Abstract
Primases are DNA-dependent RNA polymerases found in all cellular organisms. In bacteria, primer synthesis is carried out by DnaG, an essential enzyme that serves as a key component of DNA replication initiation, progression, and restart. How DnaG associates with nucleotide substrates, and how certain naturally-prevalent nucleotide analogs impair DnaG function is unknown. We have examined one of the earliest stages in primer synthesis and its control by solving crystal structures of the S. aureus DnaG catalytic core bound to metal ion cofactors and either individual nucleoside triphosphates or the nucleotidyl alarmones, pppGpp and ppGpp. These structures, together with both biochemical analyses and comparative studies of enzymes that use the same catalytic fold as DnaG, pinpoint the predominant nucleotide-binding site of DnaG, and explain how the induction of the stringent response in bacteria interferes with primer synthesis.
INTRODUCTION
DNA replication is a highly choreographed and tightly regulated event in the lifecycle of all cells (Kornberg, 1992). Carried out by a dynamic, multi-protein complex known as the replisome, the process of replication relies on the coordinated and coupled action of DNA unwinding with strand synthesis. Although many differences exist among bacterial, archaeal, and eukaryotic replisomes, they all utilize a specialized type of DNA-dependent RNA polymerase – termed primase – to synthesize short RNA oligonucleotides (Frick and Richardson, 2001). These RNA primers in turn serve as indispensible starting points for extension by DNA polymerases, which are incapable of initiating strand synthesis de novo (Frick and Richardson, 2001; Kuchta and Stengel, 2010).
In bacteria, a protein known as DnaG is responsible for catalyzing primer synthesis during DNA replication (Kitani et al., 1985; Rowen and Kornberg, 1978). DnaG was discovered through the identification of “fast-stop” mutations in the dnaG gene in early screens for temperature-sensitive replication defects (Carl, 1970). DnaG is now known to play a critical role in numerous replicative processes, including replisome assembly (Makowska-Grzyska and Kaguni, 2010), the control of fork progression (Chintakayala et al., 2009; Lee et al., 2006; Salzberg et al., 1998; Tanner et al., 2008), the regulation Okazaki fragment length (Wu et al., 1992a; Wu et al., 1992b; Zechner et al., 1992), and replication fork restart (Heller and Marians, 2006). Recent work has further shown that DnaG is a target for inhibiting DNA replication under conditions of nutrient-deprivation (Maciag et al., 2010; Wang et al., 2007), a process known as the stringent response. Thus, primer synthesis and DnaG itself are central nodes for regulating replication in bacteria as whole.
Overall, three general strategies to controlling DnaG function have been identified. One relies on the selective recruitment of DnaG to a specific site of action. For example, Escherichia coli DnaG interacts with both the replicative helicase, DnaB (Griep and Lokey, 1996; Marians, 1992; Tougu et al., 1994; Wu et al., 1992a), and the single-stranded DNA binding protein, SSB (Sun and Godson, 1996; Sun et al., 1994), interactions that allow DnaG to interface with distinct replisomal processes (Heller and Marians, 2006; Tougu and Marians, 1996a). A second strategy involves the localized control of primer synthesis. Once bound to DnaB, DnaG can self-associate as a means to both determine the starting position for primer synthesis and control primer length (Bhattacharyya and Griep, 2000; Chintakayala et al., 2009; Corn et al., 2005; Hamdan and van Oijen, 2010; Tougu and Marians, 1996b; van Oijen and Loparo, 2010). The third approach is the use of non-canonical nucleotides, such as the stringent response effectors (p)ppGpp, which directly impede primer formation (Maciag et al., 2010; Swart and Griep, 1995; Wang et al., 2007). At present, how these various protein/protein and protein/ligand interactions influence the catalytic properties of DnaG is not understood.
Although comparative studies to other DNA- and RNA-synthesizing enzymes should in principle be useful for understanding these regulatory phenomena, DnaG is unusual among replicative polymerases in that it bears no evolutionary kinship to either the archaeal/eukaryl primase, or any other polymerase lineage (Frick and Richardson, 2001; Kuchta and Stengel, 2010). Instead, the RNA-Polymerase Domain (RPD) of DnaG (Fig 1a) is related to the metal-binding center of type IA and type II topoisomerases, a region termed the TOPRIM (TOpoisomerase/PRIMase) domain (Aravind et al., 1998; Keck et al., 2000; Podobnik et al., 2000). Although several models have been proposed for how DnaG forms a productive ternary complex with template, NTPs, and/or a newly-formed primer (Corn et al., 2008; Kato et al., 2003; Keck et al., 2000; Podobnik et al., 2000), only a single partial complex – one in which single-stranded DNA is bound (Corn) – currently exists. As a consequence, the mechanisms of substrate recognition and catalysis by DnaG are ill-defined compared to other polymerases.
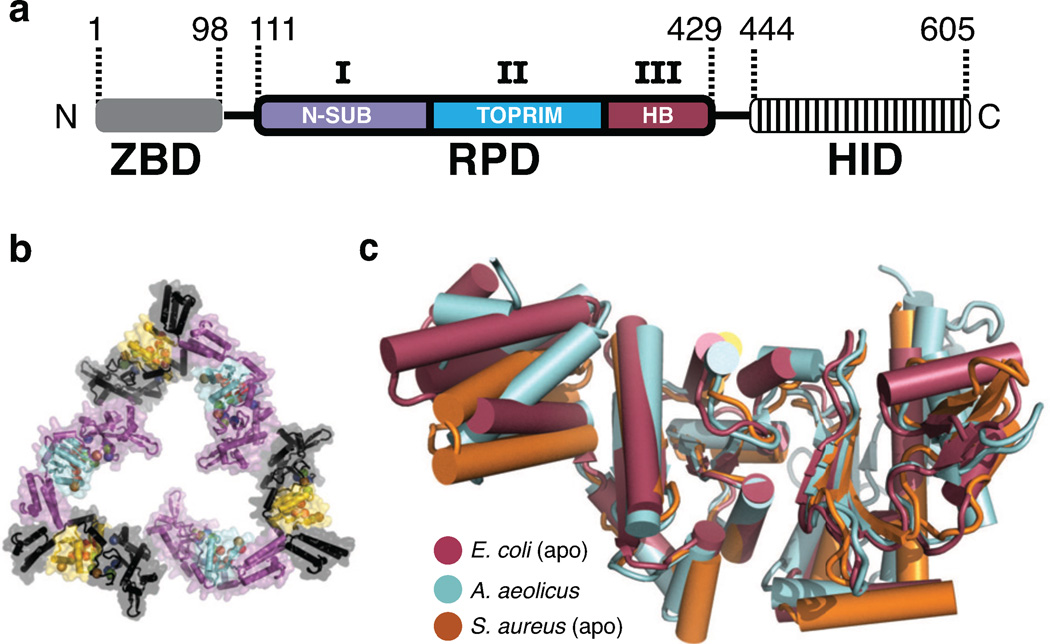
Figure 1. Structure of the S. aureus DnaG RPD.
a) Schematic of DnaG domain structure. ZBD, Zinc Binding Domain. RPD, RNA Polymerase Domain. HID, Helicase Interaction Domain. Subdomains of the RPD: I) N-SUB – N-terminal subdomain, II) TOPRIM fold, and III) HB – Helical Bundle.
b) The S. aureus DnaG RPD active site is solvent accessible. Cartoon and surface representation is shown for the crystal packing arrangement of S. aureus DnaG RPD protomers. Six protomers, related by crystal symmetry, are shown. Three are colored purple with cyan active sites; three are black with yellow active sites. Spheres denote the Cα positions of functionally important residues in the active site.
c) Superposition of several DnaG RPDs solved to date. Structures, corresponding to known apo states, are shown as cartoons and colored as follows: E. coli RPD (PDB ID: 1DDE, maroon), A. aeolicus RPD (PDB ID: 2AU3, cyan), S. aureus RPD (PDB ID: 4E2K, orange).
Barriers to conducting structural investigations of DnaG stem largely from its challenging biochemical properties. In particular, the enzyme has a relatively low affinity for substrates such as RNA•DNA heteroduplexes (~50 µM) (Mitkova et al., 2003), which has precluded the straightforward determination of pertinent co-crystal structures. However, DnaG-type primases are known to productively bind individual nucleotides in the mid-micromolar range (Mendelman et al., 1994; Swart and Griep, 1995), a property that should in principle allow for the use of high NTP concentrations to drive complex formation. Here, we have taken advantage of this property to obtain several crystal structures of the S. aureus DnaG RPD in complex with individual nucleotides and their metal-ion cofactors. These structures, together with biochemical studies and structural comparisons that validate these interactions, reveal an unexpected location and orientation for nucleotide binding within the TOPRIM region of the enzyme. The observed manner of binding in turn both highlights a mechanistic role for a number of conserved amino acids in the DnaG family and reconciles the disparate models of DnaG-substrate engagement present in the literature. Additional structures with the stringent response alarmones pppGpp (G5P) and ppGpp (G4P) further reveal an unanticipated binding modality for these agents that accounts for their mixed-inhibitor type behavior in disrupting primer synthesis.
