Abstract
Kinetochores attach the replicated chromosomes to the mitotic spindle and orchestrate their transmission to the daughter cells. Kinetochore–spindle binding and chromosome segregation are mediated by the multi-copy KNL1Spc105, MIS12Mtw1 and NDC80Ndc80 complexes that form the so-called KMN network. KMN–spindle attachment is regulated by the Aurora BIpl1 and MPS1Mps1 kinases. It is unclear whether other mechanisms exist that support KMN activity during the cell cycle. Using budding yeast, we show that kinetochore protein Cnn1 localizes to the base of the Ndc80 complex and promotes a functionally competent configuration of the KMN network. Cnn1 regulates KMN activity in a spatiotemporal manner by inhibiting the interaction between its complexes. Cnn1 activity peaks in anaphase and is driven by the Cdc28, Mps1 and Ipl1 kinases.
Kinetochores assemble from ~70 (budding yeast) to ~100 proteins (humans) on the centromeres (CEN ) of the replicated, paired chromosomes (sister chromatids)1. Kinetochores bi-orient the sister chromatids to the mitotic spindle and orchestrate their segregation1–3. Central in these activities are the conserved KNL1Spc105, MIS12Mtw1 and NDC80Ndc80 complexes that form the KMN network, which constitutes the heart of each kinetochore and extends from the centromeric chromatin to the outer kinetochore4. Erroneous activity of any KMN subunit results in chromosome missegregation and cell death. Kinetochore–microtubule attachment is mediated by the KNL1Spc105 and NDC80Ndc80 complexes, but the contribution to microtubule binding by the latter is the most essential5. The MIS12Mtw1 complex synergistically enhances the microtubule-binding activity of the KNL1Spc105 and NDC80Ndc80 complexes4. Sister-chromatid/spindle attachment and bi-orientation are controlled by the Aurora BIpl1(refs 6,7) and MPS1Mps1 (refs 8,9) kinases. If a sister chromatid is misattached to the spindle, both kinases will trigger a turnover of the sister-chromatid/spindle contact. The cells will be halted in mitosis by the spindle-assembly checkpoint10 (SAC), allowing the detached sister chromatid to establish bi-polar contact with the spindle structure. Stable kinetochore–spindle attachment is ensured by the B56-PP2A phosphatase11. When all sister chromatids are bipolarly aligned to the spindle array, the SAC becomes silenced by the PP1Glc7 phosphatase and KNL1Spc105 (refs 5,12,13). Mitosis and chromosome segregation will then proceed. Whether other mechanisms exist that help to ensure correct KMN activity is unclear. Using budding yeast, we show that inner kinetochore protein Cnn1 spatiotemporally supports the activity of the KMN network at its base. Through its amino terminus, Cnn1 interacts with the Spc24–Spc25 dimer of the Ndc80 complex, thereby preventing the Mtw1 and Spc105 complexes from binding to the Ndc80 complex. The activity of Cnn1 peaks in anaphase and depends on a phosphorylation threshold established by the Cdc28, Mps1 and Ipl1 kinases. At the end of anaphase, when all sister chromatids have segregated, the enriched Cnn1 is released from the Ndc80 complex, an event that is triggered by dephosphorylation of Cnn1. Cnn1 does not act to recruit the KMN complexes to centromeres but serves to modulate the affinities between the KMN complexes, thereby contributing to efficient KMN assembly, kinetochore–spindle binding and sister-chromatid segregation.
RESULTS
Cnn1 becomes enriched at anaphase kinetochores
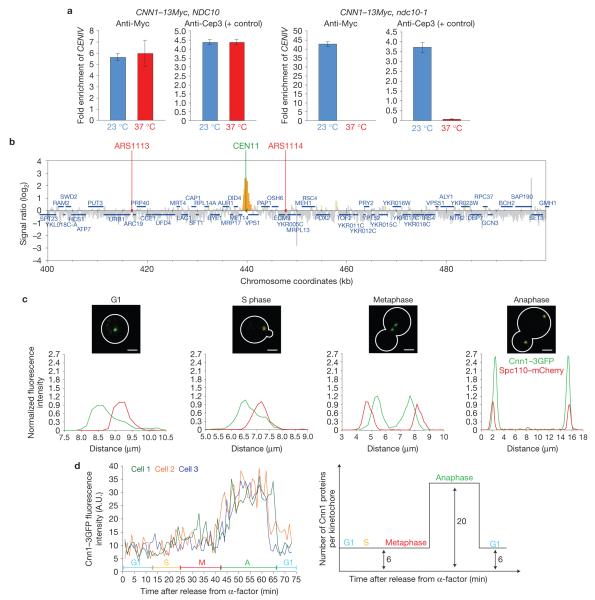
After identifying Cnn1 in the immunoprecipitate of Saccharomyces cerevisiae kinetochore protein Nnf1 (ref. 14), we examined whether Cnn1 was a new kinetochore component. Chromatin immunoprecipitation (ChIP) analysis of Cnn1–13Myc in wild-type and ndc10-1 strains showed that Cnn1 localized to centromeres in an Ndc10-dependent manner (Fig. 1a). Ndc10 is a subunit of the centromeric CBF3 complex that recruits all kinetochore proteins15. Next, ChIP of Cnn1–Pk6 followed by genome hybridization revealed that Cnn1 associates only with CEN DNA (Fig. 1b and Supplementary Fig. S1a). Next, fluorescence imaging of Cnn1–3GFP in a strain whose spindle pole protein Spc110 was marked with mCherry (the fluorescent proteins did not affect cell growth; Supplementary Fig. S6) confirmed that Cnn1 localizes to kinetochores (near the spindle poles in G1, S phase and anaphase; bi-lobed between the poles in metaphase). We did notice, however, that the intensities of the Cnn1–3GFP signals at kinetochores were the highest in anaphase (Fig. 1c). This observation was confirmed when we imaged Cnn1–3GFP Spc110–mCherry cells during a synchronous cell cycle; Cnn1–3GFP fluorescence levels were low from G1 through metaphase but then increased 3.5-fold at anaphase onset. Following exit from anaphase, the intensities of the Cnn1–3GFP kinetochore signals returned to the base level (Fig. 1d).To quantify Cnn1, we analysed the Cnn1–3GFP Spc110–mCherry strain in parallel with an Ndc80–3GFP Spc110–mCherry strain and measured the fluorescence intensities of Cnn1–3GFP and Ndc80–3GFP at kinetochores (Spc110–mCherry acted as the internal reference, Supplementary Fig. S1b). Considering that Ndc80 is present in 19 copies per anaphase kinetochore16,17, we found that Cnn1 is present in ~6 copies (5.8 ± 0.8) per kinetochore in G1, S phase and metaphase and in ~20 copies in anaphase (20 ± 2.8; Supplementary Fig. S1b). Its dynamic localization (Fig. 1d) indicated that Cnn1 might turn over at kinetochores. However, fluorescence recovery after photobleaching (FRAP) analysis showed that Cnn1–3GFP fluorescence did not recover within 2 (Supplementary Fig. S1c) or 10 min (data not shown) after photobleaching at any cell-cycle stage, indicating that Cnn1 stably associates with kinetochores.
Figure 1.
