies (Rodionov et al., 2005).
Arrow indicates the 11bp half site motif (AANATGCATTT) identified by Chip-on-chip analysis in E. coli (Partridge et al., 2009).
Promoter prediction using BPROM (Softberry) in yellow.
The putative Salmonella 27-bp NsrR consensus binding site spans the predicted promoter regions of hcp-hcr, yeaR-yoaG, STM1808, hmp, ygbA, ytfE and tehAB, but not those of dsdXA and STM1267 (Table 1). Quantitative RT-PCR analysis confirmed enhanced expression of hcp, yeaR, STM1808, hmp, ygbA and ytfE in a strain lacking NsrR (Supplemental Fig. S2). However, quantitative RT-PCR analysis found no change in STM1267 or dsdXA mRNA transcript levels in an nsrR mutant strain, indicating that these genes are not members of the NsrR regulon (Supplemental Fig. S2). Previous studies indicated that NsrR binds to the promoter region of tehAB (Bodenmiller and Spiro, 2006; Partridge et al., 2009), but NsrR regulation of tehAB was not observed (Bodenmiller and Spiro, 2006; Gilberthorpe et al., 2007). In accordance with these previous studies (Bodenmiller and Spiro, 2006; Gilberthorpe et al., 2007), we found no change in tehAB mRNA levels in our nsrR/WT microarray (Supplemental Table 1) or quantitative RT-PCR (Supplemental Fig. S2) studies.
MEME and MAST analysis of upstream DNA sequences of operons induced 4–16 fold or reduced 4-fold in an nsrR mutant compared to wild-type (Supplemental Table 1) failed to identify a NsrR consensus binding site. This suggests the regulation of these operons by NsrR is indirect. Taken together, the microarray, quantitative RT-PCR and in silico data show that the NsrR regulon in S. Typhimurium is comprised of the hcp-hcr, hmp, ygbA, ytfE and yeaR-yoaG operons, as well as the previously unidentified gene STM1808.
Hmp, STM1808 and YgbA are required for nitrosative stress resistance in S. Typhimurium
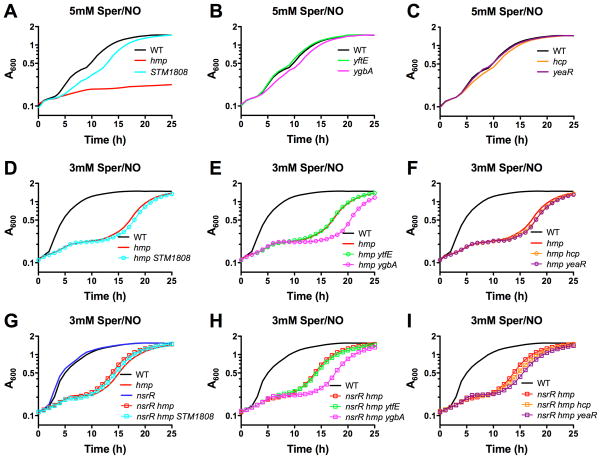
Exposure to nitric oxide (NO·) is sensed by the Fe-S cluster in NsrR (Isabella et al., 2009; Tucker et al., 2008) and leads to the derepression of NsrR-regulated genes (see previous section). Previous studies have elucidated the importance of the Hmp flavohemoglobin in NO· detoxification and redox homeostasis in Salmonella (Bang et al., 2006). To determine the contribution of other NsrR-regulated genes to nitrosative stress resistance, we constructed insertion mutations in hcp, ygbA, ytfE, STM1808 and yeaR (Experimental Procedures). The Salmonella mutant strains were monitored for growth in LB medium following the addition of the NO·-releasing compound Spermine-NONOate (Sper/NO) (Experimental Procedures). As expected, an hmp mutant strain was impaired for growth in the presence of NO· (Fig. 3A). Salmonella lacking STM1808 was also impaired for growth following the addition of NO·, although the growth defect was not as severe as in an hmp mutant strain (Fig. 3A). Mutant strains lacking hcp, ygbA, ytfE or yeaR exhibited little or no growth defect during NO· stress in comparison to wild-type (Figs. 3B and 3C). In addition, double mutant strains lacking both hmp and other NsrR-regulated genes were constructed to determine the contribution to nitrosative stress resistance in the absence of NO· detoxification by Hmp. Since hmp mutants are highly sensitive to NO· (Fig. 3A), the double mutant strains were assayed at a lower concentration of Sper/NO that resulted in only mild growth impairment of an hmp mutant (compare Figs. 3A and 3D). Mutant strains lacking hmp and hcp, ytfE or yeaR displayed no additional growth defect following the addition of Sper/NO (Figs. 3E and 3F). However, in the absence of Hmp, a ygbA mutation exhibited more pronounced growth impairment after Sper/NO treatment (Fig. 3E). When grown in M9 minimal media with glucose as a sole carbon source, the addition of NO· targets multiple sites in the tricarboxylic acid cycle of S. Typhimurium resulting in growth arrest (Richardson et al., 2011). We tested S. Typhimurium strains with single mutations in the NsrR regulon for growth in M9 glucose minimal medium following the addition of Sper/NO (Experimental Procedures). Growth defects were seen in strains lacking hmp, ygbA or hcp, whereas the growth of STM1808, yeaR and ytfE mutants was unaffected by the addition of Sper/NO (Supplemental Figure S3 A-C). Collectively these data suggest that Hmp, STM1808, YgbA and Hcp may contribute to S. Typhimurium resistance to nitrosative stress, depending upon the nutritional environment.
Figure 3. The NsrR-regulated STM1808 and ygbA genes are important for S. Typhimurium growth under nitrosative stress.
Overnight cultures of S. Typhimurium 14028s were inoculated to a cell density of 4 × 107 cfu/ml followed by addition of the NO· donor Sper/NO, and growth monitored for 25 h at 37°C. Strains carrying mutations in individual genes belonging to the NsrR regulon were compared to wild-type (WT, black) for growth in LB + 5mM Sper/NO. A. hmp (red) and STM1808 (light blue). B. ytfE (green) and ygbA (light purple). C. hcp (orange) and yeaR (dark purple). Double mutant strains containing mutations in hmp and additional NsrR-regulated genes were compared to WT (black) and hmp (red) for growth in LB + 3mM Sper/NO. D. hmp STM1808 (light blue circles). E. hmp ytfE (green circles) and hmp ygbA (light purple circles). F. hmp hcp (orange circles) and hmp yeaR (dark purple circles). Triple mutant strains containing mutations in nsrR, hmp and additional NsrR-regulated genes were compared to WT (black), hmp (red) and nsrR hmp (red squares) for growth in LB + 3mM Sper/NO. G. nsrR hmp STM1808 (light blue squares). H. nsrR hmp ytfE (green squares) and nsrR hmp ygbA (light purple squares). I. nsrR hmp hcp (orange squares) and nsrR hmp yeaR (dark purple squares).