RESULTS AND DISCUSSION
Structures of S. aureus DnaG RNA Polymerase Domain•NTP complexes
Initial efforts to soak NTP into crystals comprising the RPDs of either E. coli or Aquifex aeolicus DnaG were unsuccessful, likely due to tight packing environments that occluded their respective catalytic centers (Corn et al., 2005; Keck et al., 2000; Podobnik et al., 2000). To circumvent this problem, we surveyed the DnaG RPDs from several bacterial species for a more suitable crystal form. This investigation identified crystallization conditions for a previously uncharacterized RPD from Staphylococcus aureus (SaDnaG), which diffracted to 2 Å resolution and contained an arrangement of protomers with solvent-accessible active sites (Fig 1b). Molecular Replacement (MR) was used to generate initial phases for these data, which allowed for the subsequent building and refinement of the structure (Methods). The final model has an Rwork/Rfree of 18.4%/21.9%, with excellent geometry as judged by MolProbity (Table 1) (Chen et al., 2010). The S. aureus RPD superposes well with other RPDs solved to date, having an average Cα RMSD of ~ 1 Å (Fig 1c); only the relative orientation of a poorly-conserved C-terminal helical bundle differs appreciably among the structures.
Table 1.
Data collection and refinement statistics
| Apo SaDnaG RPD 4E2K* |
ATP Bound SaDnaG RPD 4EDG* |
CTP Bound SaDnaG RPD 4EE1* |
GTP Bound SaDnaG RPD 4EDK* |
UTP Bound SaDnaG RPD 4EDR* |
G5P Bound SaDnaG RPD 4EDV* |
G4P Bound SaDnaG RPD 4EDT* |
|
|---|---|---|---|---|---|---|---|
| Crystal Data | |||||||
| Space Group | P 61 | P 61 | P 61 | P 61 | P 61 | P 61 | P 61 |
| Cell Dimensions (a, b, c (Å), α, β, ° | 151.3,151.3,38.5 90, 90, 120 | 152.3, 152.3,38.7 90, 90, 120 | 152.2,152.2,38.8 90, 90, 120 | 152.2,152.2,38.8 90, 90, 120 | 150.6,150.6,38.6 90, 90, 120 | 152.6,152.6,38.6 90, 90, 120 | 150.2,150.2, 38.6 90, 90, 120 |
| Data Collection | Native | Native | Native | Native | Native | Native | Native |
| Wavelength (Å) | 1.115 | 1.115 | 1.115 | 1.115 | 1.115 | 1.115 | 1.115 |
| Resolution (Å) | 50.00-2.15 | 34.59-2.00 | 30.62-1.99 | 49.6-2.00 | 37.04-2.01 | 38.64-2.01 | 43.4-2.00 |
| Reflections(measure d/unique) | 248,259/28,064 | 187,094/34,499 | 368,265/32,369 | 506,322/35,005 | 183,488/33,847 | 277,309/34,211 | 284,153/35,073 |
| †Rsym (%) (highest shell) | 7.2 (64.7) | 4.6 (28.7) | 11.3 (68.2) | 9.4 (49.5) | 4.7 (17.3) | 8.3 (57.2) | 7.5 (63.0) |
| <I/σI> (highest shell) | 30.5 (3.9) | 22.2 (5.2) | 14.8 (4.1) | 21.8 (5.3) | 10.9 (3.6) | 17.8 (4.1) | 18.7 (3.2) |
| Completeness (%) (highest shell) | 100 (100) | 99.8 (98.8) | 94.7 (100) | 100 (100) | 99.9 (100) | 99.8(99.8) | 99.7(98.1) |
| Redundancy | 8.8 | 5.3 | 11.4 | 10.8 | 5.4 | 8.2 | 8.2 |
| Refinement | |||||||
| ‡Rwork/Rfree (%) | 19.1/21.7 | 16.2/19.7 | 18.2/21.6 | 16.1/20.0 | 17.2/20.5 | 16.7/20.3 | 18.0/22.0 |
| Average B factor (Å2) | |||||||
| protein | 39.4 | 34.8 | 26.9 | 32.3 | 28.8 | 36.4 | 39.4 |
| solvent | 44.8 | 46.8 | 39.6 | 43.9 | 40.9 | 47.6 | 50.3 |
| Rmsd | 0.006 | 0.02 | 0.002 | 0.002 | 0.002 | 0.003 | 0.011 |
| Bond lengths (Å) | 0.77 | 0.715 | 0.637 | 0.681 | 0.735 | 0.705 | 1.269 |
| Bond angle (°) | 319 | 321 | 321 | 321 | 321 | 321 | 321 |
| Protein residues | 240 | 355 | 348 | 360 | 377 | 338 | 299 |
| Water molecules | 0 | 3 | 3 | 3 | 3 | 3 | 2 |
| Ions | 316/3/0 | 317/4/0 | 316/5/0 | 317/4/0 | 319/2/0 | 317/4/0 | 317/4/0 |
| §Ramachandran (favored/generous/disallowed) | |||||||
| Ligand Refinement | N/A | ||||||
| Avg. B Factor (Å2) | 49.9 | 35.4 | 31.0 | 24.5 | 57.6 | 70.1 | |
| Real-space correlation coefficient | 0.887 | 0.933 | 0.957 | 0.954 | 0.948 | 0.917 | |
PDB ID.
Rsym = Σ| I−〈I〉|/ΣI, where I is the integrated intensity for a given reflection.
Rwork = Σ|Fobs − Fc|/ ΣFobs, where Fobs and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree is calculated as for Rwork, but from a subset of the data (5 %) that was withheld from crystallographic refinement.
As calculated by MolProbity (Chen, Arendall et al. 2010).
The SaDnaG RPD crystallographic system facilitated the soaking of divalent metal ions and various NTPs to obtain nucleotide-bound complexes (Methods). Manganese initially was chosen over magnesium for these studies, since primase works equally well with either cofactor (Godson et al., 2000; Rodina and Godson, 2006), and because the spectral properties of manganese enabled the unambiguous determination of its presence from anomalous diffraction data. Crystals treated in this manner diffracted to a similar resolution as apo crystals, indicating that the soaking procedure did not induce crystal damage. Structures were determined for each of the four ribonucleotide substrates using MR, and refined to a similar level of quality as the nucleotide-free model (Methods, Table 1). The resultant maps all showed a single region of strong, contiguous difference density inside the RPD active site that could be best fit by a single nucleoside triphosphate and three Mn2+ ions (Fig 2a). Anomalous difference maps confirmed the binding of manganese.
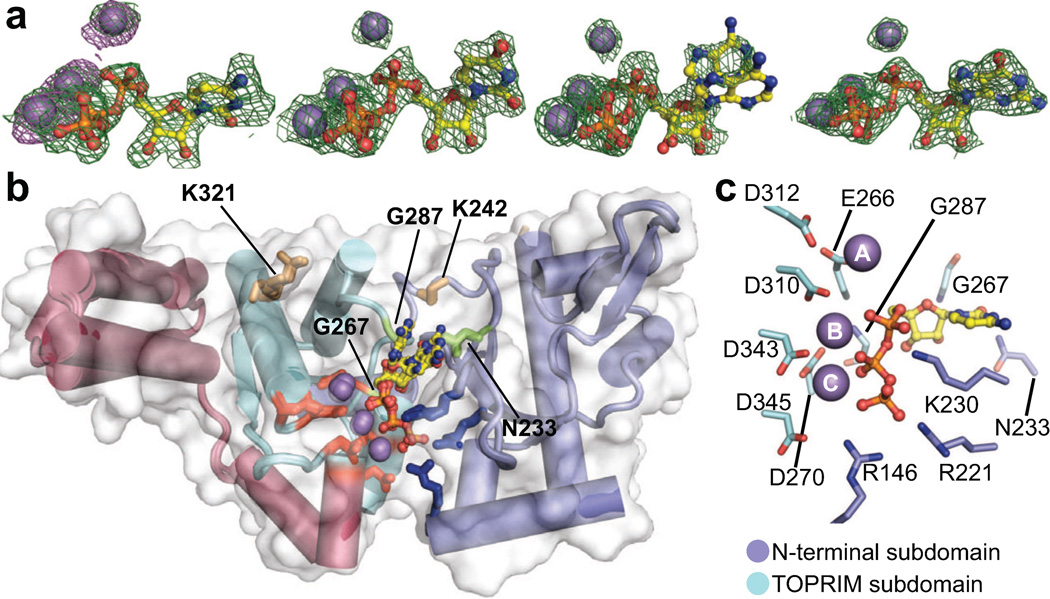
Figure 2. Overview of NTP-bound SaDnaG RPDs.
a) Difference density present in the SaDnaG RPD active site for Mn2+•NTP-soaked crystals. From left to right, density corresponds to CTP, UTP, ATP, and GTP. The resulting difference density (mFo-DFc) prior to modeling of the NTP is shown contoured at 3σ as a green mesh (Methods). Anomalous difference density maps obtained by collecting data on a CTP and Mn2+ soaked crystal at the Mn K-edge are shown contoured at 5σ as purple mesh. Individual NTPs are shown from right to left as ball-and-stick representations. Mn2+ ions are shown as gray spheres.
b) All four metal•NTP complexes bind to a common site on the SaDnaG RPD. A superposition of the apo SaDnaG RPD structure (white surface) and all four NTP bound SaDnaG RPD structures (cartoons colored by subdomain, as per Fig 1a) is shown. Sidechains of metal-binding residues are shown as red sticks, and sidechains of basic ridge amino acids as blue sticks. NTPs and Mn2+ ions are shown and colored as in (a). The sidechains of individual residues investigated here are labeled (SaDnaG numbering), and shown as stick representations.
c) Coordination of metal•NTP complexes. The CTP bound structure is displayed as a representative example. NTP and Mn2+ ions are colored as in panel (a). Sidechains of residues that contact the nucleotide and metal ions are labeled (SaDnaG numbering), shown in stick representation, and colored according subdomain.