Cnn1 is a kinetochore component whose concentration at kinetochores peaks in anaphase. (a) Cnn1–13Myc chromatin immunoprecipitates with centromeres (CEN) in a kinetochore-dependent fashion. A CNN1–13Myc NDC10 strain and the isogenic CNN1–13Myc ndc10-1 mutant were grown at 23 °C (permissive temperature, blue bars), and in parallel at 23 °C followed by a 3 h shift to 37 °C (restrictive temperature, red bars). The temperature shift inactivates Ndc10-1, which prevents kinetochore assembly following CEN replication. The bars show the amount of CENIV that was enriched in three independent ChIP experiments (n = 3) as measured relative to the input (untreated cell extract). The error bars represent s.d. In parallel, an anti-Cep3 ChIP was performed on the yeast cell extracts and served as the positive control, as Cep3 depends on Ndc10 for CEN association. A ChIP performed without antibody acted as the specificity control (no enrichment of CENIV, data not shown). (b) Cnn1 localizes to centromeres, as shown by ChIP–chip analysis45. Endogenous Cnn1 was C-terminally tagged with a Pk6 epitope and submitted to ChIP. Chromatin co-immunoprecipitated with Cnn1–Pk6 was isolated, PCR-amplified with random primers and labelled with biotin. The biotinylated DNA was then hybridized to the Saccharomyces cerevisiae genome (Tiling Array 1.0R, Affymetrix). The supernatant of the immunoprecipitate was used as the hybridization control. Data analysis was performed with GeneChip Command Console Software (Affymetrix). Cnn1–Pk6 binding (yellow) around CENXI is shown. (c) Cnn1 is a kinetochore protein. Representative cells of a Cnn1–3GFP Spc110–mCherry strain were imaged by spinning-disc confocal microscopy (top panels). The fluorescence signals were measured, normalized and plotted along the cell axis (μm; bottom panels). Scale bars,4 μm. (d) Cnn1 localizes to kinetochores in a cell-cycle-stage-dependent manner. Cnn1–3GFP Spc110–mCherry cells were synchronized in late G1 (5 μg ml−1 -factor), released into the cell cycle (2% glucose YP medium), and imaged by time-lapse wide-field Deltavision deconvolution fluorescence microscopy. Cnn1–3GFP localization during the cell cycle of three cells is plotted in the left panel (Spc110–mCherry was the internal reference) and is graphically summarized in the right panel. G1, S phase, metaphase (M) and anaphase (A) are indicated.
The anaphase enrichment of Cnn1 is phospho-driven
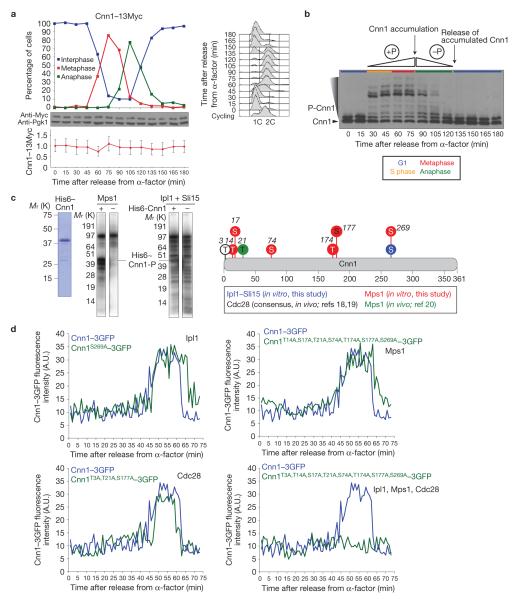
To assess whether the Cnn1 localization profile (Fig. 1d) results from a cell-cycle-stage-dependent expression or degradation of Cnn1, we tracked Cnn1–13Myc levels during a synchronous cell cycle. Cnn1–13Myc protein levels remained constant through the cell cycle (Fig. 2a), indicating that the Cnn1 recruitment dynamics resulted from post-translational regulation. Involvement of phosphorylation was examined as Cnn1 had been identified as a substrate of the Cdc28 kinase18,19. To probe Cnn1 phosphorylation, we analysed the samples from our cell-cycle experiment by PhosTag SDS–polyacrylamide gel electrophoresis (PAGE) and anti-Myc western hybridization. Although part of the Cnn1–13Myc pool was phosphorylated in G1, the level of Cnn1–13Myc phosphorylation markedly increased at S-phase entry and continued through metaphase. During anaphase, Cnn1–13Myc became dephosphorylated (Fig. 2b). Together, the phosphorylation and localization profiles indicated that the phosphorylation of Cnn1 during S phase and metaphase produces a phospho-threshold, which signals the accumulation of Cnn1 to kinetochores at anaphase entry. During anaphase, Cnn1 becomes dephosphorylated, leading to a low phospho-threshold that signals the release of the extra copies of Cnn1 at anaphase exit.
Figure 2.
The enrichment of Cnn1 at anaphase kinetochores is phospho-driven. (a) Cnn1 levels remain constant through the cell cycle. Cnn1–13Myc cells were synchronized in G1, released into the cell cycle (2% glucose YP medium) and tracked over time (DNA content, FACS; spindle morphology, anti-Tub1 immunofluorescence analysis). After 90 min, 2.5 μg ml−1 -factor was added to prevent the cells from entering a new cell cycle. The cell-cycle stages are shown in the upper plot. Cnn1–13Myc was visualized (anti-Myc western hybridization, Pgk1 was the loading control (middle blots)) and its levels during the cell cycle were quantified (relative to those of Pgk1 (lower plot)). The error bars represent the standard deviations of the measurements from three blot exposures. (b) Cnn1 is a phospho-protein. The phosphorylation state of Cnn1–13Myc was examined by PhosTag SDS–PAGE and anti-Myc western hybridization. Cell cycle stages are indicated. (c) Cnn1 is a substrate of the Mps1 and Ipl1 kinases. Recombinant His6–Cnn1 (left image) was phosphorylated by recombinant Mps1 (middle image) or by recombinant Ipl1 plus Sli15 (right image) in the presence of radiolabelled ATP. Mass spectrometric phospho-mapping of His6–Cnn1 phosphorylated by either kinase in the presence of cold ATP identified six Mps1 target residues and one Ipl1 target residue (Supplementary Fig. S2). Thr 3, Thr 21 and Ser 177 correspond to the Cdc28 recognition consensus (www.phosida.com; refs 18,19). Thr 21 was suggested to be phosphorylated by Mps1 in vivo20 (graphically summarized on the right). (d) The accumulation of Cnn1–3GFP at anaphase kinetochores is driven by Cdc28, Mps1 and Ipl1 kinase activities. Strains endogenously expressing wild-type Cnn1–3GFP (blue) or Cnn1–3GFP containing phospho-mimetic null mutations in residues targeted by each kinase or by all three kinases combined (green) were imaged during a synchronous cell cycle by wide-field Deltavision deconvolution fluorescence microscopy. Spc110–mCherry acted as the internal reference. Uncropped images of blots are shown in Supplementary Fig. S7.
Besides Cdc28 (refs 18,19), for which Cnn1 contains three phosphorylation recognition sites (Thr 3, Thr 21 and Ser 177), the Mps1 kinase was another candidate for Cnn1 phosphorylation, as Cnn1 was recently co-purified with Mps1 (ref. 20). In addition, Cnn1 contains a consensus site for the Ipl1 kinase (Ser 269; ref. 21). To confirm phosphorylation by Mps1 and Ipl1, we performed in vitro kinase assays with recombinant Cnn1, Mps1 and Ipl1–Sli15 (Fig. 2c). Mass spectrometric analysis of the phosphorylated Cnn1 identified six Mps1 target residues (Thr 14, Ser 17, Ser 74, Thr 174, Ser 177 and Ser 269) and one Ipl1 target residue (Ser 269; Fig. 2c and Supplementary Fig. S2). To examine whether the three kinases contribute to Cnn1–3GFP localization, we point-mutated the target residues of each kinase alone or in combination into alanine to mimic kinase-null states. The mutations did not affect cell fitness. The localization of each mutant Cnn1–3GFP protein was quantified during a synchronous cell cycle using time-lapse fluorescence imaging (Fig. 2d). Mutations mimicking the lack of a single kinase did not affect the kinetochore localization profile of Cnn1–3GFP. However, when the mutations were combined, we found that Cnn1–3GFP no longer accumulated at anaphase kinetochores (Fig. 2d), indicating that Cdc28, Ipl1 and Mps1 establish, in redundant fashion, a phosphorylation threshold that signals the enrichment of Cnn1 at anaphase kinetochores. The Cnn1–3GFP base levels were not affected in the triple-null mutant, indicating that its association with interphase and metaphase kinetochores is either kinase independent or depends on other kinases. Exit from mitosis requires the inactivation of mitotic kinases and the phosphatase-mediated reversal of their activities22. These activities may bring about Cnn1 dephosphorylation and establish the low phospho-threshold required for the release of Cnn1 from late-anaphase kinetochores.