STM1808 may be a zinc metalloprotein in which His32 and His82 are important for NO· resistance and zinc binding
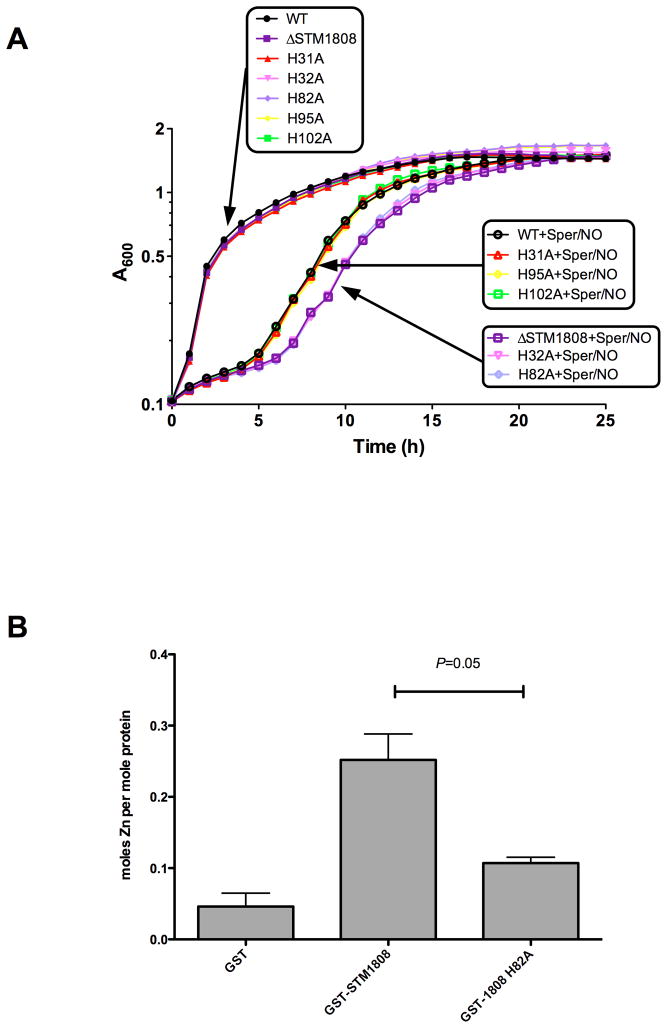
A conserved domain database search (Marchler-Bauer et al., 2011) of STM1808 revealed that the protein contains a domain of unknown function (DUF1971) commonly found in bacterial tellurite resistance proteins. In addition, Clustal W alignments (Thompson et al., 1994) and secondary structure prediction analysis using Jpred3 (Cole et al., 2008) of proteins containing DUF1971 family domains showed that His32 and His82 of STM1808 are conserved within the DUF1971 family. To determine whether individual histidine residues of STM1808 are important for NO· resistance, the histidines (H) residues at positions H31, H32, H82, H95, and H102 were individually mutagenized to alanines (A) using lambda-RED genetic engineering (Experimental Procedures). S. Typhimurium STM1808 histidine mutants were monitored for growth in the presence of Sper/NO. STM1808 H31A, H95A and H102A mutants were unaffected by Sper/NO treatment in comparison to wild-type (Fig. 4A), whereas, STM1808 H32A and H82A mutants were inhibited for growth following Sper/NO treatment to an extent similar to that of an STM1808 deletion mutant (Fig. 4A). Western blot was performed to show that the NO· sensitivity of S. Typhimurium expressing STM1808 H32A or H82A was not attributable to protein instability (data not shown). Collectively, these observations indicate that residues H32 and H82 are required for the STM1808-mediated NO· resistance.
Figure 4. Specific histidine residues are important for NO· resistance and metal binding in STM1808.
A. S. Typhimurium cultures were grown in LB with (open symbols) or without (closed symbols) the addition of 5mM Sper/NO and growth monitored by measuring OD600 for 24h. Wild-type (black circles), ΔSTM1808 (purples squares), STM1808-H31A (red up triangles), STM1808-H32A (light purple down triangles), STM1808-H82A (light blue diamonds), STM1808-H95A (yellow diamonds) and STM1808-H102A (green squares). B. Metal determination of purified GST, GST-STM1808 and GST-STM1808-H82A by ICP-MS. Values represent three independent protein isolations. P-value was calculated using the Wilcoxon Rank-Sum Test.
Structural alignment of STM1808 with the Vibrio fisheri TehB protein (3DL3-E.PDB) using Cn3D (Wang et al., 2000) suggests that H32 and H82 may form a pocket that coordinates a metal. To determine whether STM1808 is a metalloprotein, GST-fusion proteins of STM1808 and an STM1808-H82A mutant were purified and the metal content of the wild-type and mutant proteins determined by ICP-MS analysis (Experimental Procedures). Metals screened included Fe, Zn, Cu, Co, Ni, W, Mn, Mg, Mo and Se. Zinc was the only metal found to associate with GST-STM1808 25.2%+3.64% (Fig. 4B). Zinc co-purification was not attributable to the GST fusion, as only 4.6%+1.88% zinc was present in a GST-only protein sample (Fig. 4B). An H82A mutation reduced STM1808 zinc-binding by 60% (Fig. 4B), suggesting that this histidine residue participates in metal coordination.
Hcp-Hcr mediates NO· detoxification
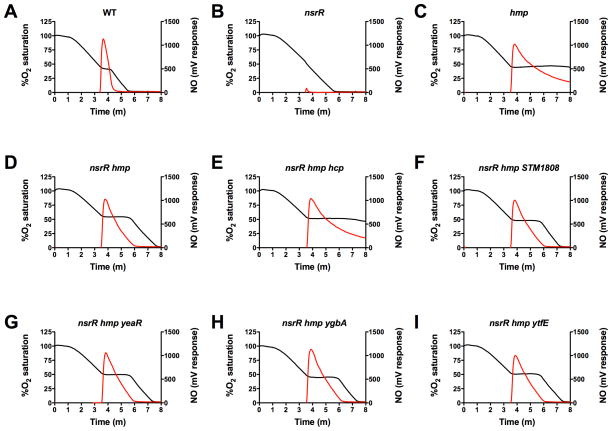
Previous studies have shown the Hmp flavohemoglobin to be the primary mediator of NO· detoxification under aerobic conditions (Gardner et al., 2002; Mills et al., 2008). Cells lacking Hmp are sensitive to growth inhibition by NO· (Fig. 3A). In the absence of Hmp, YgbA can be seen to contribute to S. Typhimurium resistance to growth inhibition following treatment with the NO· donors GSNO (Gilberthorpe et al., 2007) and Sper/NO (Figs. 3D-I). The respiratory chain has long been recognized as an important molecular target of NO·, due to reversible inhibition of the heme-containing cytochrome oxidases bo’ and bd (Stevanin et al., 2000; Yu et al., 1997). Previous studies have suggested that NsrR-regulated genes in addition to Hmp may help to defend aerobic respiration from inhibition by NO· (Gilberthorpe et al., 2007). To further investigate this possibility, hcp, STM1808, yeaR, ygbA and ytfE mutations were introduced into an nsrR hmp mutant background and the strains compared for their ability to respire following bolus NO· treatment (see Experimental Procedures). In brief, bacterial cells were grown to mid-log phase, harvested, washed and resuspended in PBS. Respiration was initiated with the addition of glucose. After 50% of the saturated oxygen was consumed, Proline NONOate (ProliNO), a rapid-releasing NO· donor, was added. Under these assay conditions, NO· reversibly inhibits S. Typhimurium respiration in wild-type cells for approximately 1 min, during which the NO· is detoxified, with subsequent resumption of oxygen consumption (Fig. 5A). In an nsrR mutant strain, expression of hmp is enhanced (Supplemental Fig. S2) and added NO· rapidly consumed with little or no effect on cellular respiration (Fig. 5B). In cells lacking Hmp, sustained inhibition of aerobic respiration is observed following the addition of NO· (Fig. 5C). As previously reported, mutant strains lacking both nsrR and hmp ultimately recover from NO· inhibition of respiration (Gilberthorpe et al., 2007), but the resumption of respiration and consumption of NO· are substantially delayed relative to wild-type (Fig. 5D). The addition of STM1808, yeaR, ygbA or ytfE mutations to an nsrR hmp strain had little effect on oxygen consumption following the addition of NO· (compare Figs. 5F,G, H and I with Figure 5D). However, an nsrR hmp hcp mutant strain displayed a respiration profile similar to that of an hmp mutant alone, which exhibited impaired NO· consumption and failed to resume respiration following NO· challenge (Compare Fig. 5C and Fig. 5E). Since hcp is co-regulated in an operon with hcr, we subsequently examined the contribution of hcr to the recovery of respiration following inhibition by NO·. An hcr and a Δhcp-hcr mutation were constructed in an nsrR hmp strain background (Experimental Procedures). These mutant strains displayed a respiration profile similar to that of an nsrR hmp hcp mutant following treatment with NO· (Supplemental Figs. S4 E and F), suggesting that both hcp and hcr are required for S. Typhimurium resistance to NO·-mediated inhibition of respiration. Resistance to NO·-mediated inhibition of respiration in nsrR hmp hcp, nsrR hmp hcr and nsrR hmp Δhcp-hcr mutants was completely restored in trans by a plasmid expressing both hcp and hcr (Supplemental Figs. S4J, K and L). Although partial restoration of resistance to NO·-mediated inhibition of respiration could be shown by expression of hcp alone (Supplemental Figs. S4G, H and I), expression of both hcp and hcr together was required for wild-type levels of NO· detoxification (Compare Supplemental Figs. S4J, K and L with S4A). Together, these observations indicate a novel role for Hcp-Hcr in NO· detoxification.
