Abstract
(G2019S) mutation of leucine-rich repeat kinase 2 (LRRK2) is the most common genetic cause of both familial and sporadic Parkinson's disease (PD) cases. Twelve- to sixteen-month-old (G2019S) LRRK2 transgenic mice prepared by us displayed progressive degeneration of substantia nigra pars compacta (SNpc) dopaminergic neurons and parkinsonism phenotypes of motor dysfunction. LRRK2 is a member of mixed lineage kinase subfamily of mitogen-activated protein kinase kinase kinases (MAPKKKs). We hypothesized that (G2019S) mutation augmented LRRK2 kinase activity, leading to overphosphorylation of downstream MAPK kinase (MKK) and resulting in activation of neuronal death signal pathway. Consistent with our hypothesis, (G2019S) LRRK2 expressed in HEK 293 cells exhibited an augmented kinase activity of phosphorylating MAPK kinase 4 (MKK4) at Ser257, and protein expression of active phospho-MKK4Ser257 was upregulated in the SN of (G2019S) LRRK2 transgenic mice. Protein level of active phospho-JNKThr183/Tyr185 and phospho-c-JunSer63, downstream targets of phospho-MKK4Ser257, was increased in the SN of (G2019S) LRRK2 mice. Upregulated mRNA expression of pro-apoptotic Bim and FasL, target genes of phospho-c-JunSer63, and formation of active caspase-9, caspase-8 and caspase-3 were also observed in the SN of (G2019S) LRRK2 transgenic mice. Our results suggest that mutant (G2019S) LRRK2 activates MKK4-JNK-c-Jun pathway in the SN and causes the resulting degeneration of SNpc dopaminergic neurons in PD transgenic mice.
Keywords: Parkinson's disease, (G2019S) LRRK2, dopaminergic neurons, MKK4, JNK
Patients with familial type 8 of Parkinson′s disease (PARK8) exhibit late-onset parkinsonism symptoms and autosomal dominant inheritance.1, 2 Initial linkage studies mapped PARK8 gene to the chromosome 12p11.2-q13.1 region.3 Subsequent sequencing of candidate genes identified missense mutations in leucine-rich repeat kinase 2 (LRRK2) as the cause of PARK8.4, 5 LRRK2 gene encodes a 2527-amino-acid protein with a predicted molecular weight of ∼280 kDa. LRRK2 consists of several functional domains, including ankyrin repeat region, leucine-rich repeat (LRR) domain, ROC (Ras of complex proteins) GTPase domain, C-terminal of ROC (COR) domain, kinase domain related to mitogen-activated protein kinase kinase kinase (MAPKKK) and C-terminal WD40 region.6, 7 LRRK2 expression is found in several brain regions implicated in the neuropathology of Parkinson′s disease (PD), such as substantia nigra (SN), caudate putamen, and cerebral cortex.8, 9
LRRK2 mutations account for 5–13% of familial PD and 1–5% of sporadic PD.6, 10 Therefore, LRRK2 mutation is the most common genetic cause of both familial and sporadic PD cases. PD-associated mutations are found in every domain of LRRK2.6, 7 For example, (I2012T), (G2019S), and (I2020T) mutations are located in the kinase domain, (I1122V) mutation in the LRR domain, (R1441C) and (R1441G) mutations in the ROC domain, (Y1699C) mutation in the COR domain, and (G2385R) in the WD 40 domain. Among LRRK2 mutations found in PD patients, G2019S is the most prevalent amino-acid substitution.10, 11, 12 Neuropathological examination showed a loss of dopaminergic neurons in the substantia nigra pars compacta (SNpc) of PD patients with (G2019S) LRRK2 mutation.11, 12, 13, 14 Thus, mutant (G2019S) LRRK2 causes neurodegeneration of SNpc dopaminergic cells and resulting in parkinsonism.
In-vitro studies demonstrated that LRRK2 induced Ser/Thr phosphorylation but not Tyr phosphorylation and that (G2019S) mutation augmented LRRK2 kinase activity.15, 16, 17 Thus, (G2019S) mutation is believed to exert a gain-of-function and activating effect on LRRK2 kinase activity. Kinase domain of LRRK2 closely resembles that of mixed lineage kinases (MLKs), which are members of a large family of MAPKKKs and mediate cellular stress responses.6, 18, 19 MLK family of MAPKKKs activates neuronal death signal pathway by phosphorylating and activating downstream MAPK kinases (MKKs), which subsequently induces the phosphorylation and activation of c-Jun N-terminal kinases (JNKs), essential mediators of neuronal death observed in various neurodegenerative diseases.18, 19 MKK4-JNK pathway has been shown to mediate the death of SNpc dopaminergic neurons caused by 6-hydroxydopamine injection and medial forebrain bundle axotomy, which are performed to prepare PD animal models.20, 21 Thus, it is possible that (G2019S) LRRK2-induced aberrant activation of MKK4-JNK pathway causes degeneration of SNpc dopaminergic neurons.
A mouse model, which replicates (G2019S) LRRK2-induced degeneration of SNpc dopaminergic neurons, should be a valuable tool to investigate pathogenic mechanism of familial and sporadic PD caused by (G2019S) LRRK2 mutation. In the present study, we prepared an animal model of PD by generating transgenic mice expressing mutant (G2019S) LRRK2. Similarly to (G2019S) LRRK2 PD patients, (G2019S) LRRK2 transgenic mice exhibited degeneration of SNpc dopaminergic neurons and PD neurological phenotypes. Our results also suggest that mutant (G2019S) LRRK2 activates MKK4-JNK pathway in the SN and causes the resulting degeneration of SNpc dopaminergic neurons in PD transgenic mice.
Results
Generation of transgenic mice expressing mutant (G2019S) orwild-typeLRRK2
In the present study, we prepared a PD animal model by generating transgenic mice expressing HA-tagged disease-causing (G2019S) LRRK2. As a control, transgenic mice expressing HA-tagged wild-type LRRK2 were also generated. Previous studies by us and others showed that a high level of PDGF-β promoter-mediated neuronal transgene expression was observed in the cerebral cortex, striatum, and SN.22, 23, 24 Furthermore, hybrid CMV enhancer/PDGF-β promoter mediated a high level of transgene expression in SNpc dopaminergic neurons.25 Therefore, the expression of (G2019S) or wild-type LRRK2HA in the transgenic mouse was under transcriptional control of CMV enhancer/PDGF-β promoter. Southern blot analysis was performed to identify founder animals expressing transgene (CMV enhancer/PDGF-β promoter-(G2019S) LRRK2HA or CMV enhancer/PDGF-β promoter-LRRK2HA) and determine approximate copy number. Two CMV enhancer/PDGF-β promoter-(G2019S) LRRK2HA founder mice with a high copy number (10 copies) of transgene were used to establish two stable lines of (G2019S) LRRK2 transgenic mice (lines 32 and 51). As a control, two lines of LRRK2 transgenic mice (lines 36 and 43) were also generated from two CMV enhancer/PDGF-β promoter-LRRK2HA founder mice with 10 copies of transgene.