Cnn1 localizes to the base of the KMN network
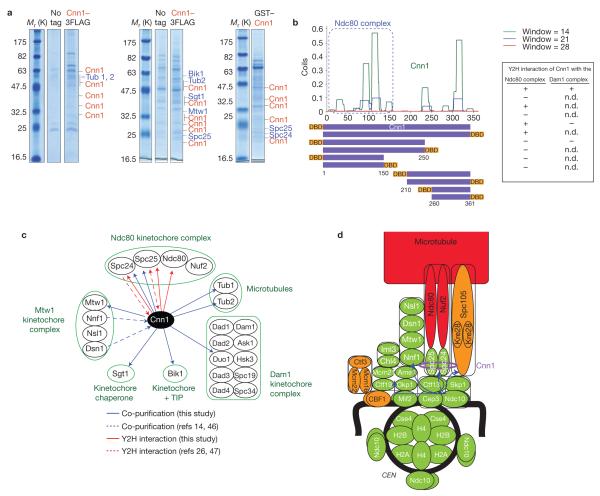
Cnn1 was originally co-purified with Nnf1 (ref. 14), a subunit of the Mtw1 complex (Mtw1, Dsn1, Nsl1 and Nnf1) that is part of the KMN network in yeast. This indicated that Cnn1 localizes to the KMN network. However, to more precisely reveal its position within the kinetochore, we affinity-purified Cnn1 from yeast cell extract (Fig. 3a). Mass spectrometric analysis showed that Mtw1, Spc24 and Spc25 (lower dimer of the KMN’s Ndc80 complex23), kinetochore chaperone Sgt1 (ref. 24), the kinetochore +TIP Bik1 (ref. 25) and tubulin (Tub1, Tub2) were specifically enriched with Cnn1 (Fig. 3a). Of note, Spc24 and Spc25 were more significantly enriched than Mtw1 (MASCOT scores of 203, 346 and 125, respectively). The KMN Spc105 complex was not co-purified with Cnn1. Yeast two-hybrid (Y2H) screens with full-length Cnn1 and with fragments chosen from the predicted secondary structure of Cnn1 (Fig. 2b), against a genomic library of confirmed open reading frames26 (ORFs), identified as interactors only once Duo1 (Dam1 kinetochore complex), but always Spc24, Spc25 and Ndc80 (Fig. 3b,c), indicating that Cnn1 binds to the Ndc80 complex. The interaction with the Ndc80 complex required the first 150 residues of Cnn1 (Fig. 3b).
Figure 3.
Cnn1 protein–protein interactions and localization to the inner kinetochore. (a) SDS–PAGE gels depicting Cnn1 and co-purifying proteins. Cnn1 was tagged with a 3FLAG or GST epitope and expressed from PGAL1, which replaced the endogenous promoter of CNN1 (PGAL1–CNN1–3FLAG) or was placed on a 2 μ vector (pPGAL1–GST–CNN1). Both strains and the untagged parent (negative control) were grown in 2% galactose YP medium (23 °C). For purification details, see Methods. The left, central and right panels indicate peptides corresponding to Cnn1 (red) and enriched proteins (blue) that were identified by mass spectrometry following five, two or three washes of the affinity beads, respectively. The number of peptides and MASCOT scores of the enriched proteins were: left panel: Cnn1 (6, 5, 4, 4, 3, 2, 3, 2, 2; 384, 272, 195, 193, 148, 141, 126, 113, 80), Tub1 (6; 394), Tub2 (5; 346); central panel: Cnn1 (15, 10, 10, 6, 6, 6, 5, 4, 3, 3; 891, 602, 500, 343, 295, 288, 199, 182, 159, 126), Tub2 (4; 174), Spc25 (2; 90), Sgt1 (3, 134), Mtw1 (3; 125), Bik1 (3; 118); and right panel: Cnn1 (7, 7, 6, 4, 4; 423, 319, 298, 223, 213), Spc24 (3; 203) and Spc25 (8; 346). (b) Identification of Cnn1 interactors in Gal4-based Y2H screens. Cnn1 fragments (purple bars) were chosen from the predicted secondary structure of Cnn1 (plotted, www.ch.embnet.org/software/COILS_form.html) and tagged N- or C-terminally with the Gal4 DNA-binding domain (DBD). The bait constructs were screened against all S. cerevisiae ORFs tagged C-terminally with the Gal4 activation domain26. The construct containing the first 150 residues of Cnn1 consistently interacted with Spc24, Spc25 and Ndc80 (Ndc80 complex). Cnn1 interacted only once with Duo1 (Dam1 complex; n.d., not determined). (c) Cnn1 interactors identified in affinity purifications (blue arrows, this study; dashed blue arrows, published14,46) and Y2H screens (red arrows: this study; dashed red arrows, published26,47). (d) Kinetochore localization of Cnn1 (purple ellipse) as revealed by CENIV ChIP (Supplementary Fig. S4). The blue arrows indicate the kinetochore recruitment pathways29. Recruitment code: green, total dependence; orange, partial dependence; red, independence.
As the proteins interacting with Cnn1 are part of well-characterized complexes and as Cnn1 was not identified as a prey in the Y2H screens, Cnn1 might exist as a monomer. We probed this possibility by gel filtration and glycerol sedimentation ultra-centrifugation analysis of Cnn1–13Myc yeast cell extract (Supplementary Fig. S3a). Calculations27 revealed that Cnn1–13Myc has a relative molecular mass of about 111,000 (~Mr 111K), reflecting that of a homodimer (Cnn1, Mr 41K; 13Myc, Mr 15K). However, its frictional coefficient was larger than 1 (1.2) and its axial ratio equalled 2, pointing to Cnn1 being elongated. As an elongated shape makes a small protein behave as if it were large in hydrodynamic studies, we could not exclude that Cnn1–13Myc existed as a monomer. To examine this possibility we tried to co-immunoprecipitate Cnn1–13Myc and Cnn1–Pk6 from the cell extract of a diploid strain; they never immunoprecipitated each other (Supplementary Fig. S3b), indicating that Cnn1 is an elongated monomer in yeast.
To determine whether the Cnn1 protein interactions (Fig. 3c) underlie its localization to kinetochores, we studied Cnn1 recruitment to CENIV by ChIP in well-characterized kinetochore mutants and in a tubulin mutant. Besides Ndc10 (Fig. 1a), Cnn1 also required the centromeric nucleosome (Cse4) and CEN-associated Mif2 to associate with kinetochores (Fig. 3d and Supplementary Fig. S4). Of the more downstream complexes (recruitment routes are indicated with blue arrows; Fig. 3d), Cnn1 depended on the centromere-associated Iml3–Chl4 dimer, the Mtw1 complex, the Spc24–Spc25 dimer of the Ndc80 complex (not on the Ndc80–Nuf2 dimer, which is absent from kinetochores in the ndc80-1 mutant (ref. 28)) and partially on the Spc105 complex (Fig. 3d and Supplementary Fig. S4). Cnn1 did not localize to kinetochores through the spindle microtubules. Taken together, our protein-interaction and CEN-recruitment data indicated that Cnn1 localizes to the base of the KMN network, and more specifically to the Spc24–Spc25 dimer of the Ndc80 complex.