Figure 5. Hcp is important for NO· detoxification and resistance to NO·-mediated inhibition of respiration.
Concentrations of O2 (black line) and NO· (red line) were measured using O2 and NO· probes as described in Experimental Procedures. Respiration was stimulated in S. Typhimurium cells by the addition of 0.1% glucose at T = 1 m. After 50% of the saturated oxygen was consumed, ProliNO, a rapidly-releasing NO· donor, was added to a concentration of 5 mM, and O2 and NO· concentrations monitored. A. Wild-type (WT). B. nsrR. C. hmp. D. nsrR hmp. E. nsrR hmp hcp. F. nsrR hmp STM1808. G. nsrR hmp yeaR. H. nsrR hmp ygbA. I. nsrR hmp ytfE.
Hmp, STM1808 and YtfE contribute to S. Typhimurium virulence in mice
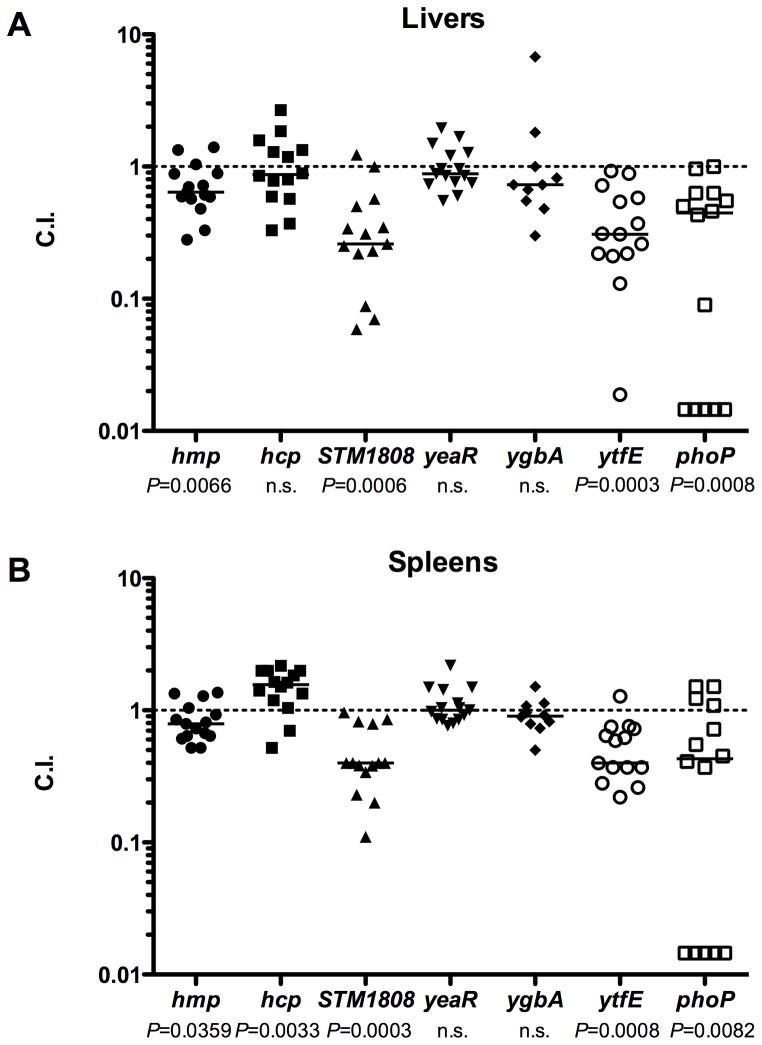
Previous studies have demonstrated that Hmp promotes S. Typhimurium survival within NO·-producing human and murine macrophages as well as in mice (Bang et al., 2006; Gilberthorpe et al., 2007; McCollister et al., 2007; Stevanin et al., 2002). Unexpectedly, S. Typhimurium SL1344 strains carrying mutations in hcp, hcr or ytfE were reported to exhibit greater survival than wild-type after oral challenge of C57Bl/6J mice (Kim et al., 2003). However, C57Bl/6J mice lack a functional Nramp1/Slc11a1 (ity/lsh/bcg) locus, which influences susceptibility to intracellular pathogens and host NO· production (Bradley, 1977; Gros et al., 1981; Plant and Glynn, 1976). We therefore determined whether mutations in individual genes of the NsrR regulon confer a competitive disadvantage compared to wild-type during co-infection of C3H/HeN mice that harbor a functional Nramp1/Slc11a1 locus. C3H/HeN mice were inoculated intraperitoneally (i.p.) with a 1:1 ratio of wild-type 14028s and mutant S. Typhimurium strains. Five days post-infection, livers and spleens were harvested and the competitive index of survival between wild-type and mutant strains determined (Experimental Procedures). Our studies found that an hcp mutant out-competes wild-type S. Typhimurium for survival in the spleens of C3H/HeN mice (Fig. 6B). In contrast, a ytfE mutant was attenuated for survival in the livers and spleens of C3H/HeN mice (Fig. 6A and 6B). In addition to ytfE, strains lacking either hmp or STM1808 were attenuated for survival in the livers and spleens of C3H/HeN mice in comparison to wild-type (Figs. 6A and 6B). These data confirm previous findings with regard to hmp, and additionally demonstrate that the NsrR-regulated ytfE and STM1808 genes contribute to Salmonella virulence.
Figure 6. Hmp, STM1808 and YtfE are required for Salmonella virulence in mice.
C3H/HeN mice (Nramp1+) were inoculated i.p. with a 1:1 ratio of wild-type and mutant S. Typhimurium strains: hmp (circles), hcp (squares), STM1808 (triangles), yeaR (inverted triangles), ygbA (diamonds), ytfE (open circles) and phoP (open squares). Competitive index (C.I.) for survival in mice was determined five days post-infection (Experimental Procedures) in livers (A) and spleens (B). Medians are indicated by horizontal bars. P-values were calculated using the Wilcoxon Rank Sum Test, n.s. denotes not significant.
DISCUSSION
The transcriptional repressor NsrR contains an NO·-sensitive Fe-S cluster and plays a central role in coordinately regulating the response of Salmonella enterica sv. Typhimurium to nitrosative stress (Bang et al., 2006; Bodenmiller and Spiro, 2006; Gilberthorpe et al., 2007; Gilberthorpe and Poole, 2008; Rodionov et al., 2005; Tucker et al., 2008; Tucker et al., 2010). The NsrR-regulated hmp gene encodes a flavohemoglobin that is capable of detoxifying NO· under both aerobic and anaerobic conditions and is required for S. Typhimurium virulence (Bang et al., 2006; Crawford and Goldberg, 1998; Stevanin et al., 2002). However, the contributions of other components of the NsrR regulon to nitrosative stress resistance and pathogenesis have not been demonstrated.