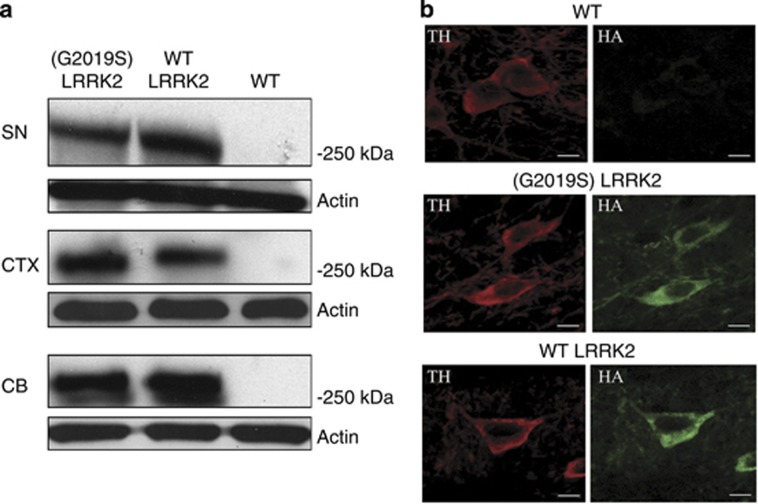
HA-tagged (G2019S) LRRK2 was expressed in various brain regions of (G2019S) LRRK2 mouse, including the cerebral cortex, cerebellum, SN, and striatum (Figure 1a). Similarly to (G2019S) LRRK2 mice, HA-tagged wild-type LRRK2 was found in several brain regions including the SN (Figure 1a) and striatum of LRRK2 transgenic mice. Protein level of wild-type LRRK2HA in the SN of LRRK2 mouse was similar to that of (G2019S) LRRK2HA in the SN of (G2019S) LRRK2 transgenic mouse (Figure 1a).
Figure 1.
Expression of mutant (G2019S) or wild-type LRRK2HA in the brain of transgenic mouse. (a) Western blot analysis using anti-HA antiserum indicated that mutant (G2019S) LRRK2HA or wild-type LRRK2HA (MW=∼280 kDa) was expressed in the SN, cerebral cortex (CTX), or cerebellum (CB) of a 10-month-old transgenic mouse expressing mutant (G2019S) (line 32) or wild-type LRRK2 (line 36). (b) Double immunofluorescence staining using anti-TH antiserum and anti-HA antibody showed that HA-tagged (G2019S) or wild-type LRRK2 was expressed in the cytoplasm of SNpc TH-positive dopaminergic neurons of a 10-month-old (G2019S) LRRK2 (line 32) and LRRK2 transgenic mouse (line 36), respectively. Scale bar is 10 μm
Double immunofluorescence staining using anti-TH antiserum and anti-HA antibody was performed to visualize the expression of HA-tagged (G2019S) or wild-type LRRK2 in SNpc dopaminergic neurons. Consistent with a previous study reporting that CMV enhancer/PDGF-β promoter mediated transgene expression in SNpc dopaminergic cells,25 confocal microscopy showed that HA-tagged (G2019S) or wild-type LRRK2 was found in the cytoplasm of about 70% of SNpc TH-positive dopaminergic neurons in 10-month-old transgenic mice expressing (G2019S) or wild-type LRRK2 (Figure 1b; n=4 animals).
(G2019S) LRRK2 transgenic mice exhibit progressive degeneration of SNpc dopaminergic neurons
Immunohistochemical TH staining was conducted to visualize degeneration of SNpc dopaminergic cells in 8- to 16-month-old (G2019S) LRRK2 transgenic mice. The number of SNpc TH-positive dopaminergic cells of 8- to 9-month-old (G2019S) LRRK2 mice (n=4 animals) was not significantly different from that of wild-type mice at the same age. Compared with 12-month-old wild-type mice, two lines of age-matched (G2019S) LRRK2 mice exhibited a significant reduction in the number of SNpc dopaminergic neurons (Figure 2a). Neuronal death of SNpc dopaminergic cells progressively deteriorated in 16-month-old (G2019S) LRRK2 transgenic mice (Figure 2a). In contrast, the number of SNpc dopaminergic neurons was not significantly altered in two lines of 12- or 16-month-old transgenic mice expressing wild-type LRRK2 (Figure 2a).
Figure 2.
(G2019S) LRRK2 transgenic mice exhibit a progressive and selective degeneration of SNpc dopaminergic neurons. (a) Immunohistochemical TH staining showed that compared with a 12-month-old wild-type mouse, age-matched (G2019S) LRRK2 mice (line 32) exhibited a reduction in the number of SNpc TH-positive dopaminergic neurons. Neuronal death of SNpc dopaminergic cells progressively deteriorated in 16-month-old (G2019S) LRRK2 transgenic mice. Note that degeneration of SNpc dopaminergic neurons was not observed in a 16-month-old transgenic mouse expressing wild-type LRRK2 (line 43). Each bar represents the mean±S.E. value of six mice. (b) Western blot analysis using AT8 monoclonal antibody demonstrated an upregulated protein expression of phospho-tauSer202/Thr205 in the SN of a 12-month-old (G2019S) LRRK2 mouse (line 32). Note that protein level of phospho-tauSer202/Thr205 was not altered in the SN of LRRK2 transgenic mouse (line 43). Each bar shows the mean±S.E. value of five mice. (c) Immunocytochemical staining of neuronal marker NeuN showed that compared with an age-matched wild-type mouse, significant neuronal death was absent in the striatum of a 12-month-old (G2019S) LRRK2 transgenic mouse (line 51). Each bar represents the mean±S.E. value of five mice. Scale bar is 100 μm. *P<0.01 compared with wild-type mice
In addition to a loss of SNpc dopaminergic neurons, another common pathological hallmark of PD is the formation of intracytoplasmic protein aggregates termed as Lewy bodies, composed of the synaptic vesicle-associated protein α-synuclein, ubiquitin, and other components, in the surviving SN neurons.26 Immunocytochemical staining using monoclonal FK2 antiserum, which recognizes ubiquitinated proteins, anti-α-synuclein antibody or anti-phospho-α-synucleinSer129 antiserum failed to observe Lewy bodies or Lewy neurites in the SN of 12- to 16-month-old (G2019S) LRRK2 transgenic mice (n=5 animals; data not shown).