Cnn1 supports sister-chromatid/spindle binding and bi-orientation
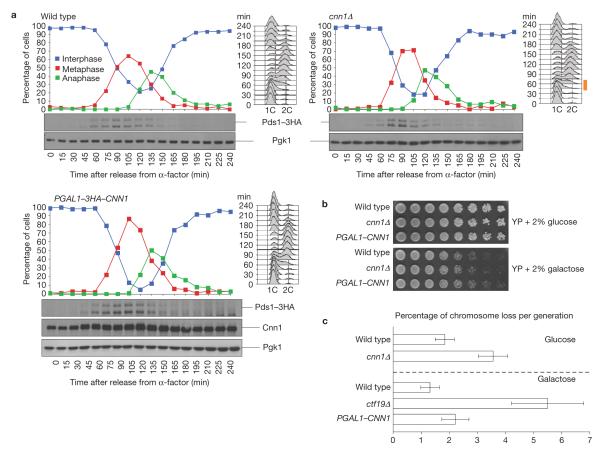
To establish how Cnn1 contributes to kinetochore activity, we first probed whether Cnn1 acts in the SAC, which is also recruited to kinetochores by the KMN complexes10,28–30. In the presence of spindle poisons, the viability of the cnn1Δ mutant was not affected (Supplementary Fig. S5a,b), indicating that Cnn1 does not contribute to SAC activity. Next, we examined how Cnn1 affected the yeast cell cycle. In the absence of Cnn1(cnn1Δ), S phase was reproducibly shortened by 15 min (indicated with an orange bar in the fluorescence-activated cell sorting (FACS) profile; Fig. 4a). The lengths of the other cell-cycle stages were not affected. This indicated that Cnn1 contributes to the timely execution of yeast S-phase activities, which include kinetochore assembly following CEN replication, sister-chromatid/microtubule binding and bi-orientation31. Slightly elevated levels of Cnn1 (pgal1–CNN1, the promoter of CNN1 was replaced with PGAL1 and the cells were grown in 2% galactose YP medium) did not affect cell-cycle dynamics (Fig. 4a). When analysed over multiple generations (2% glucose or 2% galactose YP agar), the cnn1Δ and pgal1–CNN1 cells did not exhibit reduced fitness (Fig. 4b). However, use of an ade3-2 reporter plasmid32 showed that the cnn1Δ and pgal1–CNN1 strains suffered from a 2- to 3-fold increase in chromosome loss (kinetochore mutant ctf19Δ acted as a reference; Fig. 4c).
Figure 4.
Cnn1 ensures a timely progression through S phase and promotes faithful chromosome transmission. (a) In the absence of Cnn1, S phase was reduced by 15 min (indicated by an orange bar next to the FACS profiles). Wild-type, cnn1Δ and PGAL1–CNN1 strains were grown in 2% raffinose YP medium (23 °C), synchronized in G1 (5 μg ml−1 α-factor), washed and released into 2% galactose YP medium. Cells were tracked by FACS (DNA content, right) and spindle morphology analysis (curves; anti-Tub1 immunofluorescence analysis). The levels of Pds1–3HA, whose degradation signals the metaphase–anaphase transition, were revealed by anti-HA western hybridization, as were the levels of 3HA–Cnn1 in the PGAL1–3HA–CNN1 strain. Pgk1 was the loading control (bottom blots). After 90 min, 2.5 μg ml−1 of α-factor was added to the culture to prevent the cells from entering a second cell cycle. (b) A lack or elevated levels of Cnn1 do not affect cell fitness. Wild-type, cnn1Δ and PGAL1–CNN1 strains were serially diluted on 2% glucose YP agar (represses PGAL1–CNN1) and on 2% galactose YP agar (induces PGAL1–CNN1). The plates were incubated for 2 d (glucose) or 3 d (galactose) at 23 °C. (c) A lack or slightly elevated levels of Cnn1 lead to mild chromosome loss. Chromosome missegregation was quantified by colony sectoring analysis with a centromeric ade3-2 reporter plasmid32. In the cnn1Δ and PGAL1–CNN1 strains, a 2–3-fold increase in chromosome loss was measured in five independent experiments (n = 5). Kinetochore mutant ctf19Δ acted as the reference. The error bars= represent s.d. Uncropped images of blots are shown in Supplementary Fig. S7.
Combined mutations causing additive effects on cell growth may point to a functional relationship between the mutated proteins. To determine how Cnn1 supports kinetochore activity, we probed cnn1Δ and pgal1–CNN1 for genetic interactions with all yeast deletion mutations (SGA collection; ref. 33) and with temperature-sensitive mutations in essential centromere, kinetochore and spindle components (Table 1). The screens against the non-essential mutations did not reveal a single genetic interaction, not even with any of the six SAC mutations present in the SGA collection. This indicated that the SAC is satisfied in the cnn1Δ and pgal1–CNN1 strains. In contrast, mild positive interactions (decreased fitness) were observed between cnn1Δ and ndc10-1, cse4-1 and mif2-3. Very strong positive interactions were detected between cnn1Δ and temperature-sensitive mutations in the COMA, Mtw1, Spc105, Ndc80 and Dam1 kinetochore complexes (for example, cnn1Δ nnf1-17; Fig. 5a). pgal1–CNN1 positively interacted with mutations in the Spc105 and Ndc80 complexes. No interactions were found with mutations in α- and β-tubulin, or in the centromere-associated factors cohesion and topoisomerase II (Table 1). In conclusion, Cnn1 supports the activity of the KMN network and, consequently, of the complexes that functionally interact with it (COMA, Dam1 complexes; refs 14,34,35).
Table 1.
Genetic interactions between cnn1Δ, pgal1–CNN1 and mutations in essential members of the mitotic apparatus.
| Localization | Complex | Allele | cnn1Δ | pgal1–CNN1 |
|---|---|---|---|---|
| Centromere | Topoisomerase II | top2-1 | o | o |
| top2-4 | o | o | ||
| Cohesin | scc1-73 | o | n.d. | |
| CBF3 complex | ndc10-1 | + | o | |
| CEN nucleosome | cse4-1 | + | o | |
| Mif2 dimer | mif2-3 | + | o | |
|
| ||||
| Lower kinetochore | COMA complex | okp1-5 | +++ | o |
| ame1-4 | +++ | n.d. | ||
|
| ||||
| Central kinetochore | MIND complex | nnf1-17 | ++ | o |
|
| ||||
| Outer kinetochore | Spc105 complex | spc105-15 | ++ | +++ |
| Ndc80 complex | ndc80-1 | + | ++ | |
| spc24-2 | o | o | ||
| Dam1 complex | dam1-1 | +++ | n.d. | |
| Stu2 dimer | stu2-277 | o | o | |
|
| ||||
| Mitotic spindle | α-tubulin | tub1-724 | o | n.d. |
| β-tubulin | tub2-423 | o | n.d. | |
Interactions were either positive (+, decreased fitness of the double mutant) or absent (o, unaltered fitness of the double mutant). No negative interactions (improved fitness of the double mutant) were observed. n.d., not determined. Interactions were very strong (+++), strong (++) or moderate (+). The data were obtained from growth analysis (serial dilutions) and/or tetrad dissection studies at temperatures ranging from 15 °C to 37 °C.
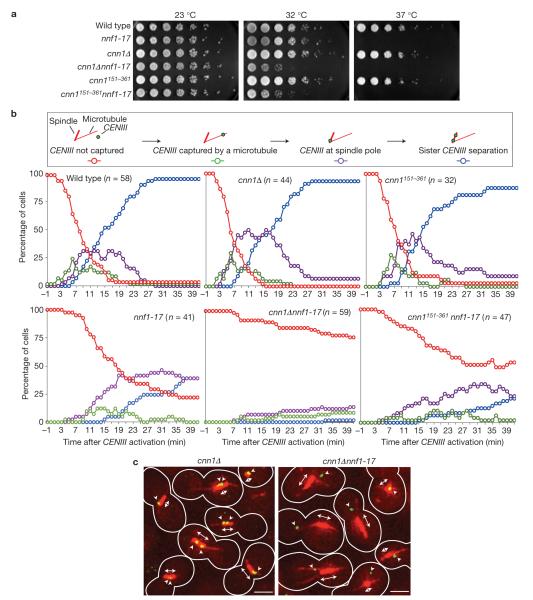
Figure 5.