In the present study, we have shown that NsrR is able to respond to very low NO· concentrations in vivo. The exquisite NO· sensitivity of NsrR relative to other NO·-responsive iron-containing transcriptional regulators is consistent with a primary role of NsrR in coordinately regulating the nitrosative stress response in Salmonella. Further, we have defined the NsrR regulon in S. Typhimurium 14028s using microarray, qRT-PCR and in silico methods. We have identified a novel NsrR-regulated gene designated STM1808, and demonstrated a role for specific NsrR-regulated genes in promoting growth during nitrosative stress in vitro (hmp, STM1808, ygbA and hcp) and during systemic infection of mice in vivo (hmp, STM1808, ytfE). ICP-MS measurements suggest that STM1808 binds zinc. Specific histidine residues important for NO· resistance, and H82 has also been implicated in zinc binding. In addition, we have obtained evidence to support a role of the hcp-hcr locus in NO· detoxification during aerobic respiration.
STM1808 expression is strongly NsrR-dependent (Figs. 2 and Supplemental Fig. S2), and an STM1808 mutant exhibits impaired growth in the presence of NO· (Fig. 3A) and reduced virulence in a competitive infection assay (Fig. 6). STM1808 and YeaR are homologs of the Haemophilus influenzae TehB protein, which has been shown to be important for tellurite and oxidative stress resistance, and virulence in rats (Whitby et al., 2010). H. influenzae TehB is bipartite in structure with an N-terminal domain of unknown function (DUF1971) and a conserved C-terminal AdoMet_MTase domain that functions as an S-adenosyl-L-methionine (SAM)-dependent methyltransferase. However, the S. Typhimurium STM1808 and YeaR proteins do not have homology to TehB in regions important for SAM binding and tellurite resistance (Liu et al., 2000). Furthermore, sequence alignments with H. influenzae TehB reveal that the S. Typhimurium and E. coli TehB proteins are truncated. This suggests that S. Typhimurium STM1808 and YeaR are related only to the N-terminal domain of H. influenzae TehB protein containing the domain DUF1971, and do not possess SAM-dependent methyltransferase activity. Interestingly, Salmonella tehB is expressed in an operon with tehA downstream of an NsrR binding site but is not transcriptionally regulated by NsrR (Bodenmiller and Spiro, 2006; Gilberthorpe et al., 2007; Partridge et al., 2009); this study), suggesting that both the regulation and function of this protein may have diverged in S. Typhimurium. In the present study, we failed to demonstrate a role for YeaR in nitrosative stress resistance or virulence, but given its close homology to STM1808, further investigation may be warranted.
Comparative genomic studies indicate that STM1808 homologs are found in Escherichia spp, Klebsiella, Citrobacter, Enterobacter, Vibrio and Photobacterium spp. (Rodionov et al., 2005). Clustal W and secondary structure prediction analysis using Jpred3 of STM1808 as well as structural alignments with Cn3D of STM1808 with Vibrio fisheri TehB (3Dl3-E.PDB) show conserved histidine residues that may be important for metal binding and function. We have found that His32 and His82 are necessary for NO· resistance, and His82 also appears to be important for zinc binding in STM1808 (Fig. 4). It is presently unknown how zinc might facilitate STM1808-mediated NO· resistance.
STM1808 and ytfE can be added to hmp as NsrR-regulated loci that contribute to Salmonella virulence (Fig. 6). YtfE is a di-iron protein that is reportedly important for iron-sulfur cluster assembly (Justino et al., 2006). A YtfE homolog in Ralstonia eutropha H16 has previously shown to bind to NO· (Strube et al., 2007), and another homolog is required for Haemophilus influenzae nitrosative stress resistance and survival in macrophages (Harrington et al., 2009). However, a Salmonella ytfE mutant survived as well as wild-type during NO· stress (Fig. 3B and Supplemental Fig. S3B) yet was defective for virulence in mice (Fig. 6). Our studies, in concert with previous observations, suggest that the pathogenic role of YtfE may be dependent on the host Nramp1 (Slc11a1) locus. We found that ytfE mutant S. Typhimurium 14028s is attenuated for virulence in C3H/HeN (Nramp1+) mice (Fig. 6). Nramp1 encodes a divalent metal transporter with pleiotropic effects on innate immunity, including the enhanced production of reactive oxygen and nitrogen species (Cellier et al., 2007; Forbes and Gros, 2001). The failure of a ytfE mutant to successfully compete with wild-type S. Typhimurium in Nramp1+ mice suggests that YtfE may be of particular importance in a cation-limited environment or during severe oxidative or nitrosative stress.
Although STM1808 is important for nitrosative stress resistance in vitro (Figs. 3A and 4A), its role in promoting Salmonella virulence is unknown. STM1808, along with other members of the NsrR regulon, is induced in macrophages, particularly during later time points (Eriksson et al., 2003; Faucher et al., 2006). NsrR-regulated genes may be particularly important for Salmonella survival in macrophages and mice during later stages of infection, when NO· expression is increased (Mastroeni et al., 2000; Vazquez-Torres and Fang, 2001; Vazquez-Torres et al., 2000).
In the absence of the major NO·-detoxifying actions of the Hmp flavohemoglobin, YgbA was found to be necessary for S. Typhimurium growth during nitrosative stress (Fig. 3E and Supplemental Fig. S3B). In E. coli, expression of ygbA is repressed by the oxygen sensitive regulator Fnr (Constantinidou et al., 2006), which is consistent with a role of YgbA under aerobic or nitrosative stress conditions (Cruz-Ramos et al., 2002).
NO· inhibits respiration by inactivating heme-containing respiratory chain enzymes (Stevanin et al., 2000; Yu et al., 1997). Hmp plays a vital role in protecting the respiratory chain from NO· inhibition (Stevanin et al., 2000). Our observations show that in the absence of Hmp, mutations in hcp or hcr prolong the half-life of NO· and the recovery of S. Typhimurium respiration following NO· treatment, suggesting that both Hcp and Hcr are required for NO· detoxification under aerobic conditions (Fig. 5 and Results). Resistance to NO·-mediated inhibition of respiration in hcp and hcr mutant strains can be restored in trans with a plasmid carrying hcp-hcr. This suggests that Hcp and Hcr comprise an Hmp-independent auxiliary mechanism of NO· detoxification under aerobic conditions. Previous studies demonstrated that hcp is induced during anaerobic growth in nitrite and nitrate (Kim et al., 2003; van den Berg et al., 2000). It has been hypothesized that Hcp may play a role in the detoxification of hydroxylamine formed during nitrite respiration or by non-enzymatic conversion of nitrogen oxides (Kuznetsova et al., 2004; Rodionov et al., 2005; Rudolf et al., 2002; Wolfe et al., 2002). However, some authors have noted that Hcp exhibits only modest hydroxylamine reductase activity in vitro, suggesting that the physiological substrate or function has yet to be identified (Aragao et al., 2003; Overeijnder et al., 2009). Structural studies show that Hcp may function to sequester oxygen and reactive nitrogen species (Aragao et al., 2003). Class I and Class II hybrid cluster proteins have been shown to reduce hydrogen peroxide, but only with low activity and in the presence of ascorbate (Almeida et al., 2006). Additional studies are required to determine whether Hcp binds and reduces NO· in vitro. Hcr is expressed in a small operon with Hcp but only in facultatively anaerobic bacteria including Enterobacteriaceae, Vibrionaceae and Shewanella spp. (Rodionov et al., 2005). Previous studies showed Hcr catalyzes the reduction of Hcp with NADH in vitro (van den Berg et al., 2000), which may imply that Hcp and Hcr function in concert in vivo. The present study has found Hcp and Hcr to have a role in NO· detoxification that serves as an additional defense against nitrosative stress during aerobic respiration. Expression of a variety of NO· detoxifying enzymes allows S. Typhimurium to neutralize NO· within a range of redox environments encountered within the host.
The influence of NsrR on SPI-1 and SPI-4 gene expression is an unexpected observation that appears to be functionally significant, as nsrR mutant S. Typhimurium exhibits reduced invasiveness of epithelial cells (Supplemental Figure S1). NO· congeners have previously been shown to reduce SPI-2 gene expression by direct nitrosylation of SsrB (Husain et al., 2010). Our observations suggest another instance in which host-derived NO· may modulate Salmonella virulence gene expression.