See also Figure S2.
Inspection of the four NTP-bound structures revealed that the first two subdomains of the RPD form the nucleotide-binding pocket (Fig 2b). One subdomain consists of the metal-binding center of the TOPRIM fold, which coordinates all three divalent metals using a constellation of six highly-conserved acidic amino acids (Fig 2c/S1/S2a). The other forms an abutting ridge that is rich in invariant, basic amino acids. One of the observable Mn2+ ions (denoted here as metal A) is offset from the other two, and makes no direct contacts with the nucleotide; by contrast, metals B and C closely associate with one side of the nucleotide triphosphate moiety, which is further sandwiched by two arginines (Arg146, Arg221) and a lysine (Lys230) from the basic ridge. Overall, the B-factors of the bound ligands (35 Å2) are close to the average B-factor for the entire protein (27 Å2), indicating that the ligands are well ordered and bound at high occupancy (Table 1). Three of the nucleotides (CTP, GTP and UTP) were seen to bind in a single conformation, whereas ATP adopted two orientations due to a rotation between its ribose and α-phosphate groups. An alignment of the GTP-bound structure with the A. aeolicus or E. coli DnaG RPD structures reveals that the nucleotide occupies a consistently solvent-exposed region in all of the structures (Fig S2b). By varying the substituents of the soaking conditions (Methods), we established that: 1) NTP binding is metal dependent (Fig S2c), 2) nucleotide is required for metal C to bind (Fig S2d), and 3) Mg2+ and Mn2+ are essentially interchangeable (Fig S2e).
The DnaG nucleotide-binding site is preserved with other TOPRIM-dependent enzymes
Having established the non-proteinaceous density associated with the SaDnaG RPD after soaking was a metal•NTP complex, we next asked if the nucleotide-binding configuration was adventitious, or if it reflected a pertinent mode of association consistent with primase function. One clue to resolving this question came from the relationship between DnaG and type IA/II topoisomerases. Both enzyme families employ a TOPRIM fold to promote metal-assisted catalysis of nucleotidyl phospho-transfer reactions – nucleotide addition in the case of DnaG, and reversible DNA strand scission through a catalytic tyrosine in topoisomerases (Aravind et al., 1998) (Fig 3a–b/S3a). The availability of multiple topoisomerase structures, in particular a crystal structure of yeast topoisomerase II (topo II) captured as a metal-associated product complex with DNA (Schmidt et al., 2010), afforded the opportunity to compare our metal-NTP SaDnaG complexes with a homologous, catalytically competent system.
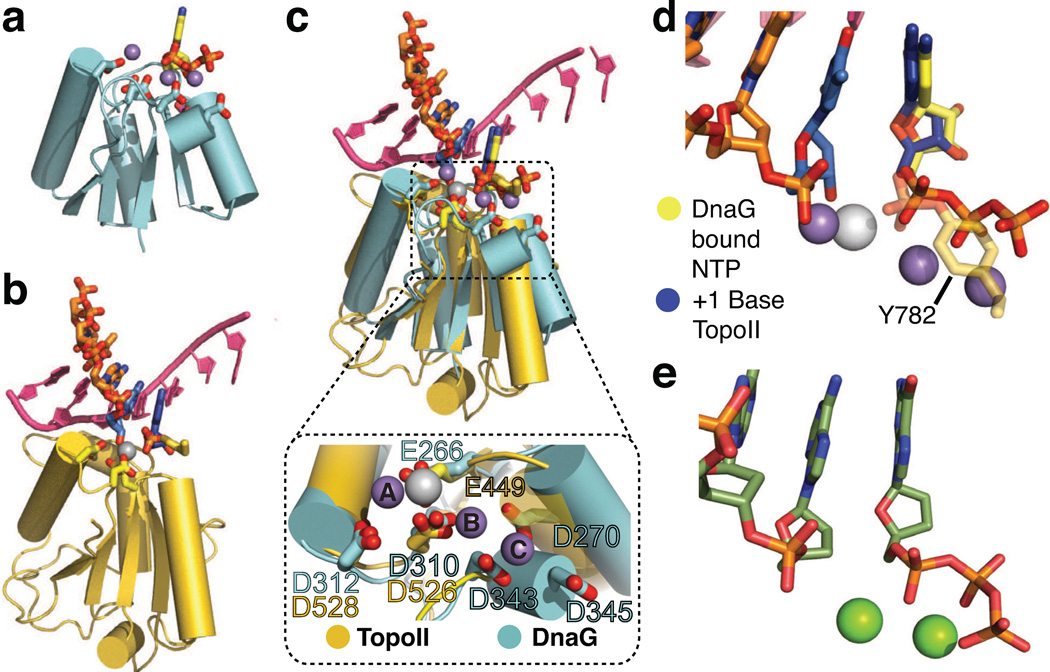
Figure 3. Alignment of nucleotide-bound structures with S. cerevisiae topoisomerase II.
a) Cartoon representation of the SaDnaG RPD TOPRIM fold bound to nucleotide (CTP). Sidechains of the conserved acidic residues in the DnaG TOPRIM fold are shown as sticks with carbon colored cyan and oxygen red. CTP and its associated Mn2+ ions are colored as in Fig 2a.
b) Cartoon representation of the S. cerevisiae topo II TOPRIM fold as found in a DNA-cleavage complex (PDB ID 3L4K). Sidechains of the conserved acidic residues and the covalently attached catalytic tyrosine (Tyr782) from the neighboring protomer are shown as sticks with carbon colored gold and oxygen in red. The scissile strand is shown as orange sticks. The −1 and +1 nucleobases at the cleavage site are colored blue. The complementary strand is shown in bright pink as a cartoon representation. The metal ion bound to the topo II catalytic center (Zn2+, in this particular complex) is shown as a white sphere.
c) Superposition of TOPRIM folds from CTP-bound SaDnaG RPD and the yeast topo II•DNA cleavage complex. The DnaG and topo II TOPRIM folds and sidechains, along with their associated substrates, are depicted as per panels (a) and (b), respectively. Boxed panel: close-up of the aligned metal binding regions (region highlighted by dashed outline in main panel) of both TOPRIM folds, with conserved acidic residues shown as stick representations and colored as per panels (a) and (b). The metal ions bound to topo II and SaDnaG are colored white and gray, respectively.
d) Close-up of substrate configuration based on a TOPRIM-fold alignment between DnaG and yeast topo II. Only the substrates from both structures are shown, and are colored as in panel (c). The covalent linkage between Tyr782 and DNA observed in topo II is shown as a semi-transparent stick representation.
e) Schematic of the active site in the T7 DNA polymerase ternary complex structure (PDB ID: 1T7P, (Doublie et al., 1998)). The incoming nucleotide and primer strand are shown in stick representations with carbon colored dark green. Mg2+ ions are shown as green spheres.
See also Figure S3.
To carry out this analysis, we superposed the catalytic centers of topo II and the SaDnaG RPD. Only conserved Cα positions within the TOPRIM folds of the two enzymes were used in the alignments, so as not to bias the relative relationship between bound substrates. Although significant variation is seen in the position and length of the outer α-helices that comprise both TOPRIM folds, the three internal β-strands and turns bearing the preserved catalytic acidic residues align closely (Fig 3c). The resulting comparison shows that the single catalytic metal of topo II, which engages both the reactive phosphate and the 3´-OH of the cleaved DNA strand, is coordinated by a pair of acidic amino acids (Glu449/Asp526), and that the homologous counterparts to these residues (Glu266/Asp310) ligand metal A in SaDnaG. The NTP bound to the SaDnaG active site also occupies the same location as the +1 nucleobase in topo II, which marks the 5´ portion of the cleaved strand resulting from DNA cleavage by this enzyme (Fig 3d). In the yeast ternary complex, the scissile phosphate associated with this nucleotide is linked covalently to the active-site tyrosine of the enzyme; superposition of the SaDnaG and topo II TOPRIM folds results in a near-exact overlap of this phosphate and the α-phosphate of the bound NTP. Thus, the NTP-binding site of the SaDnaG RPD maps to the same region of the TOPRIM domain that supports phosphoryl transfer in topo II, and further appears to pre-position the metal cofactors and reactive groups of the bound NTP in an analogous manner.