Cnn1 promotes the sister-chromatid/spindle binding and bi-orientation activity of the KMN network. (a) cnn1 genetically interacts with nnf1-17. The wild-type, nnf1-17, cnn1Δ, cnn1151–361, cnn1Δ nnf1-17 and cnn1151–361nnf1-17 strains were serially diluted on 2% glucose YP agar and incubated at 23 °C (permissive), 32 °C (semi-permissive) and 37 °C (non-permissive) for 2 days. (b) Cnn1 supports sister-chromatid/microtubule binding and bi-orientation. In wild-type, nnf1-17, cnn1Δ, cnn1151–361, cnn1Δ nnf1-17 and cnn1151–361 nnf1-17 strains, CENIII was placed under the control of PGAL1 and marked with 112 tetO2 tandem repeats. TETR–3ECFP was expressed from PURA3 to label CENIII fluorescently. Furthermore, tubulin was marked (1GFP–Tub1) and CDC20 was placed under the control of the methionine-repressible PMET3. Following growth in methionine dropout medium containing 2% raffinose, the cells were shifted to YP medium containing 2% galactose, 2% raffinose and 2 mM methionine (23 °C). Next, the temperature was set to 35 °C and the cells arrested in metaphase (owing to repression of CDC20 expression) without a kinetochore on CENIII (transcription from PGAL1 across CENIII prevented kinetochore formation on CENIII). They were immobilized in a glass-bottom dish and cultured in 2% glucose YP medium containing methionine (2 mM) to repress CENIII transcription. This triggered kinetochore formation on CENIII. Sister-chromatid-III/microtubule binding (green), its localization to the spindle pole (purple) and bi-orientation on the spindle (blue) were tracked for 41 min by time-lapse Deltavision deconvolution microscopy2.The plots show averaged values measured in the indicated number (n) of cells. Red, sister chromatid III not captured by a microtubule; green, sister chromatid III captured by a microtubule; purple, sister chromatid III at spindle pole; blue, CENIII regions separated. (c) Still images from the CENIII-activation experiment plotted in b. Images depict cells (25 min after CENIII activation) with bi-oriented, mono-oriented or unbound sister chromatid III. Red, tubulin; double-headed arrow, spindle (red); arrowheads, CENIII (green dot). Scale bars, 3 μm.
To determine how Cnn1 supports KMN activity, we phenotyped the synthetic relationship between cnn1Δ and nnf1-17 (Fig. 5a). In our study, we further included a cnn1 mutant lacking the first 150 residues (cnn1151–361) through which Cnn1 interacts with the Ndc80 complex (Fig. 3b). In a plate assay, the cnn1151–361 nnf1-17 strain showed the same growth phenotype as the cnn1Δ nnf1-17 mutant (Fig. 5a). To dissect the contribution of Cnn1 to kinetochore activity, we tracked microtubule attachment and bi-orientation of CFP-marked CENIII following CENIII activation2 in the wild-type, cnn1Δ, cnn1151–361, nnf1-17, cnn1Δ nnf1-17 and cnn1151–361nnf1-17 strains by time-lapse fluorescence imaging (Fig. 5b,c). When compared to the wild-type strain, the cnn1 and cnn1151–361 mutants did not show marked changes in CENIII–microtubule capture or sister-chromatid separation dynamics. In the nnf1-17 mutant, reduced CENIII–microtubule attachment kinetics, increased mono-orientation and, consequently, impaired sister-chromatid separation were observed. When Cnn1 activity was removed from the nnf1-17 strain, the ability of the cells to attach sister chromatid III to the spindle was almost completely abrogated. In the nnf1-17 strain lacking the N terminus of Cnn1, CENIII–microtubule capture occurred more frequently than in the cnn1Δ nnf1-17 strain. However, we observed in 11% of the cnn1151–361nnf1-17 cells that the bound sister chromatid III quickly detached from the spindle microtubules, pointing to a reduced kinetochore–microtubule affinity. Taken together, our findings indicate that Cnn1 and its N terminus support sister-chromatid/spindle attachment and bi-orientation. Although Cnn1 is not essential for viability, its activity becomes indispensable when kinetochores are structurally or functionally compromised (Table 1 and Fig. 5a,b).
Cnn1 inhibits the interaction between the KMN complexes
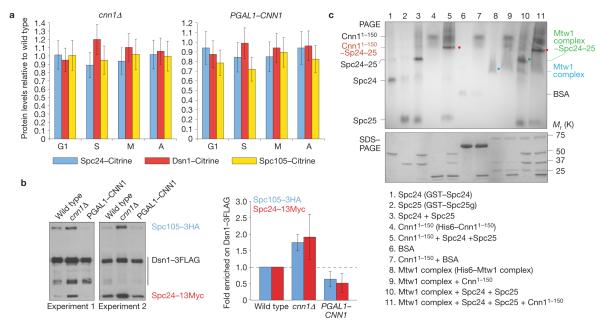
As Cnn1 localizes to the base rather than the spindle interface of the KMN network, it could support kinetochore activity by promoting KMN complex recruitment or by modulating the structural configuration of the network. To examine the first hypothesis, we measured the fluorescence levels of Dsn1–Citrine, Spc24–Citrine and Spc105–Citrine at kinetochores in wild-type, cnn1Δ and pgal1–CNN1 strains grown in 2% galactose medium. At each cell cycle stage, the kinetochore levels of each KMN component were statistically the same in the three strains (Fig. 6a), indicating that Cnn1 does not support the recruitment of KMN proteins to centromeres. Next, we examined KMN network integrity in wild-type, cnn1Δ and pgal1–CNN1 strains endogenously expressing Dsn1–3FLAG, Spc24–13Myc and Spc105–3HA. When Dsn1–3FLAG was purified from cells grown in 2% galactose medium, we reproducibly observed that in the cnn1Δ strain, the levels of Spc24–13Myc and Spc105–3HA bound to Dsn1–3FLAG were twice those bound to Dsn1–3FLAG in the wild-type. In contrast, the concentrations of Spc24–13Myc and Spc105–3HA bound to Dsn1–3FLAG in the pgal1–CNN1 extract were half those measured in the wild type (Fig. 6b). These findings indicated that Cnn1 negatively regulates the interaction between the Mtw1, Ndc80 and Spc105 complexes. To corroborate this conclusion, we examined in vitro the interaction between the N terminus of Cnn1 (first 150 residues), the recombinant Mtw1 complex36 and recombinant Spc24 and Spc25. The binding reactions were analysed by native PAGE and by denaturing SDS–PAGE to confirm proteins, complexes and protein ratios (Fig. 6c). Spc24 and Spc25 formed a dimer to which Cnn11–150 stoichiometrically bound. The Mtw1 complex stoichiometrically bound to Spc24–Spc25 but did not bind to Cnn11–150. When Cnn11–150, the Mtw1 complex, Spc24 and Spc25 were incubated in equimolar amounts, we reproducibly observed that the Mtw1 complex was excluded from a complex comprising Cnn11–150-Spc24–Spc25 (Fig. 6c). This did not change when Mtw1 levels were increased (data not shown). Taken together, our biochemical data (Fig. 6b,c) show that the binding of Cnn1 to Spc24–Spc25 prevents the Mtw1 and Spc105 complexes from binding to the Ndc80 complex.
Figure 6.