Several iron-containing transcriptional regulators, including NsrR, NorR, Fur, and SoxR, have been previously shown to respond to NO· (Crack et al., 2012; Fleischhacker and Kiley, 2011; Spiro, 2006; Zheng and Storz, 2000). NO· interactions with NsrR and SoxR are coordinated through iron-sulfur clusters (FeS), whereas NorR and Fur interact directly with Fe2+. Previous studies have shown that NsrR can contain either [4Fe-4S] as in Bacillus subtilis or [2Fe-2S] cluster found in Streptomyces coelicolor and Neisseria gonorrhoeae (Tucker et al., 2010). Damage to the Fe-S cluster inactivates NsrR-mediated repression to result in the expression of NsrR-repressed genes (Tucker et al., 2008). The type of Fe-S cluster in S. Typhimurium NsrR has not yet been determined, but the ability of NsrR to respond to very low NO· concentrations in comparison to NorR, SoxR or Fur (Fig. 1), suggests that the NsrR Fe-S cluster in NsrR is configured to optimize NO· responsiveness.
This report has expanded recognition of the contributions of the NsrR regulon to NO· detoxification, nitrosative stress resistance and bacterial virulence beyond the role of the Hmp flavohemoglobin. It will now be of considerable interest to explore the molecular mechanisms by which STM1808 helps Salmonella to resist the cytotoxic actions of NO· and by which Hcp-Hcr promotes NO· consumption.
EXPERIMENTAL PROCEDURES
Media, growth condition and chemicals
Bacteria were grown in Luria-Bertani (LB) medium at 37°C with shaking at 250 rpm unless otherwise stated. Media were supplemented with ampicillin (100 μg ml−1), kanamycin (50 μg ml−1) or chloramphenicol (20 μg ml−1) as indicated. Spermine-NONOate (Sper/NO) was purchased from Calbiochem (SanDiego, CA, USA), Proline NONOate (ProliNO) and diethylamine NONOate (DEA/NO) from A.G. Scientific Inc. (San Diego, CA, USA). The half-life of NO donors used in this study are as follows: Sper/NO (t1/2 = 39 m), ProliNO (t1/2 = 1.8 s) and DEA/NO (t1/2 = 2 m). All other chemicals used were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Bacterial strains
The bacterial strains and plasmids used in this study are listed in Supplemental Table S2. Primers used in this study are listed in Supplemental Table S3. All experiments were conducted using Salmonella enterica serovar Typhimurium ATCC 14028s or an isogenic derivative. Mutant alleles were constructed by λ-Red recombination as described (Datsenko and Wanner, 2000). After construction, all mutant alleles were transduced to isogenic wild-type 14028s background using phage P22 and mutations confirmed by PCR analysis.
Plasmid constructions
Construction of pJK693 (pRB3-Phcp-hcp-hcr) and pJK694 (pRB3-Phcp-hcp) for complementation studies were made as follows. Primer sets JKP408/JKP397 and JKP408/JKP395 were used in PCR reactions with wild-type 14028 genomic DNA template to amplify the promoter and coding regions of hcp-hcr and the promoter and coding region of hcp, respectively. PCR products were digested with BamHI-HindIII and ligated into the stable low-copy cloning vector pRB3 (Berggren et al., 1995).
Construction of pJK678 (pGEX-2T-STM1808) and pJK681 (pGEX-2T-STM1808-H82A) for isolation of GST-fusion proteins were performed as follows. Primer sets JKP341/JKP342 were used in PCR reactions with genomic DNA template isolated from either wild-type 14028s (for pJK678) or from VT86 (for pJK681). PCR products were digested with BamHI and EcoRI and ligated into pGEX-2T digested with BamHI and EcoRI.
Diethylamine NONOate (DEA/NO) treatment of S. Typhimurium
S. Typhimurium 14028s was grown overnight in LB medium at 37°C. Overnight cultures were diluted 1:100 and grown to log phase (OD600nm = 0.6). Cultures were treated for 15 m with various concentrations of diethylamine NONOate (A.G. Scientific, San Diego, CA). RNA was stabilized with RNA Protect reagent (Qiagen, Valencia, CA) and purified using the Qiagen RNeasy Mini kit. Quantitative real time reverse transcription-PCR reaction was performed as described (Experimental Procedures). Primer sets used were designed to monitor transcript levels of hmp (NsrR regulon), norV (NorR regulon), entC (Fur regulon) and soxS (SoxR regulon); rpoD was used as an internal control. Three experiments were performed per sample, with each experiment in technical triplicates. The percent (%) maximum gene expression was calculated as the ratio of fold-change in expression after DEA/NO exposure to fold-change during maximal expression of each gene under the following conditions: hmp expression in an nsrR mutant strain, peak norV expression after 3.2 mM DEA/NO treatment under aerobic conditions, entC expression in a fur mutant strain, and soxS expression after treatment with 3.2 mM paraquat.
cDNA microarray analysis
For microarray analysis, wild-type and nsrR mutant S. Typhimurium were grown aerobically to early log-phase (OD600~0.5) at 37°C in LB medium. Total RNA was isolated from three independent cultures of each strain. Each 12 ml culture was mixed with 24 ml of RNAprotect reagent (Qiagen, Valencia, CA, USA) and RNA immediately purified using the Qiagen RNeasy midi kit (Qiagen). Fifty μg of total RNA were used as a template for cDNA synthesis. A mixture of Cy3-labeled WT cDNA and Cy5-labeled nsrR mutant cDNA, and another mixture of oppositely labeled cDNAs, were separately hybridized onto slides of a Salmonella whole-ORF PCR-product microarray constructed as previously described (Porwollik et al., 2003). Array scanning and quantification were performed essentially as described previously (Navarre et al., 2005). Transcriptional profiles are provided in Supplemental Table 1.
Quantitative reverse transcription PCR (qRT-PCR)
Three independent bacterial cultures were grown to mid-log phase and total RNA isolated using the RNeasy mini kit (Qiagen). cDNA was synthesized from 500ng total RNA using a QuantiTech RT kit (Qiagen). Primers for qRT-PCR were designed using Primer3 (Rozen and Skaletsky, 2000) and are listed in Supplemental Table 3. qRT-PCR assays on cDNA were performed using the QuantiFast SYBR Green kit (Qiagen) with a Rotogene 3000 real time thermal cycler (Corbett Research, Qiagen, Valencia, CA, USA). The rpoD gene target was used as an internal control.
Sper/NO sensitivity assays
Growth kinetics of cells grown in the presence of the nitric oxide donor spermine NONOate (Sper/NO, Calbiochem) were performed as described previously (Karlinsey et al., 2010; Richardson et al., 2011).
NO dependent inhibition of respiration
NO· concentration was measured using an ISO-NOP probe with an ISO-NO Mark II meter (WPI Inc., Sarasota, FL, USA). O2 concentration was measured using an MI-730 probe with an O2-ADPT oxygen adapter (Microelectrodes, Inc. Bedford, NH, USA). Data were acquired using LabChart (ADInstruments, Colorado Springs, CO, USA). Cells were grown in LB medium at 37°C to OD600 ~1.0. The cells were washed 1× in PBS, resuspended in O2-saturated PBS warmed to 37°C, and transferred into a beaker containing the two probes fitted with a rubber stopper. Respiration was stimulated by the addition of 0.1% glucose; 5mM ProliNO (A.G. Scientific, Inc.) was added after 50% of the O2 was consumed.