A more global inspection of the superposition between the DnaG and topo II TOPRIM folds highlights additional congruencies that reinforce the similarities between the conformation of the bound nucleotide and the incoming nucleotide seen in polymerase ternary complexes (Fig 3e). For example, the orientation of the NTP in SaDnaG allows for the clash-free stacking of its base against the −1 nucleobase of the DNA bound to topo II (which bears a free 3’-OH in the cleavage complex). This orientation also positions the hydrogen-bonding groups of the NTP base in-line to engage the complementary strand of the topoisomerase-bound duplex through Watson-Crick pairing (Fig 4a). Moreover, the polarity and general position of the non-scissile DNA strand paired with the TOPRIM-bound segment in topo II overlaps with a previously-observed binding site for single-stranded template DNA on the DnaG RPD (Corn et al., 2008). Interestingly, inspection of the DNA modeled from topo II onto DnaG placed the intact DNA strand next to both an invariant lysine in subdomain I of the RPD (Lys242) and a highly-conserved, basic amino acid (Lys321) in the RPD TOPRIM fold (Fig S4a). In E. coli DnaG, the first amino acid has been shown to be important for template binding (Sun et al., 1999), while the latter has been shown to crosslink to the 3′-terminus of thiolated ssDNA oligonucleotides when mutated to cysteine (Corn and Berger, 2007). To determine if Lys321 also plays a role in primer synthesis by DnaG through template binding, we substituted the corresponding amino acid in E. coli DnaG (Arg320) with either alanine or glutamate; both mutations decreased activity in accord with the severity of the mutation (Fig 4b), and further disrupted binding to an RNA•DNA heteroduplex that mimics a preformed primer-template substrate (Fig S4b, Sup. Methods). Thus, the position of DNA binding to topo II, as defined by its interaction with the TOPRIM fold, provides a prospective model for considering how DnaG engages its own nucleic acid substrates.
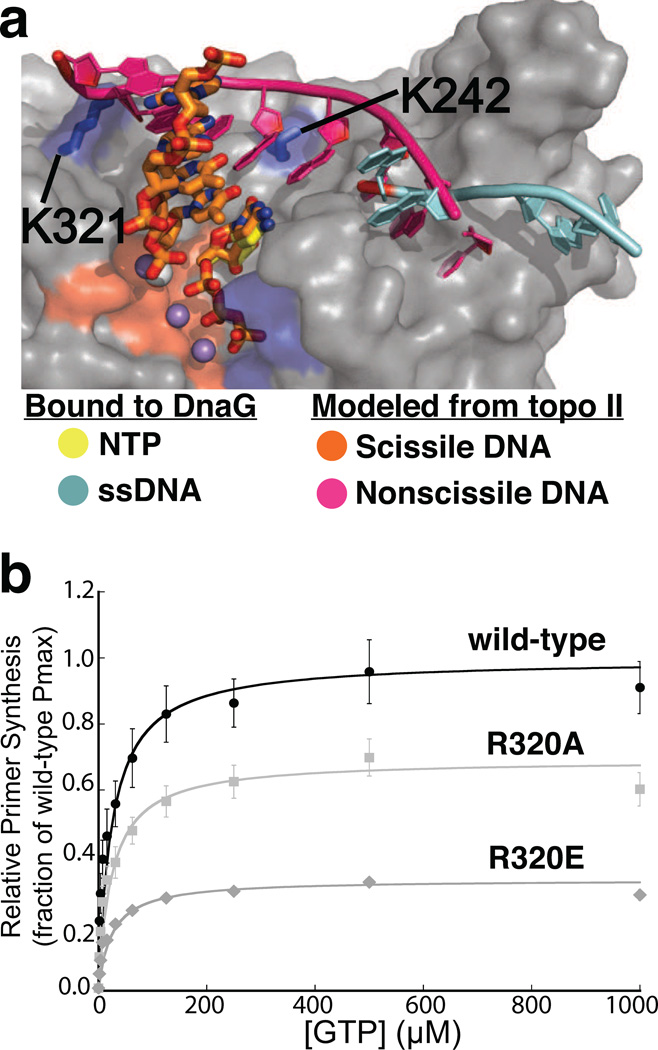
Figure 4. Congruencies between the binding sites for the complementary DNA strand in topo II DNA and SaDnaG.
a) The polarity and terminal position of the complementary DNA strand bound to the topo II TOPRIM fold lines up with the site for ssDNA binding in the DnaG RPD. A superposition of the TORPIM folds between an S. cerevisiae topo II•DNA cleavage complex and an ssDNA-bound state of the E. coli DnaG RPD (PDB ID 3B39) is shown. The scissile strand in topo II is shown as orange sticks, and its complement as a magenta cartoon. ssDNA bound to EcDnaG (cyan cartoon) is thought to mark the site of template binding (Corn and Berger, 2007; Corn et al., 2008). The protein portion of the SaDnaG RPD bound to CTP (gray surface representation) is shown, with the metal binding cluster of DnaG highlighted in red, and the basic ridge in blue. The modeling implicates residues K321 and K242 of SaDnaG (blue sticks with corresponding blue surfaces) as possibly playing a role in binding a primer•template product.
b) Arg320 of EcDnaG (corresponding to Lys321 in SaDnaG) is required for de novo primer synthesis. Helicase-stimulated, GTP-dependent primer synthesis was assayed for the wild-type enzyme (black), Arg320Ala (light gray) and Arg320Glu (gray) mutant enzymes in a fluorometric de novo primer synthesis assay. Curves were fit as described (Methods), yielding the parameters listed in Table S1; error bars represent ±SEM (Standard Error of the Mean).
See also Figure S4.
The nucleotide-binding site is critical for primase activity
Because modeling efforts such as these are by nature correlative, we next turned to mutagenesis studies to examine the manner of NTP binding. The observed NTP-binding site in SaDnaG is formed by multiple highly-conserved residues, many of which have been demonstrated previously to be critical for catalysis (Fig S1/S2a, Table 2). For example, the invariant acidic amino acids that ligand the metal ions involved in nucleotide coordination have been shown to be critical for metal binding and priming activity in both E. coli DnaG and in the related phage protein, T7 gp4 (Godson et al., 2000,; Lee and Richardson, 2005; Rodina and Godson, 2006; Urlacher and Griep, 1995). The positively-charged amino acids that emerge from the basic ridge to contact the nucleotidyl triphosphate moiety likewise have been examined, and found to be important for primase function (Keck et al., 2000; Rodina and Godson, 2006). In particular, substitutions at the amino acid equivalent to Lys229 in T7 gp4 (Lys128) interfere with both template-dependent and template-independent synthesis (Lee and Richardson, 2001), directly implicating this residue in nucleotide binding.
Table 2.
Conserved residues references or investigated in this study
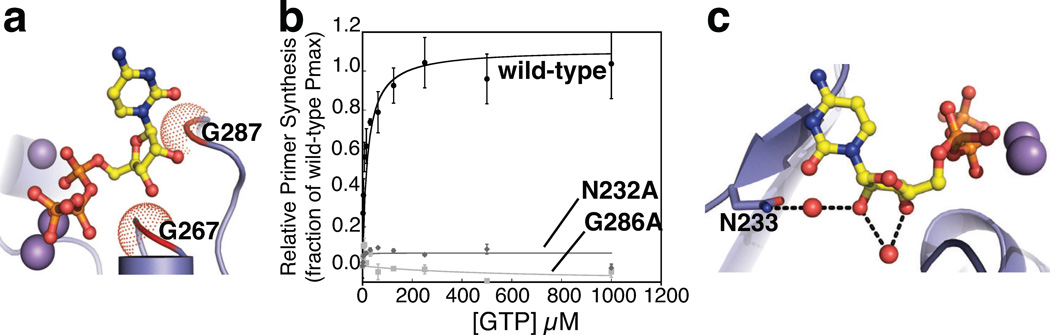
The portion of the RPD seen to surround the sugar and base of the bound nucleotide in SaDnaG has received somewhat less attention. Two conserved loops in this region each bear an invariant glycine that buttresses the ribose (Gly267 and Gly287 in SaDnaG) (Fig 5a). The close contacts in this interaction suggested that substitution of these amino acids with larger sidechains should occlude binding of the nucleotide and abrogate primer synthesis. Mutation of the more N-terminal glycine to alanine in E. coli DnaG (Gly266) has been shown previously to disrupt priming activity (Rodina and Godson, 2006); using the same system, we investigated the role of the second glycine (Gly286), and found that it, too, is required for priming activity (Fig 5b). By contrast, neither glycine proved important for binding to an RNA•DNA heteroduplex (Sup. Methods, Fig S5), indicating that disruption of primer synthesis arises from a defect in the catalytic center. Further examination of the NTP binding environment identified an invariant asparagine (Asn233) in subdomain I that forms a water-mediated hydrogen bond with the ribose 2′-OH (Fig 5c). As with the glycine substitutions, changing the corresponding residue in E. coli DnaG (Asn232) to alanine abrogated primer synthesis, but had little effect on heteroduplex binding (Fig S5).