Cnn1 inhibits the interaction between the KMN complexes. (a) Cnn1 does not recruit KMN complexes to centromeres. Endogenously expressed Dsn1–Citrine, Spc24–Citrine and Spc105–Citrine were quantified by spinning-disc confocal fluorescence imaging in asynchronous cultures of wild-type, cnn1Δ and PGAL1–CNN1 strains (2% galactose minimal medium, 23 °C). Cell-cycle stages are indicated; G1, S phase, metaphase (M) and anaphase (A). Fluorescence levels were normalized to those measured in the wild-type strain (value = 1.0). Spc110–mCherry acted as the internal reference. For each measurement, 50 kinetochores were analysed (n = 50). The error bars represent s.d. (b) Cnn1 inhibits the binding between the Mtw1, Spc105 and Ndc80 complexes. Spc105–3HA and Spc24–13Myc co-purifying with Dsn1–3FLAG from wild-type, cnn1Δ and PGAL1–CNN1 stains (2% galactose YP medium, 23 °C) were identified by anti-FLAG, anti-HA and anti-Myc western hybridization (left panels). The levels of Spc105–3HA and Spc24–13Myc co-purifying with Dsn1–3FLAG were quantified (ImageJ 1.43u) and normalized to those of Dsn1–3FLAG (right plot). The error bars represent s.d. (c) Cnn11–150 prevents the Mtw1 complex (Dsn1, Mtw1, Nsl1 and Nnf1) from binding to the Spc24–Spc25 dimer. Epitope-tagged recombinant proteins were incubated in various combinations as indicated in the rectangle (Spc25g, global domain of Spc25; residues 128–222). The single proteins and reaction mixtures were analysed by non-denaturing PAGE to visualize the relative positions of input proteins and formed complexes (indicated), and by denaturing SDS–PAGE to confirm protein purity and concentrations. Proteins were visualized with Gelcode blue. BSA, bovine serum albumin. Uncropped images of blots and gels are shown in Supplementary Fig. S7.
DISCUSSION
Advanced bioinformatics have indicated that Cnn1 is the yeast orthologue of metazoan kinetochore protein CENP-T (ref. 37). CENP-T is a subunit of the CCAN protein assembly that is constitutively associated with centromeric chromatin38. CENP-T and the CCAN member CENP-W form a dimer of histone-fold proteins that together with CENP-S and CEN-X associate into a nucleosome39 that is located next to the centromere-defining CENP-A-containing nucleosome1. CENP-T and, in parallel, CENP-C orchestrate kinetochore assembly downstream of CENP-A. CENP-T, through its N terminus, recruits the NDC80Ndc80 complex40, and CENP-CMif2 recruits the MIS12Mtw1 complex41,42. In metazoans, the NDC80Ndc80 and KNL1Spc105 complexes require the MIS12Mtw1 complex to localize to kinetochores43. Without CENP-T or CENP-CMif2, no functional kinetochores are formed40. In budding yeast, however, the Mtw1, Ndc80 and Spc105 complexes are recruited in parallel by the CBF3 complex29.
We show that, similarly to CENP-T, Cnn1 localizes to the lower region of the KMN network. Cnn1 and CENP-T share a carboxy-terminal histone-fold motif through which CENP-T localizes to centromeres39,40. Through their N termini, CENP-T and Cnn1 bind to the Ndc80 complex. In yeast, this interaction does not serve to recruit the Ndc80 complex, as its levels at kinetochores are not affected in the absence of Cnn1. This is consistent with Cnn1 not being essential for viability. Indeed, in budding yeast, essential kinetochore proteins localize to kinetochores through other essential kinetochore proteins14,29. Rather than serving to recruit the Ndc80 complex, we find that the Cnn1–Ndc80 complex interaction serves to prevent the Mtw1 and Spc105 complexes from binding to the Ndc80 complex. We found no evidence that Cnn1 directly binds to the Mtw1 or Spc105 complexes. We believe that the negative activity of Cnn1 towards the Mtw1 and Spc105 complexes serves to regulate the formation, structural configuration and, consequently, activity of the KMN network in a spatiotemporal manner.
From interphase to metaphase, yeast kinetochores contain Cnn1, the Mtw1, Spc105 and Ndc80 complexes in 6, 15, 15 and 19 copies, respectively (the levels of the yeast KMN complexes do not change during the cell cycle; A. Oldani and P. De Wulf, unpublished observations). Hence, Cnn1 probably has a minor inhibitory effect on the association of the KMN complexes during these cell-cycle stages. This may promote kinetochore assembly, sister-chromatid/spindle binding and bi-orientation. At anaphase onset, when cells have irrevocably committed to segregating their chromatids, Cnn1 (20 copies per kinetochore) accumulates to levels equalling those of the Ndc80 complex (19 copies per kinetochore), thereby exceeding those of the Mtw1 and Spc105 complexes (15 copies each)16,17, allowing for a competitive binding of Cnn1 to the Ndc80 complex. This could translate to a less firm configuration of the KMN network, possibly allowing for an optimal kinetochore–microtubule coupling and force transduction from microtubule depolymerization through the Dam1 complex, as required for chromosome segregation34,35.
A negative effect on kinetochore complex–complex interactions, similar to that observed for Cnn1, was recently demonstrated for the kinetochore protein Ybp2, whose absence from yeast promoted the binding between the Ndc80 and COMA complexes44. This observation indicates that Cnn1 and Ybp2 may act in overlapping pathways and that negative regulation of complex interactions within kinetochores may be a common strategy of yeast to modulate kinetochore structure and activity during the cell cycle. Whether the kinetochore localization of Ybp2 changes during the cell cycle is unknown.
The importance of a timely activity of Cnn1 at KMN is underlined by the Cdc28, Mps1 and Ipl1 kinases acting on Cnn1. Their combined activity establishes a Cnn1 phospho-threshold that triggers the enrichment of Cnn1 in anaphase. The subsequent dephosphorylation and dissociation of Cnn1 from KMN at mitotic exit may lead to a more rigid KMN network in kinetochores that no longer undergo spindle forces. The N terminus of CENP-T, similarly to Cnn1, is subject to phosphorylation by CDK1Cdc28, an event that supports its interaction with the NDC80 complex40.
Our data and those recently published in vertebrates39,40 indicate that conserved CCAN members such as CENP-T and Cnn1 may have evolved from having a leading role in orchestrating kinetochore recruitment (metazoans) to mediating a supporting role in yeast, aimed at regulating the interactions between already recruited complexes, thereby fine-tuning kinetochore structure and activity in time and space.
METHODS
Yeast strains and manipulations
All strains have a W303a background and are MATa, unless stated otherwise. Genetic manipulations and growth conditions were as described previously14. The Cnn1–3GFP phospho-mutants and cnn1151–361 constructs were made on an integrative vector and recombined in single copy at the promoter region of CNN1 in a cnn1Δ strain.
ChIP and ChIP–chip
ChIPs were performed as described previously14. Temperature-sensitive mutants were grown at 23 °C, possibly followed by a 3 h shift to 37 °C or an overnight shift to 15 °C (tub1-724). Immunoprecipitations were performed with an anti-Myc monoclonal antibody (9E11, Covance) and an anti-Cep3 polyclonal antibody (P. K. Sorger, Harvard Medical School, USA). CENIV was amplified by PCR (ExTaq polymerase, Takara) with oligomers 5′-GCGCAAGCTTGCAAAAGGTCACATG-3′ and 5′-CGAATTCATTTTGGCCGCTCCTAGGTA-3′ visualized by GelRed agarose gel electrophoresis and quantified with ImageJ 1.43u (NIH).
ChIP–chip analysis was performed as described previously45. Amplified chromatin was hybridized to the S. cerevisiae Genome Tiling Array 1.0R (Affymetrix), analysed with GeneChip Command Console software (Affymetrix) and deposited to the Gene Expression Omnibus (GSE31567).
Fluorescence microscopy
Spinning-disc confocal fluorescence imaging was performed with an UltraVIEW VoX spinning-disc confocal unit (PerkinElmer), equipped with an Eclipse Ti inverted microscope (Nikon) and a C9100-50 electron-multiplying CCD (charge-coupled device) camera (Hamamatsu). All components were controlled by Volocity software (PerkinElmer). Images (×100 immersion oil objective, NA 1.49) were acquired as Z stacks (21 slices of 0.2 μm) at 488 nm (GFP; 527/55 nm band-pass filter) and at 561 nm (mCherry; 615/70 nm band-pass filter). Following maximum axial projection of the stacks, signal intensities were quantified after subtracting the background signal (ImageJ 1.43u, NIH).
FRAP experiments were performed using a spinning-disc confocal fluorescence microscope connected to a FRAP Photokinesis Unit (PerkinElmer). Z stacks were collected every 1 s (2 min tracking) or every 10 s (10 min tracking) using the set-up described above.