Purification of GST, GST-STM1808 and GST-STM1808-H82 for Metal Determination
Strains JK953, JK954 and JK962 were grown in 1 liter LB supplemented with ampicillin (100 μg μl−) at 37oC to OD600 = 1.0. IPTG was added to 1 mM and incubated for 30 min. Cells were centrifuged and cell pellets lysed with 35ml P-BER reagent (Thermo Scientific). The lysate was sonicated briefly and clarified by centrifugation at 14,000 RCF for 1 hr. 100 ml of MTPBS (16 mM Na2HPO4/4 mM NaH2PO4/150 mM NaCl, pH = 7.3) + 1% Triton X-100 was added to the clarified lysate and run over a 3 ml glutathione column (Thermo Scientific). The column was washed and proteins eluted in MTPBS+ 3 mg ml−1 reduced glutathione. The purified GST-proteins were dialyzed against 25 mM Tris-HCl pH7.5/150 mM NaCl, then concentrated using Amicon Ultra 3,000 MWCO centrifugation filters. ICP-MS analysis for metal determination was performed by the Environmental Health Laboratory and Trace Organics Analysis Center, Department of Environmental and Occupational Health Sciences, University of Washington. Three independent GST-protein purifications of each sample and filtrate controls were resuspended in 10% trace metal grade nitric acid (Fisher Chemical) and submitted for ICP-MS analysis. Screening was performed for the presence of the following metals: Fe, Zn, Cu, Co, Ni, W, Mn, Mg, Mo and Se.
Competitive infections
Competitive infections were performed as described (Richardson et al., 2011), except C3H/HeN mice were acquired from Charles River Laboratories.
Epithelial cell invasion assays
Invasion of Salmonella strains into HeLa cells were performed as described (Karlinsey et al., 2010).
Accession numbers
The GEO database reference number for microarrays reported in the paper is GSE32585.
Supplementary Material
Acknowledgments
This work was supported by research grants (AI39557, AI77629) from the National Institutes of Health. The authors thank Pierre Moënne-Loccoz for informative discussions.
References
- Almeida CC, Romao CV, Lindley PF, Teixeira M, Saraiva LM. The role of the hybrid cluster protein in oxidative stress defense. J Biol Chem. 2006;281:32445–32450. doi: 10.1074/jbc.M605888200. [DOI] [PubMed] [Google Scholar]
- Altier C. Genetic and environmental control of Salmonella invasion. J Microbiol. 2005;43(Spec No):85–92. [PubMed] [Google Scholar]
- Aragao D, Macedo S, Mitchell EP, Romao CV, Liu MY, Frazao C, Saraiva LM, Xavier AV, LeGall J, van Dongen WM, Hagen WR, Teixeira M, Carrondo MA, Lindley P. Reduced hybrid cluster proteins (HCP) from Desulfovibrio desulfuricans ATCC 27774 and Desulfovibrio vulgaris (Hildenborough): X-ray structures at high resolution using synchrotron radiation. J Biol Inorg Chem. 2003;8:540–548. doi: 10.1007/s00775-003-0443-x. [DOI] [PubMed] [Google Scholar]
- Arendsen AF, Hadden J, Card G, McAlpine AS, Bailey S, Zaitsev V, Duke EHM, Lindley PF, Kröckel M, Trautwein AX, Feiters MC, Charnock JM, Garner CD, Marritt SJ, Thomson AJ, Kooter IM, Johnson MK, van den Berg The “prismane” protein resolved: X-ray structure at 1.7 Å and multiple spectroscopy of two novel 4Fe clusters. JBIC. 1998;3:81–95. [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Bang IS, Liu L, Vazquez-Torres A, Crouch ML, Stamler JS, Fang FC. Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin hmp. J Biol Chem. 2006;281:28039–28047. doi: 10.1074/jbc.M605174200. [DOI] [PubMed] [Google Scholar]
- Berggren RE, Wunderlich A, Ziegler E, Schleicher M, Duke RC, Looney D, Fang FC. HIV gp120-specific cell-mediated immune responses in mice after oral immunization with recombinant Salmonella. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;10:489–495. [PubMed] [Google Scholar]
- Bodenmiller DM, Spiro S. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J Bacteriol. 2006;188:874–881. doi: 10.1128/JB.188.3.874-881.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley DJ. Regulation of Leishmania populations within the host. II. genetic control of acute susceptibility of mice to Leishmania donovani infection. Clin Exp Immunol. 1977;30:130–140. [PMC free article] [PubMed] [Google Scholar]
- Bush M, Ghosh T, Tucker N, Zhang X, Dixon R. Transcriptional regulation by the dedicated nitric oxide sensor, NorR: a route towards NO detoxification. Biochem Soc Trans. 2011;39:289–293. doi: 10.1042/BST0390289. [DOI] [PubMed] [Google Scholar]
- Cabello P, Pino C, Olmo-Mira MF, Castillo F, Roldan MD, Moreno-Vivian C. Hydroxylamine assimilation by Rhodobacter capsulatus E1F1. requirement of the hcp gene (hybrid cluster protein) located in the nitrate assimilation nas gene region for hydroxylamine reduction. J Biol Chem. 2004;279:45485–45494. doi: 10.1074/jbc.M404417200. [DOI] [PubMed] [Google Scholar]
- Cellier MF, Courville P, Campion C. Nramp1 phagocyte intracellular metal withdrawal defense. Microbes Infect. 2007;9:1662–1670. doi: 10.1016/j.micinf.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Chan MK, Mukund S, Kletzin A, Adams MW, Rees DC. Structure of a hyperthermophilic tungstopterin enzyme, aldehyde ferredoxin oxidoreductase. Science. 1995;267:1463–1469. doi: 10.1126/science.7878465. [DOI] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinidou C, Hobman JL, Griffiths L, Patel MD, Penn CW, Cole JA, Overton TW. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J Biol Chem. 2006;281:4802–4815. doi: 10.1074/jbc.M512312200. [DOI] [PubMed] [Google Scholar]
- Cooper SJ, Garner CD, Hagen WR, Lindley PF, Bailey S. Hybrid-cluster protein (HCP) from Desulfovibrio vulgaris (Hildenborough) at 1.6 A resolution. Biochemistry. 2000;39:15044–15054. doi: 10.1021/bi001483m. [DOI] [PubMed] [Google Scholar]
- Crack JC, Green J, Hutchings MI, Thomson AJ, Le Brun NE. Bacterial Iron-Sulfur Regulatory Proteins As Biological Sensor-Switches. Antioxid Redox Signal. 2012 doi: 10.1089/ars.2012.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford MJ, Goldberg DE. Role for the Salmonella flavohemoglobin in protection from nitric oxide. J Biol Chem. 1998;273:12543–12547. doi: 10.1074/jbc.273.20.12543. [DOI] [PubMed] [Google Scholar]
- Cruz-Ramos H, Crack J, Wu G, Hughes MN, Scott C, Thomson AJ, Green J, Poole RK. NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 2002;21:3235–3244. doi: 10.1093/emboj/cdf339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Autreaux B, Touati D, Bersch B, Latour JM, Michaud-Soret I. Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc Natl Acad Sci U S A. 2002;99:16619–16624. doi: 10.1073/pnas.252591299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Demple B. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc Natl Acad Sci U S A. 2000;97:5146–5150. doi: 10.1073/pnas.97.10.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- Fang FC. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- Faucher SP, Porwollik S, Dozois CM, McClelland M, Daigle F. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc Natl Acad Sci U S A. 2006;103:1906–1911. doi: 10.1073/pnas.0509183103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filenko N, Spiro S, Browning DF, Squire D, Overton TW, Cole J, Constantinidou C. The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J Bacteriol. 2007;189:4410–4417. doi: 10.1128/JB.00080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–22. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischhacker AS, Kiley PJ. Iron-containing transcription factors and their roles as sensors. Curr Opin Chem Biol. 2011;15:335–341. doi: 10.1016/j.cbpa.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes JR, Gros P. Divalent-metal transport by NRAMP proteins at the interface of host-pathogen interactions. Trends Microbiol. 2001;9:397–403. doi: 10.1016/s0966-842x(01)02098-4. [DOI] [PubMed] [Google Scholar]