Tau pathology, caused by accumulation of hyperphosphorylated microtubule-associated protein tau with numerous Ser/Thr phosphorylation sites including Ser202 and Thr205, was found in PARK8 patients with (G2019S) LRRK2 mutation.13 Western blot analysis with AT8 monoclonal antibody, which recognizes a tau epitope phosphorylated at Ser202 and Thr205, was performed to visualize phospho-tauSer202/Thr205 expression in the brain of wild-type, LRRK2, or (G2019S) LRRK2 mouse. Protein level of phospho-tauSer202/Thr205 was increased in the SN of 12-month-old (G2019S) LRRK2 transgenic mice (Figure 2b). In contrast, protein expression of phospho-tauSer202/Thr205 was not altered in the SN of transgenic mice expressing wild-type LRRK2 (Figure 2b). Protein level of phospho-tauSer202/Thr205 was not significantly altered in the cerebellum, cerebral cortex, and striatum of 12-month-old (G2019S) LRRK2 mice (n=5 animals; data not shown).
Consistent with previous neuropathological studies of (G2019S) LRRK2 PD patients,12, 14, 15, 16 immunohistochemical staining of neuronal marker NeuN showed the absence of significant neuronal death in the cerebral cortex, hippocampus, and striatum (Figure 2c) of 12- or 16-month-old (G2019S) LRRK2 transgenic mice (n=5 animals).
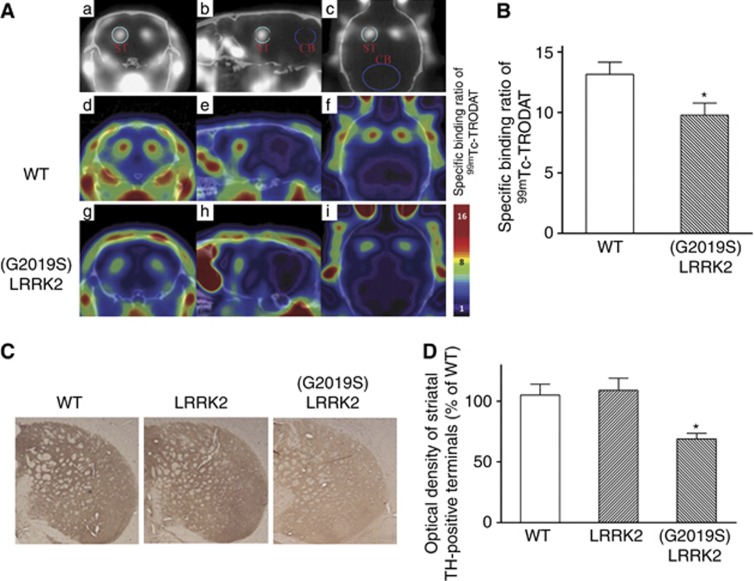
(G2019S) LRRK2-induced neuronal death of SNpc dopaminergic cells is expected to cause the degeneration of nigrostriatal dopaminergic terminals. Therefore, microSPECT imaging of 99mTc-TRODAT, a tracer of presynaptic dopamine transporter (DAT), was performed to visualize in-vivo DAT density in the striatum of mouse. Single-photon emission computed tomography (SPECT) imaging studies demonstrated that striatal 99mTc-TRODAT uptake was significantly decreased in 12-month-old (G2019S) LRRK2 transgenic mice as compared with age-matched wild-type mice (Figures 3A and B). Compared with 16-month-old control mice or transgenic mice expressing wild-type LRRK2, density of TH-immunoreactive staining was also decreased in the striatal sections of (G2019S) LRRK2 mice (Figures 3C and D).
Figure 3.
The density of dopamine transporters or TH staining is reduced in the striatum of (G2019S) LRRK2 transgenic mice. (A) In-vivo 99mTc-TRODAT microSPECT imaging was performed to visualize the density of dopamine transporters in the striatum. The uptake of 99mTc-TRODAT is normal (red color scale) in the striatum (ST) of a wild-type mouse (d–f), but it is markedly reduced (green color scale) in a 12-month-old (G2019S) LRRK2 transgenic mouse (line 32) (g–i) (a–c: VOI definition of bilateral striatum and cerebellum (CB); d–i: fusion imaging of microCT and microSPECT; a, d, g: coronal section; b, e, h: sagittal section; c, f, i: horizontal section). (B) Compared with 12-month-old wild-type mice, specific uptake ratios of 99mTc-TRODAT were significantly reduced in (G2019S) LRRK2 transgenic mice at the same age. Each bar shows the mean±S.E. value of eight mice. *P<0.01. (C) Compared with 16-month-old control mouse or transgenic mouse expressing wild-type LRRK2 (line 43), density of TH-immunoreactive staining was decreased in the striatum of a (G2019S) LRRK2 mouse (line 32). (D) Optical density of striatal TH-positive terminals was significantly reduced in (G2019S) LRRK2 transgenic mice. Each bar represents the mean±S.E. value of four mice. *P<0.01
(G2019S) LRRK2 transgenic mice display progressive parkinsonism phenotypes
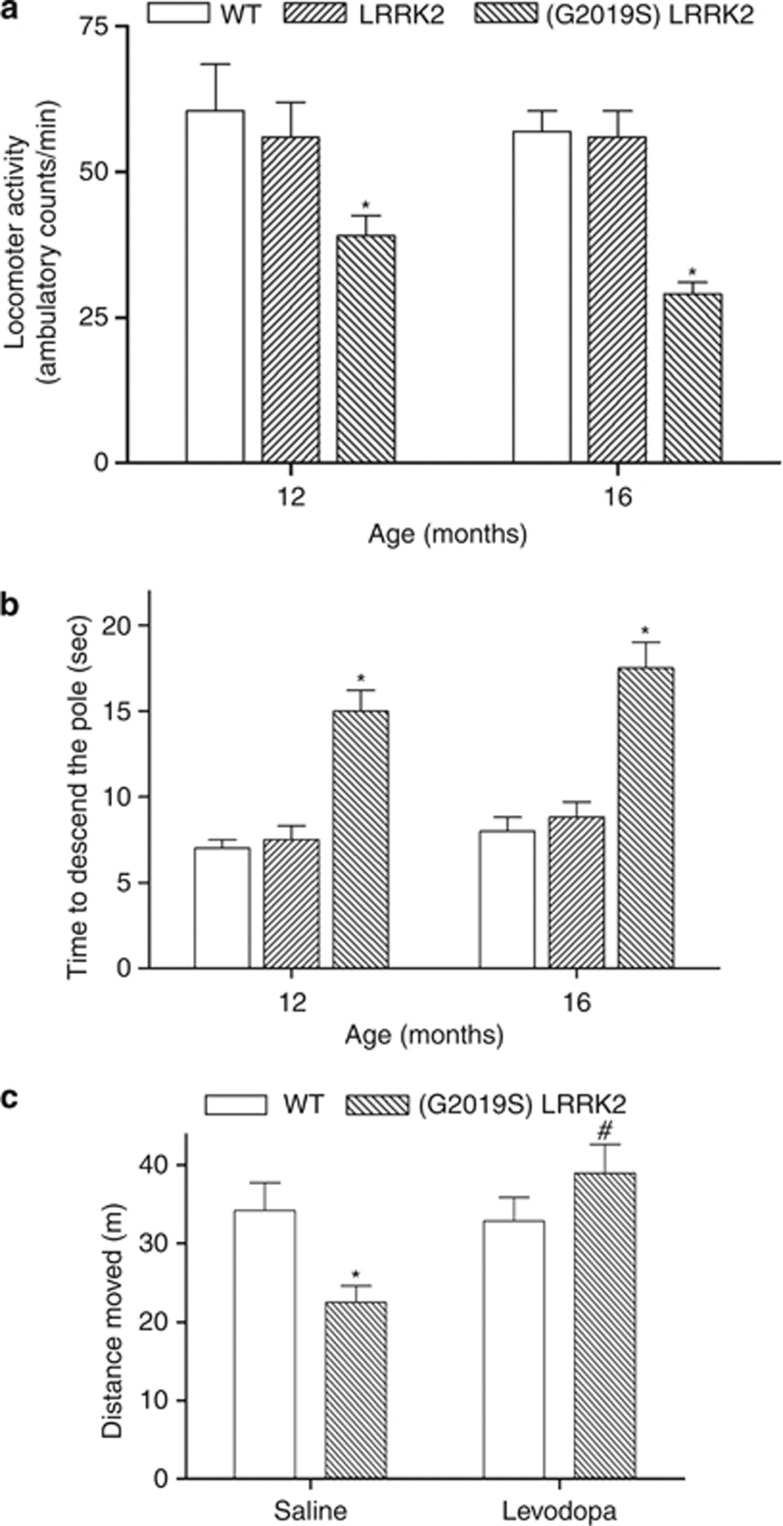
Similarly to hypokinesia exhibited by PD patients, 12-month-old (G2019S) LRRK2 transgenic mice were less active, which was indicated by a significant reduction in measured spontaneous ambulatory activity (Figure 4a). The severity of reduced locomotor activity progressively increased in (G2019S) LRRK2 mice at 16 months of age (Figure 4a). In contrast, 12- or 16-month-old LRRK2 transgenic mice did not display the symptom of hypoactivity (Figure 4a).
Figure 4.