Figure 5. Analysis of conserved residues that engage the ribose of bound nucleotide.
a) Views of the region near the ribose in the CTP-bound SaDnaG RPD. The protein portion of the RPD is shown as a cartoon. Conserved residues Gly267 and Gly287 (corresponding to Gly266 and Gly286 in EcDnaG) are highlighted in red, with their Cα van der Waals radii represented by red dots.
b) Ribose-binding amino acids are critical for DnaG function. The results of a GTP-dependent, helicase-stimulated de novo primer synthesis assay are shown for wild-type (black circles), Gly286Ala (gray diamonds), and Asn232Ala (light-gray squares) E. coli DnaG. Curves were fit as described (Methods), yielding the parameters listed in Table S1; error bars represent ±SEM.
c) Asn233 (Asn232 in EcDnaG) associates with the 2´-OH of the bound nucleotide through a water-mediated hydrogen bond. Dashed lines represent hydrogen bonds.
See also Figure S5.
In aggregate, the available biochemical data, combined with the comparative studies detailed above, strongly argue that the manner of Mn2+•NTP binding to SaDnaG is representative of the site and configuration used to coordinate the incoming nucleotide during strand synthesis. They also suggest that, despite their distinct active-site architecture, DnaG-type primases may employ a nucleotide-addition strategy that reflects the well-established two-metal mechanism seen in other polymerases (Doublie and Ellenberger, 1998; Kiefer et al., 1998; Steitz et al., 1994). In this vein, we speculate that metal A would serve to coordinate and activate a 3´ acceptor hydroxyl for nucleophilic attack the α-phosphate of the incoming NTP, whereas metal B would promote dissociation of the pyrophosphate leaving group and aid metal A in transition state stabilization. Metal C, which is generally not seen in other polymerase structures, would be a distinctive feature of DnaG that could play a role in binding avidity and/or nucleotide positioning. We note that metal C is coordinated in part by two acidic amino acids (Asp343 and Asp345 in SaDnaG) that are known to be important for synthesis (Godson, Schoenich et al. 2000), but that also comprise a unique “DPD” signature motif to DnaG-family TOPRIM folds (Fig S3A) (Aravind et al., 1998; Godson et al., 2000).
Mechanism of primase inhibition by (p)ppGpp
The stringent response inhibits DNA replication in Gram-positive bacteria by two alarmones, pppGpp (G5P) and ppGpp (G4P) (Jain et al., 2006) (Fig 6a). Long known to block translation and transcription (Potrykus and Cashel, 2008), both nucleotides (collectively known as (p)ppGpp) have been shown recently to additionally impede primer synthesis by inhibiting DnaG through an unknown mechanism (Maciag et al., 2010; Wang et al., 2007). To understand this phenomenon, we initially attempted to model the two modified nucleotides into our NTP-bound structures; however, these efforts indicated that neither G5P nor G4P could bind to the SaDnaG RPD in a manner similar to that of Mn2+•NTP without introducing steric clashes between the 3′-phosphates of the alarmones and the protein.
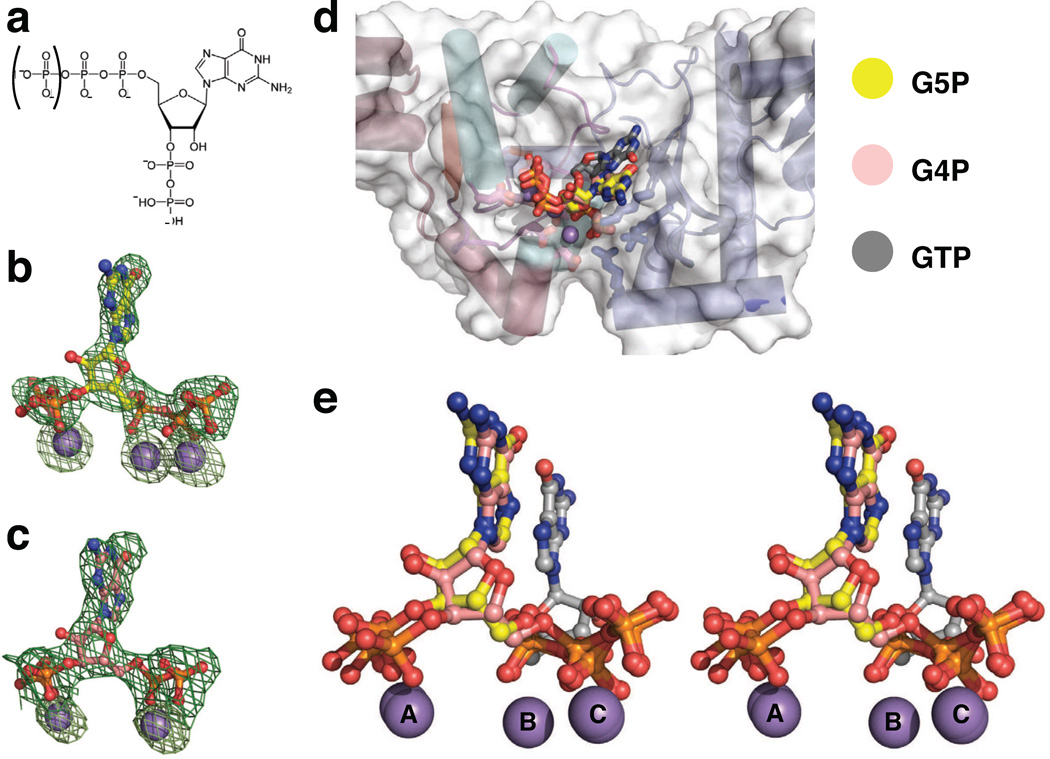
Figure 6. Binding of the S. aureus DnaG RPD to the stringent response alarmones, pppGpp and ppGpp.
a) The chemical structure of (p)ppGpp. The phosphate enclosed in parentheses is present in pppGpp (G5P), but not ppGpp (G4P).
b,c) Difference density present in the SaDnaG RPD active site generated from crystals soaked with b) G5P or c) G4P. Difference density (green mesh, mFo-DFc) is contoured at 3σ prior to modeling of the nucleotide. Anomalous difference density generated from data collected from the same crystal at the Mn-K edge is shown as light green mesh, and is contoured at 8σ. Ball-and-stick models represent G5P (carbon in yellow) and G4P (carbon in salmon), with Mn2+ ions as gray spheres.
d) Alignment of the G5P and G4P bound structures with each other and with the GTP-bound SaDnaG RPD structure. GTP is shown as a ball-and-stick model with carbon colored gray. G5P and G4P are colored as in panels (b) and (c), respectively. Gray spheres represent Mn2+ ions.
e) Stereo image of the alignment shown in panel (d) (protein models omitted).
To establish the location and orientation of G5P and G4P binding to the DnaG RPD more directly, we collected diffraction data and determined structures for crystals soaked with either G5P or G4P in the presence of divalent metal ions (Methods). Clear difference density for both molecules (Fig 6b–c) revealed that the two guanosine analogs associate with the SaDnaG RPD in a similar manner. Interestingly, comparison with the nucleotide-bound SaDnaG complexes shows that the site of binding for G5P and G4P overlaps with the binding locus seen for the metal•NTP complexes, but only partly (Fig 6d–e). The 5′-phosphates and associated metal ions of both inhibitors are coordinated in an identical manner as standard nucleotides, with the exception that metal B does not appear to bind with G4P, leading its diphosphate moiety to shift over and ligand metal C. By contrast, the 3′-phosphates of both alarmones directly engage metal A, an interaction that reorients the base and ribose into a configuration distinct from that seen with NTPs.