- Galan JE, Curtiss Rr. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner AM, Helmick RA, Gardner PR. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J Biol Chem. 2002;277:8172–8177. doi: 10.1074/jbc.M110471200. [DOI] [PubMed] [Google Scholar]
- Gerlach RG, Jackel D, Stecher B, Wagner C, Lupas A, Hardt WD, Hensel M. Salmonella Pathogenicity Island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell Microbiol. 2007;9:1834–1850. doi: 10.1111/j.1462-5822.2007.00919.x. [DOI] [PubMed] [Google Scholar]
- Gilberthorpe NJ, Lee ME, Stevanin TM, Read RC, Poole RK. NsrR: a key regulator circumventing Salmonella enterica serovar Typhimurium oxidative and nitrosative stress in vitro and in IFN-gamma-stimulated J774.2 macrophages. Microbiology. 2007;153:1756–1771. doi: 10.1099/mic.0.2006/003731-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilberthorpe NJ, Poole RK. Nitric oxide homeostasis in Salmonella typhimurium: roles of respiratory nitrate reductase and flavohemoglobin. J Biol Chem. 2008;283:11146–11154. doi: 10.1074/jbc.M708019200. [DOI] [PubMed] [Google Scholar]
- Gros P, Skamene E, Forget A. Genetic control of natural resistance to Mycobacterium bovis (BCG) in mice. J Immunol. 1981;127:2417–2421. [PubMed] [Google Scholar]
- Harrington JC, Wong SM, Rosadini CV, Garifulin O, Boyartchuk V, Akerley BJ. Resistance of Haemophilus influenzae to reactive nitrogen donors and gamma interferon-stimulated macrophages requires the formate-dependent nitrite reductase regulator-activated ytfE gene. Infect Immun. 2009;77:1945–1958. doi: 10.1128/IAI.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausladen A, Gow A, Stamler JS. Flavohemoglobin denitrosylase catalyzes the reaction of a nitroxyl equivalent with molecular oxygen. Proc Natl Acad Sci U S A. 2001;98:10108–10112. doi: 10.1073/pnas.181199698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausladen A, Gow AJ, Stamler JS. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc Natl Acad Sci U S A. 1998;95:14100–14105. doi: 10.1073/pnas.95.24.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabella VM, Lapek JDJ, Kennedy EM, Clark VL. Functional analysis of NsrR, a nitric oxide-sensing Rrf2 repressor in Neisseria gonorrhoeae. Mol Microbiol. 2009;71:227–239. doi: 10.1111/j.1365-2958.2008.06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AW, Todd JD, Curson AR, Lei S, Nikolaidou-Katsaridou N, Gelfand MS, Rodionov DA. Living without Fur: the subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other alpha-proteobacteria. Biometals. 2007;20:501–511. doi: 10.1007/s10534-007-9085-8. [DOI] [PubMed] [Google Scholar]
- Justino MC, Almeida CC, Goncalves VL, Teixeira M, Saraiva LM. Escherichia coli YtfE is a di-iron protein with an important function in assembly of iron-sulphur clusters. FEMS Microbiol Lett. 2006;257:278–284. doi: 10.1111/j.1574-6968.2006.00179.x. [DOI] [PubMed] [Google Scholar]
- Karlinsey JE, Maguire ME, Becker LA, Crouch ML, Fang FC. The phage shock protein PspA facilitates divalent metal transport and is required for virulence of Salmonella enterica sv. Typhimurium. Mol Microbiol. 2010;78:669–685. doi: 10.1111/j.1365-2958.2010.07357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CC, Monack D, Falkow S. Modulation of virulence by two acidified nitrite-responsive loci of Salmonella enterica serovar Typhimurium. Infect Immun. 2003;71:3196–3205. doi: 10.1128/IAI.71.6.3196-3205.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SO, Orii Y, Lloyd D, Hughes MN, Poole RK. Anoxic function for the Escherichia coli flavohaemoglobin (Hmp): reversible binding of nitric oxide and reduction to nitrous oxide. FEBS Lett. 1999;445:389–394. doi: 10.1016/s0014-5793(99)00157-x. [DOI] [PubMed] [Google Scholar]
- Kletzin A, Mukund S, Kelley-Crouse TL, Chan MK, Rees DC, Adams MW. Molecular characterization of the genes encoding the tungsten-containing aldehyde ferredoxin oxidoreductase from Pyrococcus furiosus and formaldehyde ferredoxin oxidoreductase from Thermococcus litoralis. J Bacteriol. 1995;177:4817–4819. doi: 10.1128/jb.177.16.4817-4819.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova S, Knaff DB, Hirasawa M, Setif P, Mattioli TA. Reactions of spinach nitrite reductase with its substrate, nitrite, and a putative intermediate, hydroxylamine. Biochemistry. 2004;43:10765–10774. doi: 10.1021/bi048826r. [DOI] [PubMed] [Google Scholar]
- Lin HY, Bledsoe PJ, Stewart V. Activation of yeaR-yoaG operon transcription by the nitrate-responsive regulator NarL is independent of oxygen- responsive regulator Fnr in Escherichia coli K-12. J Bacteriol. 2007;189:7539–7548. doi: 10.1128/JB.00953-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Turner RJ, Winstone TL, Saetre A, Dyllick-Brenzinger M, Jickling G, Tari LW, Weiner JH, Taylor DE. Escherichia coli TehB requires S-adenosylmethionine as a cofactor to mediate tellurite resistance. J Bacteriol. 2000;182:6509–6513. doi: 10.1128/jb.182.22.6509-6513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lostroh CP, Lee CA. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 2001;3:1281–1291. doi: 10.1016/s1286-4579(01)01488-5. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–9. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni P, Grant AJ. Spread of Salmonella enterica in the body during systemic infection: unravelling host and pathogen determinants. Expert Rev Mol Med. 2011;13:e12. doi: 10.1017/S1462399411001840. [DOI] [PubMed] [Google Scholar]
- Mastroeni P, Vazquez-Torres A, Fang FC, Xu Y, Khan S, Hormaeche CE, Dougan G. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J Exp Med. 2000;192:237–248. doi: 10.1084/jem.192.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollister BD, Myers JT, Jones-Carson J, Husain M, Bourret TJ, Vazquez-Torres A. N(2)O(3) enhances the nitrosative potential of IFNgamma-primed macrophages in response to Salmonella. Immunobiology. 2007;212:759–769. doi: 10.1016/j.imbio.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SI, Kukral AM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills CE, Sedelnikova S, Soballe B, Hughes MN, Poole RK. Escherichia coli flavohaemoglobin (Hmp) with equistoichiometric FAD and haem contents has a low affinity for dioxygen in the absence or presence of nitric oxide. Biochem J. 2001;353:207–213. doi: 10.1042/0264-6021:3530207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills PC, Richardson DJ, Hinton JC, Spiro S. Detoxification of nitric oxide by the flavorubredoxin of Salmonella enterica serovar Typhimurium. Biochem Soc Trans. 2005;33:198–199. doi: 10.1042/BST0330198. [DOI] [PubMed] [Google Scholar]
- Mills PC, Rowley G, Spiro S, Hinton JC, Richardson DJ. A combination of cytochrome c nitrite reductase (NrfA) and flavorubredoxin (NorV) protects Salmonella enterica serovar Typhimurium against killing by NO in anoxic environments. Microbiology. 2008;154:1218–1228. doi: 10.1099/mic.0.2007/014290-0. [DOI] [PubMed] [Google Scholar]