(G2019S) LRRK2 transgenic mice display motor deficit of parkinsonism, which is rescued by ℒ-DOPA. (a) Compared with 12-month-old wild-type mice, (G2019S) LRRK2 transgenic mice at the same age exhibited a significant decrease in spontaneous ambulatory activity. Severity of hypoactivity progressively increased in (G2019S) LRRK2 mice at 16 months of age. In contrast, 12- or 16-month-old LRRK2 transgenic mice did not display the symptom of hypoactivity. Each bar shows the mean±S.E. value of 10–12 mice. (b) Pole test indicated that compared with wild-type mice or transgenic mice expressing wild-type LRRK2, 12- or 16-month-old (G2019S) LRRK2 transgenic mice took a significantly longer time to perform the pole test and exhibited an impaired motor performance. Each bar represents the mean±S.E. value of eight mice. *P<0.01 compared with wild-type mice. (c) Compared with saline-treated wild-type mice, 12-month-old (G2019S) LRRK2 mice injected with saline displayed a significant reduction in open-field locomotor activity measured by the distance traveled during the test period. Methyl ℒ-DOPA (2 mg/kg of body weight) was intraperitoneally injected into animals, and locomotor activity was measured 40 min after the injections. Note that methyl ℒ-DOPA rescued the hypoactivity exhibited by (G2019S) LRRK2 transgenic mice. Each bar represents the mean±S.E. value of seven mice. *P<0.01 compared with saline-injected wild-type mice. #P<0.01 compared with saline-injected (G2019S) LRRK2 mice
The pole test was also conducted to investigate the motor performance of mouse. Similarly to bradykinesia displayed by PD patients, animal with a lesion of SNpc dopaminergic neurons takes a longer time to perform the pole test.27 Twelve- or sixteen-month-old (G2019S) LRRK2 transgenic mice exhibited an impaired motor performance and the phenomenon of bradykinesia, which was indicated by a significant increase in time required to perform the pole test (Figure 4b). The performance of LRRK2 transgenic mice in the pole test was similar to that of wild-type mice (Figure 4b).
(G2019S) LRRK2 PD patients had a good response to ℒ-DOPA treatment.6 Therefore, we tested the possible beneficial effect of ℒ-DOPA on PD phenotype displayed by (G2019S) LRRK2 transgenic mice. Compared with saline-treated wild-type mice, 12-month-old (G2019S) LRRK2 mice injected with saline displayed a significant reduction in open-field locomotor activity measured by the distance traveled during the test period (Figure 4c). Similarly to PD patients with (G2019S) LRRK2 mutation, intraperitoneal administration of methyl ℒ-DOPA (2 mg/kg of body weight) reversed the hypoactivity exhibited by (G2019S) LRRK2 transgenic mice (Figure 4c). Thus, (G2019S) LRRK2 mice prepared by us recapitulated (G2019S) LRRK2 PD patient′s clinical characteristics, progressive motor deficit, and responsiveness to ℒ-DOPA.
(G2019S) LRRK2 exhibits an augmented kinase activity of phosphorylating MAPK kinase 4 at Ser 257
(G2019S) LRRK2 mutation resulted in an augmented in-vitro kinase activity.15, 16, 17 Thus, it is very likely that (G2019S) LRRK2 mutation causes overphosphorylation of downstream substrates, aberrant activation of neuronal death signal pathway and resulting in degeneration of SNpc dopaminergic neurons in (G2019S) LRRK2 transgenic mice.
Recently, we performed quantitative phosphoproteomic analysis by quantifying differentially labeled phosphopeptides isolated from the SN of wild-type or (G2019S) LRRK2 mice and identified upregulated phosphoproteins, which are likely to be (G2019S) LRRK2 substrates, in the (G2019S) LRRK2 transgenic mice. Interestingly, one of the identified phosphoproteins upregulated in the SN of (G2019S) LRRK2 mice was MAPK kinase 4 (MKK4) phosphorylated at Ser257 (Chen et al., unpublished results). LRRK2 is believed to be a member of MLK subfamily of MAPKKKs.18, 19 MLKs are known to activate neuronal death signal pathway by phosphorylating downstream MKK4 at Ser257. Therefore, we hypothesized that (G2019S) LRRK2 displays an augmented kinase activity of phosphorylating MKK4 at Ser257.