Time-lapse fluorescence imaging of the Cnn1–3GFP Spc110–mCherry strains was performed with a Deltavision RT microscope (Applied Precision), Olympus UPlanSApo (×100 immersion oil objective lens, NA 1.40), SoftWoRx software (Applied Precision) and a CoolSnap HQ (Photometrics, USA) CCD camera. Cells were imaged at 488 nm (Cnn1–3GFP) and 561 nm (Spc110–mCherry) through seven Z stacks (0.7 μm each). The stacks were deconvoluted and analysed with Volocity and ImageJ 1.43u.
Real-time CENIII-reactivation analysis2 was performed by Deltavision RT deconvolution fluorescence imaging (see above). CENIII was placed under the control of PGAL1 and marked by TetR–ECFP (constitutively expressed from PURA3) that bound to a CENIII-flanking × 112tetO2 array. Tubulin was labelled (1GFP–Tub1) and CDC20 was placed under the control of the methionine-repressible PMET3. After growth in 2% raffinose-based methionine dropout medium, the cells were shifted to YP medium complemented with methionine (2 mM), 2% galactose and 2% raffinose, leading to a metaphase arrest of the cells, which lacked a kinetochore on CENIII (23 °C). After a shift to 2 mM methionine 2% glucose YP medium (35 °C), kinetochore assembly on CENIII was induced and CENIII–microtubule binding and sister-chromatid-III bi-orientation were tracked (41 min).
Hydrodynamics
Size-exclusion chromatography (Superose 6 10/300, GE Healthcare) and 10–40% glycerol sedimentation velocity ultracentrifugation analysis (SW41Ti, Beckman) of Cnn1–13Myc yeast cell extract and protein standards with known Stokes radii and Svedberg coefficients were performed as described previously27. Cnn1–13Myc was identified by anti-Myc (9E10, Covance) western hybridization. The mass, frictional coefficient and axial ratio of Cnn1–13Myc were calculated from its Stokes radius and Svedberg constant27.
Cell-cycle analysis
Yeast cultures were synchronized in late G1 with 5 μg ml−1 α-factor (PrimmBiotech). The cells were then filtered, washed and transferred to medium free of α-factor. Ninety minutes after the release, 2.5 μg ml−1 α-factor was added to prevent the cells from entering a second cell cycle. Cell-cycle progression was assayed by DNA content (FACS, Becton Dickinson FACScan) and spindle morphology (rat anti-Tub1 (Oxford Biotechnology) indirect immunofluorescence).
Protein interaction analysis
For affinity purifications, CNN1–3FLAG and GST–CNN1 were expressed from PGAL1 in 2% galactose YP medium. Following cell extraction, Cnn1 was purified with anti-FLAG M2 agarose resin (Sigma) or 50% glutathione Sepharose 3 Fast Flow beads (GE Healthcare). Cnn1 and interactors were released with 3FLAG peptide (Sigma-Aldrich) or by cleavage with Precision protease (Cogentech), separated by 12.5% SDS–PAGE, stained with colloidal blue (Invitrogen) and identified by microcapillary liquid chromatography–tandem mass spectrometry (Cogentech).
Y2H screens were performed with ten CNN1 constructs cloned into the NcoI/PvuII sites (N-terminal Gal4 BD fusion) or into the NruI site (C-terminal fusion) of pBDC (ref. 26). The constructs were screened against a genomic library of ORFs C-terminally tagged with the Gal4 DNA activation domain26.
To study the affinity between Dsn1, Spc105 and Spc24, the endogenous proteins were provided with a C-terminal 3FLAG, 3HA and 13Myc epitope, respectively, in the wild-type, cnn1Δ and pgal1–CNN1 strains. Extracts (glass beads; FastPrep, MP Biomedicals) of exponentially growing cells (D595 nm = 1.0) were incubated (2 h, 4 °C) with anti-FLAG M2 agarose resin (Sigma-Aldrich) = to purify Dsn1–3FLAG. Following five washes with detergent and salt solutions, Dsn1–3FLAG, and co-purifying Spc105–3HA and Spc24-13Myc were separated by 8% SDS–PAGE and visualized by anti-FLAG (Cogentech), anti-HA (MMS-101R, Covance) and anti-Myc western hybridization. Signals were quantified with ImageJ 1.43u.
Co-immunoprecipitations of Cnn1–13Myc and Cnn1–Pk6 from whole-cell yeast extract and subsequent western hybridization analyses were performed with anti-Myc and anti-Pk6 (MCA1360, AbD Serotec) antibodies (1/50).
To study the interaction between recombinant Cnn11–150, Spc24, Spc25 and the Mtw1 complex, we cloned His6–CNN11–150 in pET28b (EMD Bioscience), GST–SPC24 and GST–SPC25 (globular domain; residues 128–222) in pGEX-6P-1 (GE Healthcare) and transformed each into Escherichia coli BL21–DE3. Following induction (0.2 mM IPTG, overnight, 25 °C) and cell lysis (sonication), cell extracts were incubated with Ni-NTA affinity agarose resin (Qiagen) or glutathione agarose beads (Thermo Scientific). Bound proteins were eluted with 200 mM imidazole or 10 mM reduced glutathione (Thermo Scientific). Purified Mtw1 complex was provided by M. Singleton36. The purity of the recombinant proteins was verified by SDS–PAGE and staining with Gelcode blue (ThermoFischer Scientific). Protein concentrations were measured with the BCA Protein Assay Kit (ThermoFischer Scientific). After incubation (10 mM Tris–HCl at pH 8.0 and 150 mM NaCl), the proteins were analysed using native PAGE (4–12% Bis–TrisNuPAGE, Invitrogen) or SDS–PAGE (4–15% Tris–HCl, Biorad). Proteins were visualized with GelCode blue.
Recombinant production, in vitro phosphorylation and phospho-mapping of Cnn1
His6–CNN1 was expressed from pET43 (Novagen) in E. coli Rosetta 2 (0.5 mM IPTG, 15 h, 18 °C) and purified from cell extract with Ni-NTA affinity agarose resin. Following elution (100 mM imidazole) and anion-exchange chromatography (MonoQ HR 5/5, GE Healthcare), the purity of His6–Cnn1 was confirmed by SDS–PAGE and Coomassie blue staining. GST–MPS1 was expressed in E. coli BL21 (DE3)plysS cells; GST–IPL1 and GST–SLI15 were expressed in E. coli C43plysS cells. The proteins were purified using glutathione Sepharose 4 Fast Flow affinity agarose (GE Healthcare). In the radioactive kinase assays, His6–Cnn1 (1 μg) was incubated (30 °C, 30 min) with Mps1 (0.1 μg), Ipl1 (0.2 μg) plus Sli15 (0.4 μg) and γ32P-ATP (1 μM) in 50 mM Tris–HCl at pH 7.5, 1 mM dithiothreitol, 5 mM MgCl2, and 25 mM -glycerophosphate. The proteins were separated by SDS–PAGE and detected on a FLA-5100 phosphor screen (Fujifilm). In the non-radioactive kinase assay, His6–Cnn1 (2 μg) was incubated (30 °C, 2 h) with Mps1 (0.2 μg), or Ipl1 (0.4 μg) plus Sli15 (0.8 μg) and ATP (1 μM). Next, His6–Cnn1 was reduced (45 mM dithiothreitol), alkylated (10 mM iodoacetamide) and trypsin-digested (37 °C, 16 h). The peptides were desalted (HYPERSEP C18, ThermoFisher Scientific), mixed with 10 mM NaH2PO4 (pH 3) and 25% acetonitrile, and passed through a HYPERSEP SCX column (ThermoFisher Scientific). The column was washed with 10 mM NaH2PO4 (pH 3) and 25% acetonitrile. Bound peptides were eluted (10 mM NaH2PO4 at pH 3, 25% acetonitrile and 350 mM KCl) and the eluate plus flow-through fractions then desalted (HYPERSEP C18 column) and mixed with Titanspheres (GL Sciences) in 80% acetonitrile, 2% trifluoroacetic acid and 200 mg ml−1 dihydroxybenzoate (23 °C, 1 h). The beads were washed three times with this buffer and three times with 80% acetonitrile and 2% trifluoroacetic acid. Peptides were eluted (400 mM ammonia) and the pH of the eluate was adjusted to pH 2 (formic acid). SCX and Titansphere eluates were analysed by LTQ-Orbitrap Velos (ThermoFisher Scientific) by multi-stage activation (analysis of neutral-loss masses of 24.4941, 32.6588, 48.9882 and 97.9763 m/z). Raw data were analysed by MaxQuant48, allowing one trypsin miscleavage, serine/threonine phosphorylation, methionine oxidation and N-terminal acetylation as modifications.