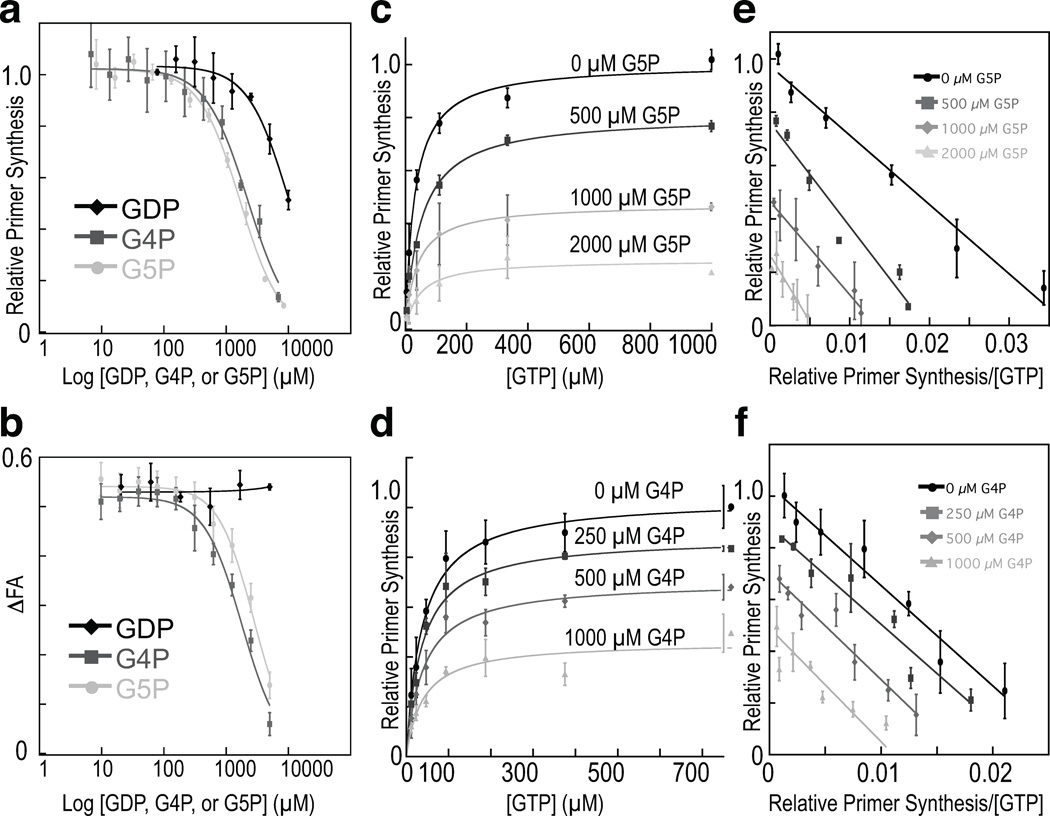
The location and arrangement of G5P and G4P in our structures suggested that the two nucleotides might impede primase activity not only by blocking entry of an incoming NTP, but by also interfering with the binding of either an initiating 5′-NTP, or the extensible 3′ end of an RNA•DNA heteroduplex. To test this idea, we examined the concentration-dependent effects of both G4P and G5P on various activities of E. coli DnaG. As a control, we first assessed the ability of our alarmone preparations to inhibit de novo primer synthesis by full-length DnaG, using an established helicase-stimulated assay (Koepsell et al., 2005). Consistent with previously published findings (Maciag et al., 2010), we found that both nucleotides were weak inhibitors, having IC50 values in the low mM range (Fig 7a). We next asked if (p)ppGpp could interfere with heteroduplex binding, as might be expected if the 3′-phosphates of the inhibitors were to clash with an acceptor 3′-OH associated with metal A. Using the change in fluorescence anisotropy of a fluorescein-labeled DNA•RNA heteroduplex as a readout for binding to a primer-template substrate by the EcDnaG RPD (Methods), we observed that increasing concentrations of either G5P or G4P progressively impeded the protein from associating with the oligo (Fig 7b). Finally, we assessed the ability of (p)ppGpp to compete with GTP in a total primer synthesis assay using full-length E. coli DnaG. Since GTP cannot provide an initiating 3′-OH due to the start site preferences of the enzyme (5’-CTG and 5’-CAG, where the T or A position marks the 5’ terminal site of the primer), it acts as a specific probe for the accessibility of the incoming nucleotide binding-site, which both alarmones block partially in our structures (Khopde et al., 2002). This experiment revealed that G4P and G5P qualitatively exhibited mixed-type inhibitor behavior in blocking the ability of DnaG to synthesize primers in a GTP-concentration dependent manner (Fig. 7c–d), with G5P causing a larger increase in the apparent pseudo-KM value (Methods, Table S1) compared to G4P (Fig 7e–f). Taken together, these data are consistent with the hybrid mode of binding for (p)ppGpp observed crystallographically, and indicate that, rather than acting at a distal allosteric locus, which is thought to occur during transcriptional inhibition (Srivatsan and Wang, 2008), these mediators of the stringent response take advantage of their nucleotidyl properties to directly obstruct the primase active site.
Figure 7. The biochemical response of the E. coli DnaG to G5P and G4P.
a) Concentration-dependent inhibition of EcDnaG by (p)ppGpp in a helicase-stimulated de novo primer synthesis assay. G5P (light-gray circles), G4P (dark-gray squares), and GDP (black diamonds) were titrated into reactions containing full-length E. coli DnaG. Curves are fit to a standard IC50 model (Methods); error bars represent ±SEM (Methods).
b) (p)ppGpp competes for product binding. Competition assays are shown for the binding of the EcDnaG RPD to a fluorescently-labeled heteroduplex in the presence of increasing amounts of G5P (light-gray circles), G4P (dark-gray squares), and GDP (black diamonds). Disruption of binding was determined by measuring the change in the Fluorescence Anisotropy (ΔFA) of the labeled heteroduplex. Curves are fit to a standard IC50 model (Methods); error bars represent ±SEM (Methods).
c,d) GTP-dependent inhibition of primer synthesis by G5P and G4P. GTP-dependent, helicase-stimulated de novo primer synthesis activity was measured (Methods) against increasing concentrations of c) G5P or d) G4P. Points represent averages of duplicate reactions. Primer synthesis extent is reported as relative primer synthesis (Methods). Curves were fit as described in (Methods), yielding the parameters listed in Table S1.
e,f) GTP-dependent, helicase-stimulated primer synthesis activity from (c) and (d), respectively, treated in a manner akin to an Eadie-Hofstee plot (Eadie, 1942; Hofstee, 1952), with Relative Primer Synthesis (RPS) on the y-axis and RPS/[GTP] on the x-axis. Linear fits to the data yield Pmax and KM,pseudo values similar to those determined by nonlinear regression for panels (c) and (d) (Table S1).
Concluding remarks
The mechanisms by which bacterial, DnaG-type primases synthesize primers in support of DNA replication has long remained enigmatic. By determining how bacterial primases engage nucleotide substrates, this work helps clarifies one of the first steps that occurs during RNA strand polymerization. We find that NTPs bind to the active site of DnaG in an unanticipated manner that reconciles prior structural studies with biochemical work in both bacterial and phage systems, and accounts for the roles of several highly conserved sequence motifs in primase function. The similarities between the active sites of DnaG and type IA/II topoisomerases, together with mutagenesis efforts, support the mode of binding we observe and implicate the NTP-binding locus as the site of nucleotide addition. Interestingly, our comparisons with topo II support earlier proposals suggesting that the newly-formed heteroduplex is guided into a shallow, positively charged basin adjacent to the C-terminal helical subdomain as polymerization occurs (Kato et al., 2003; Keck et al., 2000; Podobnik et al., 2000), rather than past the basic ridge, which actually serves as a site for triphosphate binding. This study also reinforces recent findings showing that two small molecule alarmones produced upon induction of the stringent response, pppGpp and ppGpp, inhibit DnaG, and provides a physical explanation for the mechanism by which these agents block primase function. Future efforts, aimed at imaging higher-order ternary complexes between DnaG and distinct primer•template complexes, will be necessary to better define the chemical basis for primer initiation, elongation and termination, as well as to more thoroughly compare the catalytic approaches employed by DnaG to those used in other polymerase systems.
METHODS
Cloning, expression, purification
EcDnaG full-length and RPD constructs were generated previously (Keck et al., 2000). The SaDnaG RPD (residues 111–437) was cloned into a pET28b (Novagen) derivative with a tobacco etch virus (TEV) protease-cleavable, N-terminal hexahistidine tag. Mutant E. coli proteins were generated using QuikChange (Stratagene) site-specific mutagenesis. The coding DNA sequence was verified for all constructs (Elim Biopharmaceuticals). All proteins were expressed in BL21 codon+ cells and purified by affinity and size exclusion chromatography. For details, see Supplemental Methods.
Crystallization of the SaDnaG RPD
Final crystallization conditions for the SaDnaG RPD employed a well solution of 150 mM sodium thiocyanate, 100 mM Bis-tris pH 6.5, 13% PEG 3350, and 2% benzamidine and protein at 5 mg/mL. Crystals were soaked in well solution containing 5 mM MnCl2 and 2.5 mM nucleotide, or 10 mM MnCl2, 2.5 mM G4P or G5P and 1 mM MgCl2, and incubated overnight. Crystals were cryoprotected, flash-frozen and stored in liquid nitrogen prior to data collection at 100 K. For details, see Supplemental Methods.