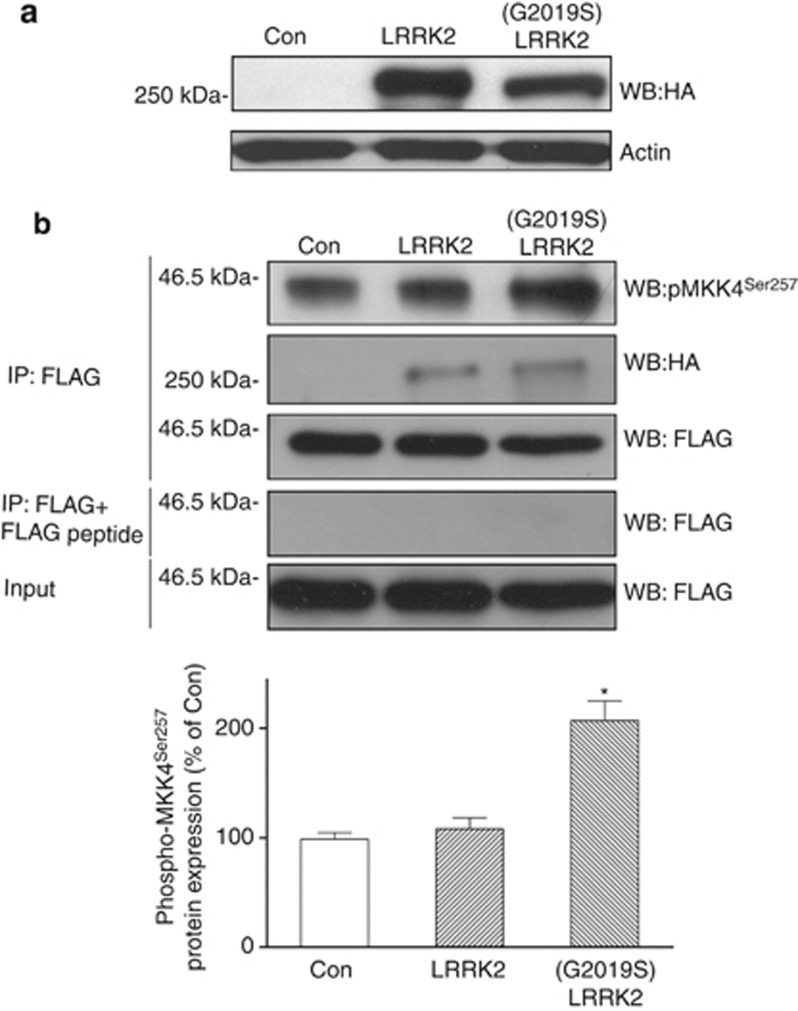
To provide evidence that (G2019S) LRRK2 mutation augments kinase activity and causes aberrant phosphorylation of MKK4 at Ser257, cDNA of FLAG-tagged MKK4 was transfected into HEK 293 cells stably expressing HA-tagged wild-type or (G2019S) LRRK2 (Figure 5a). Subsequent immunoprecipitation using anti-FLAG antibody and western blot analysis using anti-phospho-MKK4Ser257 antiserum showed that when expressed in 293 cells expressing wild-type LRRK2, FLAG-tagged MKK4 was not significantly phosphorylated at Ser257 (Figure 5b). On the other hand, immunoprecipitated protein level of phospho-MKK4Ser257 was greatly increased in 293 cells expressing FLAG-tagged MKK4 and (G2019S) LRRK2 (Figure 5b).
Figure 5.
(G2019S) LRRK2 enhances the phosphorylation of MKK4 at Ser257. (a) Western blot study using anti-HA antiserum showed that a similar protein level of HA-tagged wild-type or mutant (G2019S) LRRK2 was stably expressed in HEK 293 cells. (b) FLAG-tagged MKK4 was transiently transfected into control 293 cells or HEK293 cells stably expressing HA-tagged wild-type or mutant (G2019S) LRRK2. Then, FLAG-tagged MKK4 was immunoprecipitated by anti-FLAG antibody, and phosphorylated MKK4 at Ser257 serine was visualized by performing immunoblotting analysis using anti-phospho-MKK4Ser257 antibody. As a negative control, addition of FLAG peptide completely blocked immunoprecipitation of FLAG-tagged MKK4. Compared with control 293 cells or HEK 293 cells expressing wild-type LRRK2, immunoprecipitated protein level of phospho-MKK4Ser257was significantly increased in 293 cells expressing mutant (G2019S) LRRK2. Note that anti-FLAG antibody also co-immunoprecipitated HA-tagged wild-type or (G2019S) LRRK2. Each bar shows the mean±S.E. value of five experiments. *P<0.01 compared with control 293 cells
(G2019S) LRRK2 activates MKK4-JNK-cJun pathway and promotes formation of active caspases in the SN of (G2019S) LRRK2 transgenic mice
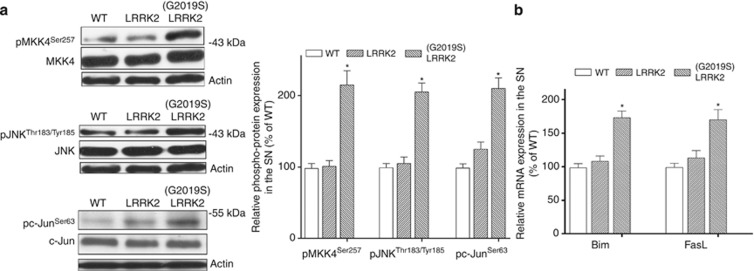
Consistent with results observed in HEK 293 cells, immunoblotting study using anti-phospho-MKK4Ser257 antibody showed that protein level of active phospho-MKK4Ser257 was greatly increased in the SN of (G2019S) LRRK2 mice at the age of 12 months (Figures 6a and b). Protein expression of phospho-MKK4Ser257 was not significantly altered in the SN of transgenic mice expressing wild-type LRRK2 (Figure 6a).
Figure 6.