Serial dilution growth analysis
Cells grown overnight were back-diluted to D595 nm = 1.0, diluted (1:4.5) in multi-well plates and transferred onto appropriate agar medium using a replica plater. The plates were incubated at the appropriate temperature for 2–3 d.
Chromosome-loss analysis
Chromosome loss was measured with the ade3-2 reporter plasmid32. Three to six colonies of each strain were sonicated and spread at 200 cells per plate on 10 plates of chromosome-loss media32. After growth (23 °C, 5 d) colonies ≥50% white were counted as having lost the plasmid in the first generation following plating. The percentage of chromosome loss per generation was calculated as the percentage of sectored colonies of the total number of colonies.
Genetic screens
cnn1::NatMX6 and pgal1–CNN1–NatMX6 were screened against the yeast SGA deletion collection as described previously33. Growth of the double mutants was scored at 23 °C and 37 °C on 2% glucose and 2% galactose YP agar media containing or lacking 8 μg ml−1 benomyl. Screens against mutations in essential kinetochore proteins, tubulin, cohesin and topoisomerase II were performed by mating and tetrad dissection. Double mutants were grown on 2% glucose or 2% galactose YP agar medium at temperatures between 15 °C and 37 °C to identify semi-permissive growth conditions.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank A. Amon (MIT, USA), S. Biggins (University of Washington, USA), D. Koshland (University of California, Berkeley, USA), M. Singleton (Cancer Research UK), F. Solomon (MIT, USA) and P. K. Sorger (Harvard Medical School, USA) for strains or reagents. We also thank M. Foiani, S. Ghandi, D. Lamont, J. Lechner, C. Lucca, J. Ortiz, S. ten Have and R. Visintin for technical support or discussions. We thank A. Schleiffer and S. Westermann for communicating results before publication. P.D.W. acknowledges financial support from the Italian Association for Cancer Research (grant 8840). T.R.H. recognizes support from the N.I.H. (grant GM087461) and the American Cancer Society (grant IRG 58-006-50). T.U.T. acknowledges a Cancer Research U.K. senior fellowship and Wellcome Trust program grant. L.J.B. acknowledges a doctoral fellowship from the European School of Molecular Medicine.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Note: Supplementary Information is available on the Nature Cell Biology website
References
- 1.Lampert F, Westermann S. A blueprint for kinetochores—new insights into the molecular mechanics of cell division. Nat. Rev. Mol. Cell Biol. 2011;12:407–412. doi: 10.1038/nrm3133. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka K, et al. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka TU. Kinetochore-microtubule interactions: steps toward bi-orientation. EMBO J. 2010;29:4070–4082. doi: 10.1038/emboj.2010.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 5.Espeut J, et al. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J. Cell Biol. 2012;196:469–482. doi: 10.1083/jcb.201111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka TU, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 7.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 8.Maure J-F, Kitamura E, Tanaka TU. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Curr. Biol. 2007;17:2175–2182. doi: 10.1016/j.cub.2007.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santaguida S, et al. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maresca TJ, Salmon ED. Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. J. Cell Sci. 2010;123:825–835. doi: 10.1242/jcs.064790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foley EA, Maldonado M, Kapoor TM. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 2011;13:1265–1271. doi: 10.1038/ncb2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meadows JC, et al. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev. Cell. 2011;20:739–750. doi: 10.1016/j.devcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg JS, Cross FR, Funabiki H. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr. Biol. 2011;21:942–947. doi: 10.1016/j.cub.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 16.Coffman VC, Wu P, Parthun MR, Wu J-Q. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J. Cell Biol. 2011;195:563–572. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ubersax JA, et al. Targets of the cyclin dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 19.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 20.Breitkreutz A, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheeseman IM, et al. Phospho-regulation of kinetochore-microtubule attachments by the aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 22.De Wulf P, Montani F, Visintin R. Protein phosphatases take the mitotic stage. Curr. Opin. Cell Biol. 2009;21:806–815. doi: 10.1016/j.ceb.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc. Natl Acad. Sci. USA. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lingelbach LB, Kaplan KB. The interaction between Sgt1p and Skp1p is regulated by HSP90 chaperones and is required for proper CBF3 assembly. Mol. Cell. Biol. 2004;24:8938–8950. doi: 10.1128/MCB.24.20.8938-8950.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blake-Hodek KA, Cassimeris L, Huffaker TC. Regulation of microtubule dynamics by Bim1 and Bik1, the budding yeast members of the EB1 and CLIP-170 families of plus-end tracking proteins. Mol. Biol. Cell. 2010;21:2013–2023. doi: 10.1091/mbc.E10-02-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong J, et al. A protein interaction map of the mitotic spindle. Mol. Biol. Cell. 2007;18:3800–3809. doi: 10.1091/mbc.E07-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuyler SC, Pellman D. Analysis of the size and shape of protein complexes from yeast. Meth. Enzymol. 2002;351:150–168. doi: 10.1016/s0076-6879(02)51845-0. [DOI] [PubMed] [Google Scholar]
- 28.Gillett ES, Espelin CW, Sorger PK. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pagliuca C, Draviam VM, Marco E, Sorger PK, De Wulf P. Roles for the conserved Spc105p-Kre28p complex in kinetochore-microtubule binding and the spindle assembly checkpoint. PLoS ONE. 2009;4:e7640. doi: 10.1371/journal.pone.0007640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krenn V, et al. Structural analysis reveals features of the spindle checkpoint kinase Bub1-kinetochore subunit Knl1 interaction. J. Cell Biol. 2012;196:451–467. doi: 10.1083/jcb.201110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koshland D, Hieter P. Visual assay for chromosome ploidy. Meth. Enzymol. 1987;155:351–372. doi: 10.1016/0076-6879(87)55024-8. [DOI] [PubMed] [Google Scholar]
- 33.Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Meth. Mol. Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- 34.Tien JF, et al. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 2010;189:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maskell DP, Hu X-W, Singleton MR. Molecular architecture and assembly of the yeast kinetochore MIND complex. J. Cell Biol. 2010;190:823–834. doi: 10.1083/jcb.201002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schleiffer A, et al. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 2012 doi: 10.1038/ncb2493. http://dx.doi.org/10.1038/ncb2493. [DOI] [PubMed]
- 38.Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- 39.Nishino T, et al. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 2012;148:487–501. doi: 10.1016/j.cell.2011.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gascoigne KE, et al. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Przewloka MR, et al. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 42.Screpanti E, et al. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu S-T, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohkuni K, Abdulle R, Tong AH, Boone C, Kitagawa K. Ybp2 associates with the central kinetochore of Saccharomyces cerevisiae and mediates proper mitotic progression. PLoS ONE. 2008;3:e1617. doi: 10.1371/journal.pone.0001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bermejo R, Katou YM, Shirahige K, Foiani M. ChIP-on-chip analysis of DNA topoisomerases. Meth. Mol. Biol. 2009;582:103–118. doi: 10.1007/978-1-60761-340-4_9. [DOI] [PubMed] [Google Scholar]
- 46.Akiyoshi B, et al. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–579. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho RJ, et al. Parallel analysis of genetic selections using whole genome oligonucleotide arrays. Proc. Natl. Acad. Sci. USA. 1998;95:3752–3757. doi: 10.1073/pnas.95.7.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.