Data collection and refinement
Diffraction data were collected at the Advanced Light Source Beamline 8.3.1 at Lawrence Berkeley National Laboratory (MacDowell et al., 2004). Diffraction data were indexed and integrated using XDS (Kabsch, 2010) and scaled with SCALA (Winn et al., 2011). Phases and an initial model were obtained by MR using a polyalanine model of the E. coli DnaG RPD (PDB code 1DDE) wherein residues 115–249, residues 260–363, and residues 366–427 were each searched for separately using PHENIX AutoMR (Adams et al., 2011). For soaked SaDnaG RPD crystals, phases were obtained using the apo SaDnaG RPD model. Rfree flags for data collected from soaked crystals were copied from the apo data set. Refinement consisted of multiple rounds of manual rebuilding and real-space refinement with Coot (Emsley et al., 2010) and the PHENIX software suite (Adams et al., 2011); real-space refinement was carried out using electron density maps calculated without the Rfree set. NTPs and metals were not added to models until the Rwork/Rfree of protein portion of the model remained stable for two rounds of refinement.
De novo primer synthesis assays
For all de novo synthesis experiments, measurements are reported here as an average of three reactions. Primer synthesis assays are based on the method described in (Koepsell et al., 2005). All reactions were carried out in a total volume of 20 µL consisting of 100 mM potassium glutamate, 20 mM HEPES (pH 7.5), 0.2 mg/mL BSA, 20 mM MgOAc and 1 mM DTT. Reactions included 0.1 mM ATP and UTP, and 500 nM full-length EcDnaG and EcDnaB (expressed and purified as in (Corn et al., 2008)). A 3′-phosphorylated, single-stranded oligonucleotide (5′-CACACACACACACACTGAAAGCCAAAAG-3′) was used as a template DNA at 600 nM-.
GTP dilutions (from 4 mM to 3.9 µM) were prepared by serially diluting GTP stock (4 mM, in TE) 1:1 into TE. Reaction mixtures (10 µL) were incubated at room-temperature (RT) for 10 min prior to addition of the GTP dilution (10 µL) Inhibitors were included in the incomplete reaction mixture. Complete reactions were incubated in a sealed 384-well plate (Bio-Rad) for 1 h at 37 °C, and then stopped by addition of 10 µL of a 1:67 dilution of PicoGreen stock solution (Invitrogen) in 20 mM Tris, 50 mM EDTA, pH 7.5, giving a final stain dilution of 1:200, as per the manufacturer’s instructions. Stopped reactions were incubated in the dark for 5 min, spun at 3000 × g for 2 min, and raw fluorescence measured in a PerkinElmer Victor3V multilabel plate reader. Raw fluorescence was background corrected with a no-NTP control to give Fluorescence Intensity (FI). The maximal amount of product produced under these conditions (Pmax) was calculated by fitting resultant curves using nonlinear regression to a form of the Michaelis-Menten equation that accounts for the total amount of product produced by the enzyme, as opposed to the reaction rate (Eqn. 1). All data within a given experiment were normalized to the wild-type Pmax (Figs 4, 5), or the Pmax of inhibitor-free reactions (Fig 7), and reported in terms of Relative Primer Synthesis.
| Equation 1 |
where Pmax is the maximal amount of product formed, and KM,pseudo is a pseudo-apparent Michaelis constant that corresponds to the concentration of GTP that yields a total amount of primer synthesis equal to one-half Pmax.
For inhibitor titrations, G5P and G4P were prepared as described (Mechold et al., 2002), and stored in desiccated form until just prior to use. Dry pellets were resuspended in 1× Tris-EDTA (TE), and the concentration measured by absorbance at 258 nm using the extinction coefficient of 13,700 M−1cm−1. GDP was prepared by dissolving GDP powder (SigmaAldrich) in TE to 100 mM, and the pH adjusted to 7.5. Inhibitor dilutions (7.5 µL) were prepared in TE, and mixed with an equal volume of an incomplete reaction mixture containing 2.6× Reaction Buffer, DnaG, DnaB, ATP, UTP and template, and allowed to incubate at room temperature for 10 minutes prior to addition of GTP solution (5 µL at 400 µM). Reactions were incubated at 37 °C for 1 h, and stopped, stained and measured using the same protocol as the standard GTP titrations. G5P was titrated from 8.4 mM to 8.2 µM, G4P was titrated from 6.9 mM to 6.8 µM, and GDP from 10 mM to 78 µM. The data were normalized to the average maximum fluorescence intensity to give RPS, and fit to Eqn. 2 (Copeland, 2005) by nonlinear regression,
| Equation 2 |
in which [I] is the inhibitor concentration, IC50 is the concentration of inhibitor the results in 50% inhibition, and h is the hill coefficient.
Heteroduplex competition assays
E. coli DnaG RPD (500 µM) was incubated with 100 nM of a 5′-fluorescein labeled RNA•DNA heteroduplex (DNA strand (IDT) sequence: 5′-AAAACTGCCGCCGC-3′, RNA strand (Dharmacon) sequence: 5′ -GCGGCGGCA-3′) and 2× Reaction Buffer conditions at RT for 10 min. Inhibitor at 10 mM was serially diluted 1:1 into TE; dilutions were then mixed 1:1 into the 2× RPD and labeled heteroduplex mixture. All inhibitors were titrated from 5 mM to 10 µM. Reactions were incubated at RT for 30 min; Fluorescent Polarization (FP) was measured with a PerkinElmer Victor3V multi-label plate reader, and converted to Fluorescence Anisotropy (FA) prior to background correction with a no-protein control to obtain ΔFA values. The data were fit by nonlinear regression to Eqn. 3 (Copeland, 2005),
| Equation 3 |
in which ΔFAmin and ΔFAmax refer to the signal minimum and signal maximum, respectively, [I] is the concentration of inhibitor, IC50 is the concentration of inhibitor that results in 50% displacement, and h is the Hill coefficient.
Visualization and data analysis methods
Structures were visualized and figures generated using PyMOL (Schrodinger, 2010). Data for activity and binding assays were processed in Microsoft Excel. Plots and corresponding fits were generated with Kaleidagraph. Sequences for sequence alignments were obtained through BLAST (Altschul et al., 1990). Alignments were calculated with MAFFT (Katoh et al., 2002), and visualized and edited with JalView (Waterhouse et al., 2009). Single position conservation analysis was carried out in Microsoft Excel.