(G2019S) LRRK2 activates MKK4-JNK-cJun pathway and upregulates mRNA expression of Bim or FasL in the SN of PD transgenic mice. (a) Western blot study showed an upregulated expression of active phospho-MKK4Ser257 in the SN of a 12-month-old (G2019S) LRRK2 mouse (line 51). Protein levels of phospho-JNKThr183/Tyr185 and phospho-c-JunSer63, which act as downstream effectors of active MKK4, were also greatly increased in the SN of (G2019S) LRRK2 transgenic mouse. Note that protein expression of phospho-MKK4Ser257, phospho-JNKThr183/Tyr185 or phospho-c-JunSer63 was not altered in the SN of a 12-month-old transgenic mouse expressing wild-type LRRK2 (line 36). Each bar represents the mean±S.E. value of six 12-month-old mice. (b) Real-time RT-PCR assay showed that compared with wild-type mice or transgenic mice expressing wild-type LRRK2, Bim or FasL mRNA expression was greatly upregulated in the SN of 12-month-old (G2019S) LRRK2 transgenic mice. Each bar shows the mean±S.E. value of 5–7 experiments. *P<0.01 compared with wild-type mice
Activated phospho-MKK4Ser257 phosphorylates and activates downstream JNKs. Western blot analysis using anti-phospho-JNKThr183/Tyr185 antiserum, which recognizes phosphorylated and activated JNKs, was performed to visualize JNK activation in the SN. Compared with wild-type mice, protein expression of active phospho-JNKThr183/Tyr185 was significantly upregulated in the SN of 12-month-old (G2019S) LRRK2 mice (Figure 6a).
A principal target of activated JNK is transcription factor c-Jun. The transcriptional activity of c-Jun is stimulated by JNK-mediated phosphorylation at Ser63, leading to increased expression of target genes that initiate the apoptotic cascade and caspase activation.18, 19 Western blot analysis using anti-phospho-c-JunSer63 antibody demonstrated the expected upregulated expression of active phospho-c-JunSer63 in the SN of (G2019S) LRRK2 transgenic mice at 12 months of age (Figure 6a).
Active phospho-c-JunSer63 causes neuronal death by enhancing the transcription of Bim and FasL.28, 29, 30 Bim, a member of pro-apoptotic BH3-only proteins, promotes cell death of CNS neurons including SNpc dopaminergic neurons by binding to Bcl-2 family pro-survival proteins and facilitating activation of Bax/Bak, which subsequently induces mitochondrial release of cytochrome-c and caspase-9 activation.31 By interacting with its cell surface receptor Fas, FasL promotes neuronal apoptosis via activating caspase-8.29 Consistent with our hypothesis that (G2019S) LRRK2 induces the activation and phosphorylation of c-Jun, mRNA expression of Bim or FasL was significantly upregulated in the SN of 12-month-old (G2019S) LRRK2 mice (Figure 6b).
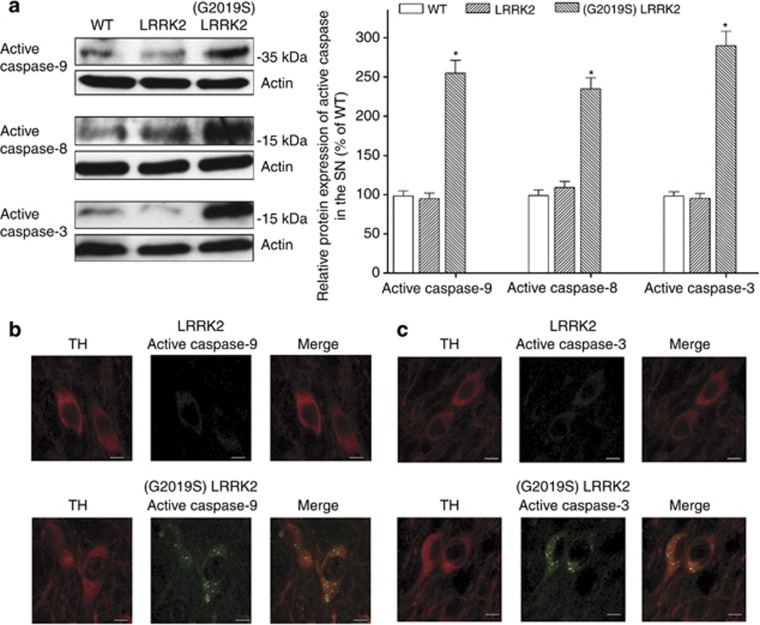
JNK-c-Jun-Bim/FasL cascade induces the formation of active caspase-9 and caspase-8 and causes neuronal death by activating intrinsic and extrinsic apoptotic pathways.29, 30, 31, 32 Therefore, it was hypothesized that (G2019S) LRRK2 causes degeneration of SNpc dopaminergic neurons in (G2019S) LRRK2 mice by promoting the formation of active caspase-9, caspase-8, and caspase-3. In accordance with this hypothesis, protein level of cleaved active caspase-9, active caspase-8, or active caspase-3 was greatly increased in the SN of 12-month-old (G2019S) LRRK2 mice (Figure 7a). Wild-type LRRK2 failed to induce the formation of active caspase-3, caspase-8, or caspase-9 in the SN of LRRK2 transgenic mice (Figure 7a). Confocal immunofluorescence imaging study further showed that active caspase-9 (Figure 7b; 27±3% TH-positive neurons; n=4 animals) and active caspase-3 (Figure 7c; 32±4% TH-positive cells; n=4 animals) were found in the cytoplasm of TH-positive SNpc dopaminergic neurons of 12-month-old (G2019S) LRRK2 mice. In contrast, the formation of active caspase-9 or caspase-3 was not observed in TH-positive SNpc dopaminergic cells of LRRK2 transgenic mice at the same age (Figures 7b and c; n=4 animals).
Figure 7.
Active caspase-9, caspase-8 and caspase-3 were expressed in the SN of (G2019S) LRRK2 transgenic mice. (a) Western blot analysis showed that protein expression of cleaved active caspase-9 (MW=37 kDa), active caspase-8 (MW=18 kDa) or active caspase-3 (MW=17 kDa) was significantly upregulated in the SN of a 12-month-old (G2019S) LRRK2 mice (line 32). Note that protein level of active caspase-3, caspase-8, or caspase-9 was not altered in the SN of LRRK2 transgenic mice (line 36) at the same age. Each bar shows the mean±S.E. value of six mice. *P<0.01 compared with wild-type mice. Double immunofluorescence staining was performed with anti-TH antiserum and anti-active caspase-9 (b) or anti-active caspase-3 (c) antibody. Confocal microscopy showed that active caspase-9 (b) or caspase-3 (c) was found in TH-positive SN dopaminergic neurons of a 12-month-old (G2019S) LRRK2 mouse (line 32). In contrast, active caspase-9 (b) or caspase-3 (c) was not observed in TH-positive SN dopaminergic cell of LRRK2 transgenic mice (line 36) at the age of 12 months. Scale bar is 10 μm
Discussion
LRRK2 is the most frequent causative gene in both familial and sporadic PD cases.6, 7, 10 Among LRRK2 mutations found in PD patients, G2019S is the most common amino-acid substitution.6, 7, 10 PD patients with (G2019S) LRRK2 mutation displayed a loss of SNpc dopaminergic neurons,11, 12, 13, 14 indicating that (G2019S) LRRK2 causes neurodegeneration of SNpc dopaminergic cells and resulting in parkinsonism. A mouse model, which replicates (G2019S) LRRK2-induced degeneration of SNpc dopaminergic neurons, should be a valuable tool to investigate the pathogenic mechanism of (G2019S) LRRK2-induced PD.