- Morgan E, Bowen AJ, Carnell SC, Wallis TS, Stevens MP. SiiE is secreted by the Salmonella enterica serovar Typhimurium pathogenicity island 4-encoded secretion system and contributes to intestinal colonization in cattle. Infect Immun. 2007;75:1524–1533. doi: 10.1128/IAI.01438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre WW, Halsey TA, Walthers D, Frye J, McClelland M, Potter JL, Kenney LJ, Gunn JS, Fang FC, Libby SJ. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol Microbiol. 2005;56:492–508. doi: 10.1111/j.1365-2958.2005.04553.x. [DOI] [PubMed] [Google Scholar]
- Overeijnder ML, Hagen WR, Hagedoorn PL. A thermostable hybrid cluster protein from Pyrococcus furiosus: effects of the loss of a three helix bundle subdomain. J Biol Inorg Chem. 2009;14:703–710. doi: 10.1007/s00775-009-0483-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JD, Bodenmiller DM, Humphrys MS, Spiro S. NsrR targets in the Escherichia coli genome: new insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mol Microbiol. 2009;73:680–694. doi: 10.1111/j.1365-2958.2009.06799.x. [DOI] [PubMed] [Google Scholar]
- Plant J, Glynn AA. Genetics of resistance to infection with Salmonella typhimurium in mice. J Infect Dis. 1976;133:72–78. doi: 10.1093/infdis/133.1.72. [DOI] [PubMed] [Google Scholar]
- Poock SR, Leach ER, Moir JW, Cole JA, Richardson DJ. Respiratory detoxification of nitric oxide by the cytochrome c nitrite reductase of Escherichia coli. J Biol Chem. 2002;277:23664–23669. doi: 10.1074/jbc.M200731200. [DOI] [PubMed] [Google Scholar]
- Porwollik S, Frye J, Florea LD, Blackmer F, McClelland M. A non-redundant microarray of genes for two related bacteria. Nucleic Acids Res. 2003;31:1869–1876. doi: 10.1093/nar/gkg298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AR, Payne EC, Younger N, Karlinsey JE, Thomas VC, Becker LA, Navarre WW, Castor ME, Libby SJ, Fang FC. Multiple Targets of Nitric Oxide in the Tricarboxylic Acid Cycle of Salmonella enterica Serovar Typhimurium. Cell Host Microbe. 2011;10:33–43. doi: 10.1016/j.chom.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Dubchak IL, Arkin AP, Alm EJ, Gelfand MS. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput Biol. 2005;1:e55. doi: 10.1371/journal.pcbi.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Rudolf M, Einsle O, Neese F, Kroneck PM. Pentahaem cytochrome c nitrite reductase: reaction with hydroxylamine, a potential reaction intermediate and substrate. Biochem Soc Trans. 2002;30:649–653. doi: 10.1042/bst0300649. [DOI] [PubMed] [Google Scholar]
- Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, Beinert H, Kiley PJ. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc Natl Acad Sci U S A. 2001;98:14895–14900. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro S. Nitric oxide-sensing mechanisms in Escherichia coli. Biochem Soc Trans. 2006;34:200–202. doi: 10.1042/BST0340200. [DOI] [PubMed] [Google Scholar]
- Stevanin TM, Ioannidis N, Mills CE, Kim SO, Hughes MN, Poole RK. Flavohemoglobin Hmp affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo’ or bd, from nitric oxide. J Biol Chem. 2000;275:35868–35875. doi: 10.1074/jbc.M002471200. [DOI] [PubMed] [Google Scholar]
- Stevanin TM, Poole RK, Demoncheaux EA, Read RC. Flavohemoglobin Hmp protects Salmonella enterica serovar typhimurium from nitric oxide-related killing by human macrophages. Infect Immun. 2002;70:4399–4405. doi: 10.1128/IAI.70.8.4399-4405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strube K, de Vries S, Cramm R. Formation of a dinitrosyl iron complex by NorA, a nitric oxide-binding di-iron protein from Ralstonia eutropha H16. J Biol Chem. 2007;282:20292–20300. doi: 10.1074/jbc.M702003200. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker NP, D’autreaux B, Spiro S, Dixon R. Mechanism of transcriptional regulation by the Escherichia coli nitric oxide sensor NorR. Biochem Soc Trans. 2006;34:191–194. doi: 10.1042/BST0340191. [DOI] [PubMed] [Google Scholar]
- Tucker NP, Hicks MG, Clarke TA, Crack JC, Chandra G, Le Brun NE, Dixon R, Hutchings MI. The transcriptional repressor protein NsrR senses nitric oxide directly via a [2Fe-2S] cluster. PLoS One. 2008;3:e3623. doi: 10.1371/journal.pone.0003623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker NP, Le Brun NE, Dixon R, Hutchings MI. There’s NO stopping NsrR, a global regulator of the bacterial NO stress response. Trends Microbiol. 2010;18:149–156. doi: 10.1016/j.tim.2009.12.009. [DOI] [PubMed] [Google Scholar]
- van den Berg WA, Hagen WR, van Dongen WM. The hybrid-cluster protein (‘prismane protein’) from Escherichia coli. Characterization of the hybrid-cluster protein, redox properties of the [2Fe-2S] and [4Fe-2S-2O] clusters and identification of an associated NADH oxidoreductase containing FAD and [2Fe-2S] Eur J Biochem. 2000;267:666–676. doi: 10.1046/j.1432-1327.2000.01032.x. [DOI] [PubMed] [Google Scholar]
- van Wonderen JH, Burlat B, Richardson DJ, Cheesman MR, Butt JN. The nitric oxide reductase activity of cytochrome c nitrite reductase from Escherichia coli. J Biol Chem. 2008;283:9587–9594. doi: 10.1074/jbc.M709090200. [DOI] [PubMed] [Google Scholar]
- Vazquez-Torres A, Fang FC. Oxygen-dependent anti-Salmonella activity of macrophages. Trends Microbiol. 2001;9:29–33. doi: 10.1016/s0966-842x(00)01897-7. [DOI] [PubMed] [Google Scholar]
- Vazquez-Torres A, Jones-Carson J, Mastroeni P, Ischiropoulos H, Fang FC. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J Exp Med. 2000;192:227–236. doi: 10.1084/jem.192.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Torres A, Vallance BA, Bergman MA, Finlay BB, Cookson BT, Jones-Carson J, Fang FC. Toll-like receptor 4 dependence of innate and adaptive immunity to Salmonella: importance of the Kupffer cell network. J Immunol. 2004;172:6202–6208. doi: 10.4049/jimmunol.172.10.6202. [DOI] [PubMed] [Google Scholar]
- Vine CE, Justino MC, Saraiva LM, Cole J. Detection by whole genome microarrays of a spontaneous 126-gene deletion during construction of a ytfE mutant: confirmation that a ytfE mutation results in loss of repair of iron-sulfur centres in proteins damaged by oxidative or nitrosative stress. J Microbiol Methods. 2010;81:77–79. doi: 10.1016/j.mimet.2010.01.023. [DOI] [PubMed] [Google Scholar]
- Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem Sci. 2000;25:300–302. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- Whitby PW, Seale TW, Morton DJ, VanWagoner TM, Stull TL. Characterization of the Haemophilus influenzae tehB gene and its role in virulence. Microbiology. 2010;156:1188–1200. doi: 10.1099/mic.0.036400-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe MT, Heo J, Garavelli JS, Ludden PW. Hydroxylamine reductase activity of the hybrid cluster protein from Escherichia coli. J Bacteriol. 2002;184:5898–5902. doi: 10.1128/JB.184.21.5898-5902.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Sato EF, Nagata K, Nishikawa M, Kashiba M, Arakawa T, Kobayashi K, Tamura T, Inoue M. Oxygen-dependent regulation of the respiration and growth of Escherichia coli by nitric oxide. FEBS Lett. 1997;409:161–165. doi: 10.1016/s0014-5793(97)00494-8. [DOI] [PubMed] [Google Scholar]
- Yukl ET, Elbaz MA, Nakano MM, Moenne-Loccoz P. Transcription Factor NsrR from Bacillus subtilis Senses Nitric Oxide with a 4Fe-4S Cluster. Biochemistry. 2008;47:13084–13092. doi: 10.1021/bi801342x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Storz G. Redox sensing by prokaryotic transcription factors. Biochem Pharmacol. 2000;59:1–6. doi: 10.1016/s0006-2952(99)00289-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.