Supplementary Material
Acknowledgements
We thank Karl Drlica and Ken Dong for assistance with the manuscript, the staff at Beamline 8.3.1 for help with data collection, and the QB3 Macrolab for help generating site-directed mutants. This work was supported by the NIGMS (GM084003, to JW and GM071747, to JMB) and the Welch Foundation (Grant Q-1698, to JW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aravind L, Leipe DD, Koonin EV. Toprim - a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Research. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Griep MA. DnaB helicase affects the initiation specificity of Escherichia coli primase on single-stranded DNA templates. Biochemistry. 2000;39:745–752. doi: 10.1021/bi991555d. [DOI] [PubMed] [Google Scholar]
- Carl PL. Escherichia coli mutants with temperature-sensitive synthesis of DNA. Mol Gen Genet. 1970;109:107–122. doi: 10.1007/BF00269647. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintakayala K, Machon C, Haroniti A, Larson MA, Hinrichs SH, Griep MA, Soultanas P. Allosteric regulation of the primase (DnaG) activity by the clamp-loader (tau) in vitro. Mol Microbiol. 2009;72:537–549. doi: 10.1111/j.1365-2958.2009.06668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland RA. Evaluation of enzyme inhibitors in drug discovery. A guide for medicinal chemists and pharmacologists. Methods Biochem Anal. 2005;46:1–265. [PubMed] [Google Scholar]
- Corn JE, Berger JM. FASTDXL: a generalized screen to trap disulfide-stabilized complexes for use in structural studies. Structure. 2007;15:773–780. doi: 10.1016/j.str.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corn JE, Pease PJ, Hura GL, Berger JM. Crosstalk between primase subunits can act to regulate primer synthesis in trans. Molecular Cell. 2005;20:391–401. doi: 10.1016/j.molcel.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Corn JE, Pelton JG, Berger JM. Identification of a DNA primase template tracking site redefines the geometry of primer synthesis. Nature Structural & Molecular Biology. 2008;15:163–169. doi: 10.1038/nsmb.1373. [DOI] [PubMed] [Google Scholar]
- Doublie S, Ellenberger T. The mechanism of action of T7 DNA polymerase. Curr Opin Struct Biol. 1998;8:704–712. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]
- Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- Eadie GS. The inhibition of cholinesterase by physostigmine and prostigmine. Journal of Biological Chemistry. 1942;146:85–93. [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick DN, Richardson CC. DNA primases. Annu Rev Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- Godson GN, Schoenich J, Sun W, Mustaev AA. Identification of the magnesium ion binding site in the catalytic center of Escherichia coli primase by iron cleavage. Biochemistry. 2000;39:332–339. doi: 10.1021/bi9916628. [DOI] [PubMed] [Google Scholar]
- Griep MA, Lokey ER. The role of zinc and the reactivity of cysteines in Escherichia coli primase. Biochemistry. 1996;35:8260–8267. doi: 10.1021/bi952948p. [DOI] [PubMed] [Google Scholar]
- Hamdan SM, van Oijen AM. Timing, coordination, and rhythm: acrobatics at the DNA replication fork. J Biol Chem. 2010;285:18979–18983. doi: 10.1074/jbc.R109.022939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- Hofstee BH. On the evaluation of the constants Vm and KM in enzyme reactions. Science. 1952;116:329–331. doi: 10.1126/science.116.3013.329. [DOI] [PubMed] [Google Scholar]
- Jain V, Kumar M, Chatterji D. ppGpp: stringent response and survival. J Microbiol. 2006;44:1–10. [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Ito T, Wagner G, Richardson CC, Ellenberger T. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol Cell. 2003;11:1349–1360. doi: 10.1016/s1097-2765(03)00195-3. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck JL, Roche DD, Lynch AS, Berger JM. Structure of the RNA polymerase domain of E coli primase. Science. 2000;287:2482–2486. doi: 10.1126/science.287.5462.2482. [DOI] [PubMed] [Google Scholar]
- Khopde S, Biswas EE, Biswas SB. Affinity and sequence specificity of DNA binding and site selection for primer synthesis by Escherichia coli primase. Biochemistry. 2002;41:14820–14830. doi: 10.1021/bi026711m. [DOI] [PubMed] [Google Scholar]
- Kiefer JR, Mao C, Braman JC, Beese LS. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- Kitani T, Yoda KY, Ogawa T, Okazaki T. EVIDENCE THAT DISCONTINUOUS DNA-REPLICATION IN ESCHERICHIA-COLI IS PRIMED BY APPROXIMATELY 10 TO 12 RESIDUES OF RNA STARTING WITH A PURINE. Journal of Molecular Biology. 1985;184:45–52. doi: 10.1016/0022-2836(85)90042-7. [DOI] [PubMed] [Google Scholar]
- Koepsell SA, Hanson S, Hinrichs SH, Griep MA. Fluorometric assay for bacterial primases. Anal Biochem. 2005;339:353–355. doi: 10.1016/j.ab.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Kornberg AB, Tania A. DNA Replication. 2 edn. New York: W. H. Freeman and Company; 1992. [Google Scholar]
- Kuchta RD, Stengel G. Mechanism and evolution of DNA primases. Biochim Biophys Acta. 2010;1804:1180–1189. doi: 10.1016/j.bbapap.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Richardson CC. Essential lysine residues in the RNA polymerase domain of the gene 4 primase-helicase of bacteriophage T7. J Biol Chem. 2001;276:49419–49426. doi: 10.1074/jbc.M108443200. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Richardson CC. Acidic residues in the nucleotide-binding site of the bacteriophage T7 DNA primase. J Biol Chem. 2005;280:26984–26991. doi: 10.1074/jbc.M504817200. [DOI] [PubMed] [Google Scholar]
- MacDowell AA, Celestre RS, Howells M, McKinney W, Krupnick J, Cambie D, Domning EE, Duarte RM, Kelez N, Plate DW, et al. Suite of three protein crystallography beamlines with single superconducting bend magnet as the source. J Synchrotron Radiat. 2004;11:447–455. doi: 10.1107/S0909049504024835. [DOI] [PubMed] [Google Scholar]
- Maciag M, Kochanowska M, Lyzen R, Wegrzyn G, Szalewska-Palasz A. ppGpp inhibits the activity of Escherichia coli DnaG primase. Plasmid. 2010;63:61–67. doi: 10.1016/j.plasmid.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Makowska-Grzyska M, Kaguni JM. Primase directs the release of DnaC from DnaB. Mol Cell. 2010;37:90–101. doi: 10.1016/j.molcel.2009.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marians KJ. PROKARYOTIC DNA-REPLICATION. Annual Review of Biochemistry. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- Mechold U, Murphy H, Brown L, Cashel M. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J Bacteriol. 2002;184:2878–2888. doi: 10.1128/JB.184.11.2878-2888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelman LV, Beauchamp BB, Richardson CC. Requirement for a zinc motif for template recognition by the bacteriophage T7 primase. EMBO J. 1994;13:3909–3916. doi: 10.1002/j.1460-2075.1994.tb06702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitkova AV, Khopde SM, Biswas SB. Mechanism and stoichiometry of interaction of DnaG primase with DnaB helicase of Escherichia coli in RNA primer synthesis. J Biol Chem. 2003;278:52253–52261. doi: 10.1074/jbc.M308956200. [DOI] [PubMed] [Google Scholar]
- Podobnik M, McInerney P, O'Donnell M, Kuriyan J. A TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases. J Mol Biol. 2000;300:353–362. doi: 10.1006/jmbi.2000.3844. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Rodina A, Godson GN. Role of conserved amino acids in the catalytic activity of Escherichia coli primase. J Bacteriol. 2006;188:3614–3621. doi: 10.1128/JB.188.10.3614-3621.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowen L, Kornberg A. Primase, the dnaG protein of Escherichia coli. An enzyme which starts DNA chains. J Bacteriol. 1978;253:758–764. [PubMed] [Google Scholar]
- Salzberg SL, Salzberg AJ, Kerlavage AR, Tomb JF. Skewed oligomers and origins of replication. Gene. 1998;217:57–67. doi: 10.1016/s0378-1119(98)00374-6. [DOI] [PubMed] [Google Scholar]
- Schmidt BH, Burgin AB, Deweese JE, Osheroff N, Berger JM. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature. 2010;465:641–644. doi: 10.1038/nature08974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger L. The PyMOL Molecular Graphics System, Version 1.3. 2010 [Google Scholar]
- Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Steitz TA, Smerdon SJ, Jager J, Joyce CM. A unified polymerase mechanism for nonhomologous DNA and RNA polymerases. Science. 1994;266:2022–2025. doi: 10.1126/science.7528445. [DOI] [PubMed] [Google Scholar]
- Sun W, Godson GN. Interaction of Escherichia coli primase with a phage G4ori(c)-E. coli SSB complex. J Bacteriol. 1996;178:6701–6705. doi: 10.1128/jb.178.23.6701-6705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Schoneich J, Godson GN. A mutant Escherichia coli primase defective in elongation of primer RNA chains. J Bacteriol. 1999;181:3761–3767. doi: 10.1128/jb.181.12.3761-3767.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Tormo J, Steitz TA, Godson GN. Domains of Escherichia coli primase: functional activity of a 47-kDa N-terminal proteolytic fragment. Proc Natl Acad Sci U S A. 1994;91:11462–11466. doi: 10.1073/pnas.91.24.11462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart JR, Griep MA. Primer synthesis kinetics by Escherichia coli primase on single-stranded DNA templates. Biochemistry. 1995;34:16097–16106. doi: 10.1021/bi00049a025. [DOI] [PubMed] [Google Scholar]
- Tanner NA, Hamdan SM, Jergic S, Loscha KV, Schaeffer PM, Dixon NE, van Oijen AM. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat Struct Mol Biol. 2008;15:170–176. doi: 10.1038/nsmb.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tougu K, Marians KJ. The extreme C terminus of primase is required for interaction with DnaB at the replication fork. J Biol Chem. 1996a;271:21391–21397. doi: 10.1074/jbc.271.35.21391. [DOI] [PubMed] [Google Scholar]
- Tougu K, Marians KJ. The interaction between helicase and primase sets the replication fork clock. J Biol Chem. 1996b;271:21398–21405. doi: 10.1074/jbc.271.35.21398. [DOI] [PubMed] [Google Scholar]
- Tougu K, Peng H, Marians KJ. Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J Biol Chem. 1994;269:4675–4682. [PubMed] [Google Scholar]
- Urlacher TM, Griep MA. Magnesium acetate induces a conformational change in Escherichia coli primase. Biochemistry. 1995;34:16708–16714. doi: 10.1021/bi00051a020. [DOI] [PubMed] [Google Scholar]
- van Oijen AM, Loparo JJ. Single-molecule studies of the replisome. Annu Rev Biophys. 2010;39:429–448. doi: 10.1146/annurev.biophys.093008.131327. [DOI] [PubMed] [Google Scholar]
- Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CA, Zechner EL, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork I Multiple effectors act to modulate Okazaki fragment size. J Biol Chem. 1992a;267:4030–4044. [PubMed] [Google Scholar]
- Wu CA, Zechner EL, Reems JA, McHenry CS, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork V Primase action regulates the cycle of Okazaki fragment synthesis. J Biol Chem. 1992b;267:4074–4083. [PubMed] [Google Scholar]
- Zechner EL, Wu CA, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. II. Frequency of primer synthesis and efficiency of primer utilization control Okazaki fragment size. J Biol Chem. 1992;267:4045–4053. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.