PDGF-β promoter mediated a high level of neuronal transgene expression in the brain.22, 23, 24 Furthermore, hybrid CMV enhancer/PDGF-β promoter drives a highly efficient transgene expression in SNpc dopaminergic neurons.25 Thus, we prepared (G2019S) LRRK2-induced PD mouse model expressing transgene CMV enhancer/PDGF-β promoter-(G2019S) LRRK2. Mutant (G2019S) LRRK2 protein was expressed in several PD-associated brain regions, including the SN, striatum, and cerebral cortex. Double immunofluorescence staining also demonstrated a high level of (G2019S) LRRK2 expression in SNpc dopaminergic neurons of (G2019S) LRRK2 transgenic mouse. Consistent with previous studies showing a late-onset degeneration of SNpc dopaminergic neurons in (G2019S) LRRK2 PD patients,11, 12, 13, 14 12-month-old (G2019S) LRRK2 transgenic mice prepared by us exhibited a significant neuronal death of SNpc dopaminergic cells, which progressively deteriorated in the following months. A reduction in striatal 99mTc-TRODAT uptake of (G2019S) LRRK2 mice, visualized by microSPECT imaging, provided a direct in-vivo evidence of (G2019S) LRRK2-induced degeneration of nigrostriatal dopaminergic terminals. (G2019S) LRRK2 transgenic mice at the ages of 12–16 months displayed progressive parkinsonism phenotypes including reduced locomotor activity and impaired motor performance, which resulted from (G2019S) LRRK2-induced degeneration of nigrostriatal dopaminergic pathway. Consistent with clinical studies showing that (G2019S) LRRK2 PD patients exhibited a good response to ℒ-DOPA treatment,6 intraperitoneal administration of methyl ℒ-DOPA rescued the hypoactivity exhibited by (G2019S) LRRK2 mice. Therefore, we, for the first time, generated transgenic mouse that recapitulates (G2019S) LRRK2 PD patient′s clinical characteristics, neuropathological and neurological phenotypes. In the present study, (G2019S) LRRK2 transgenic mice prepared by us were used to study molecular pathogenic mechanisms of (G2019S) LRRK2-induced degeneration of SN dopaminergic neurons and resulting in parkinsonism.
Post-mortem analysis of most PD patients observed the presence of intracytoplasmic protein aggregates termed as Lewy bodies and Lewy neurites, which are formed as the result of the aberrant aggregation of α-synuclein, in the surviving neurons.26 Previous neuropathological examination reported the presence of α-synuclein-positive Lewy bodies in the brain of most (G2019S) LRRK2 PD patients.11, 12, 14 However, SN neuronal loss in the absence of Lewy bodies was also found in some PD patients with (G2019S) LRRK2 mutation.13, 33 In the present study, we did not observe Lewy bodies or Lewy neurites in the SN of 12- to 16-month-old (G2019S) LRRK2 transgenic mice, which exhibit a significant neuronal death of SNpc dopaminergic cells. These results suggest that there is no direct correlation between (G2019S) LRRK2-induced degeneration of SNpc dopaminergic neurons and the formation of α-synuclein-positive Lewy body.
Consistent with a previous study reporting that tau pathology, caused by accumulation of hyperphosphorylated tau, in the absence of Lewy bodies occurred in (G2019S) LRRK2 PD patients,13 immunoblotting study with AT8 monoclonal antibody, which recognizes phosphorylated tau at Ser202 and Thr205, showed an upregulated expression of phospho-tauSer202/Thr205 in the SN of (G2019S) LRRK2 transgenic mice but not in transgenic mice expressing wild-type LRRK2. This finding suggests that mutant (G2019S) LRRK2 causes a gain-of-function tau hyperphosphorylation in the SN. Interestingly, western blot analysis using AT8 antiserum also demonstrated the hyperphosphorylation of tau in the brain of (R1441G) LRRK2 transgenic mice.34 The contribution of tau hyperphosphorylation to PARK8 mutant LRRK2-mediated neurotoxicity remains to be determined.
Similarly to the expression pattern of endogenous LRRK2 in the brain,8, 9 (G2019S) LRRK2 was widely expressed in the cerebral cortex, cerebellum, hippocampus, striatum, and SN of (G2019S) LRRK2 transgenic mice. Previous studies reported the absence of significant neuronal death in the cerebral cortex, hippocampus, and striatum of (G2019S) LRRK2 PD patients.12, 33 Consistent with the results observed from PD patients, 12- to 16-month-old (G2019S) LRRK2 transgenic mice did not exhibit a significant death of neocortical, hippocampal, and striatal neurons. In contrast with widespread expression of mutant (G2019S) LRRK2 in the brain, selective vulnerability of SNpc dopaminergic neurons is likely to result from (G2019S) LRRK2-mediated region-specific protein-protein interaction(s) or protein phosphorylation(s) in the SN. (G2019S) LRRK2 transgenic mice prepared by us can be used to identify SN protein interactor(s) or substrate(s) of (G2019S) LRRK2 and investigate molecular pathogenic mechanism underlying (G2019S) LRRK2-induced selective loss of SNpc dopaminergic cells.
Previous studies showed that (G2019S) substitution led to an enhancement of LRRK2 Ser/Thr kinase activity, which is required for (G2019S) LRRK2-mediated in-vitro neurotoxicity.15, 16, 17 LRRK2 is believed to be a member of MLK subfamily of MAPKKKs,6, 7 and MLKs activate neuronal death signal pathway by phosphorylating downstream MKKs.18, 19 Previous in-vitro kinase assays reported that recombinant wild-type or (G2019S) LRRK2 phosphorylated MKK3, MKK4, MKK6, or MKK7 and that (G2019S) mutation augmented LRRK2-mediated phosphorylating of MKK6 or MKK7.35, 36 In the present study, our quantitative phosphoproteomic analysis identified MKK4 phosphorylated at Ser257 residue as one of the upregulated phosphoproteins in the SN of (G2019S) LRRK2 mice, which are likely to be substrates of (G2019S) LRRK2-mediated phosphorylation in vivo (Chen et al., unpublished results). Thus, we hypothesized that (G2019S) LRRK2 mutation exerts a gain-of-function and enhancing effect on LRRK2 kinase activity, leading to overphosphorylation of MKK4Ser257, abnormal activation of MKK4-mediated cell death pathway and degeneration of SNpc dopaminergic cells in (G2019S) LRRK2 transgenic mice. In accordance with this hypothesis, our in-vivo phosphorylation assays showed that when co-expressed with MKK4 in HEK 293 cells, mutant (G2019S) LRRK2, but not wild-type LRRK2, significantly promoted MKK4 phosphorylation at Ser257. Furthermore, protein expression of active phospho-MKK4Ser257 was upregulated in the SN of (G2019S) LRRK2 mice but not in the SN of transgenic mice expressing wild-type LRRK2. Therefore, our study provides in-vivo evidence that mutant (G2019S) LRRK2 exhibits an augmented kinase activity of phosphorylating MKK4 at Ser257 in the SN.
Activated phospho-MKK4Ser257 induces the phosphorylation and activation of downstream JNKs. Then, phospho-JNKThr183/Tyr185 stimulates the transcriptional activity of transcription factor c-Jun by phosphorylating c-Jun at Ser63,18, 19, 28 Subsequently, active phospho-c-JunSer63 enhances the transcription of pro-apoptotic Bim and FasL, which causes the activation of caspase-9, caspase-8, and caspase-3.28, 29, 30 Our results showed that protein level of active phospho-JNKThr183/Tyr185 and active phospho-c-JunSer63 was significantly increased in the SN of (G2019S) LRRK2 mice. The mRNA expression of pro-apoptotic Bim and FasL was greatly upregulated in the SN of (G2019S) LRRK2 transgenic mice. Furthermore, protein level of cleaved active caspase-8 or caspase-9 was significantly increased in the SN of (G2019S) LRRK2 mice, and formation of cleaved active caspase-9 or active caspase-3 was observed in SNpc dopaminergic neurons of (G2019S) LRRK2 transgenic mice. These findings suggest that mutant (G2019S) LRRK2, which promotes the formation of active phospho-MKK4Ser257, causes the death of SNpc dopaminergic neurons of (G2019S) LRRK2 mice by activating JNK-c-Jun signaling pathway. When expressed in neuroblastoma cell line or cultured cortical neurons, mutant (G2019S) LRRK2 has been shown to induce apoptotic cell death by activating caspase-3 and caspase-8.37, 38, 39 Our results provide the first in-vivo evidence that mutant (G2019S) LRRK2 causes the cell death of SN dopaminergic neurons by activating caspase-9 and caspase-8 through MKK4-JNK-c-Jun pathway.
In summary, we have successfully generated PD animal model by preparing transgenic mice expressing mutant (G2019S) LRRK2. Similarly to PD patients with (G2019S) LRRK2 mutation, (G2019S) LRRK2 transgenic mice exhibited degeneration of SNpc dopaminergic neurons and PD neurological phenotypes. Our results also suggest that mutant (G2019S) LRRK2 activates MKK4-JNK-c-Jun pathway in the SN and causes the resulting degeneration of SNpc dopaminergic neurons in PD transgenic mice.
Materials and methods
Generation of transgenic mice expressing wild-type or mutant (G2019S) LRRK2
HA epitope was added to the C-terminus of human wild-type LRRK2 by PCR amplification. Oligonucleotide-directed mutagenesis was performed to prepare cDNA encoding human (G2019S) LRRK2, and DNA mutation of LRRK2 was confirmed by DNA sequencing. According to our previous studies,23, 24 transgene construct was prepared by inserting cDNA of HA-tagged wild-type or mutant (G2019S) LRRK2 into CMV enhancer/platelet-derived growth factor (PDGF)-βchain expression vector. An ∼11-kb transgene fragment contained CMV enhancer, PDGF-βchain promoter, cDNA of wild-type or mutant (G2019S) LRRK2 and a 3' SV40pA sequence. Linearized transgene construct was purified and injected into male pronuclei of fertilized oocytes prepared from FVB/N mice, which were then implanted into pseudopregnant mice. Southern blot analysis was performed to identify founders and copy number of transgene. Founder transgenic mice were mated with wild-type FVB/N mice and bred into stable transgenic lines. Animals were handled according to protocols approved by Animal Care and Use Committee of Chang Gung University.
Behavioral tests
Spontaneous locomotor activity was measured in an activity cage (30 × 45 × 45 cm; Med Associates Inc., St. Albans, VT, USA) constructed of opaque plastic walls and four photocells positioned 1 cm above the floor and spaced evenly along the longitudinal axis of each chamber. When animal moved, separate interruptions of photocell beams were detected via an electrical interface of a computer. Locomotor activity was recorded for a total session of 1 h. For measuring locomotive distance, animal was placed in an open field of acrylic box (45 × 45 × 45 cm) for 20 min. The movement of mouse was filmed with a video camera installed above and analyzed by a video tracking software TopScan (Clever Sys., Inc., Reston, VA, USA). The pole test was performed to determine motor performance of mouse. Briefly, animal was placed on the top of a pole, and the base of pole was placed in the home cage. Mice oriented themselves downward, descended along the pole and returned to their home cage. Animal received 2 days of training that was consisted of five trials for each day. On the test day, mice performed five trials, and the time required for orienting downward and descending along the pole was measured.
Western blot analysis
Frozen brain tissues were homogenized with SDS sample buffer supplemented with cocktail of protease inhibitors (Sigma, St. Louis, MO, USA). Protein samples were fractionated on SDS-polyacrylamide gel and transferred onto PVDF membrane. Then, membrane was incubated overnight with one of the following diluted primary antibodies: (1) Anti-HA monoclonal antiserum (Cell Signaling Technology, Beverly, MA, USA); (2) Monoclonal anti-LRRK2 antibody (Millipore, Bedford, MA, USA); (3) Rabbit monoclonal anti-phospho-MKK4Ser257 antiserum (Cell Signaling Technology); (4) Anti-MKK4 polyclonal antiserum (Cell Signaling Technology); (5) Monoclonal anti-phospho-JNKThr183/Tyr185 antibody (Cell Signaling Technology); (6) Anti-JNK polyclonal antibody (Cell Signaling Technology); (7) Polyclonal anti-phospho-c-JunSer63 antibody (Cell Signaling Technology); (8) Anti-c-Jun rabbit monoclonal antibody (Cell Signaling Technology); (9) Anti-cleaved active caspase-3 polyclonal antiserum (Cell Signaling Technology); (10) Monoclonal anti-caspase-8 antibody (Santa Cruz, Santa Cruz, CA, USA); (11) Anti-cleaved active caspase-9 polyclonal antiserum (Cell Signaling Technology); and (12) Monoclonal anti-phospho-Tau antibody (AT8; Thermo Scientific, Waltham, MA, USA). After being washed, the membrane was incubated with sheep anti-mouse or donkey anti-rabbit horseradish peroxidase-linked secondary antibody (Amersham, Piscataway, NJ, USA). Then, immunoreactive proteins on the membrane were visualized by using enhanced chemiluminescence protocol (ECL Kit; Amersham).
Immunohistochemical and confocal immunofluorescence staining
Animal was anesthetized and perfused transcardially with 4% paraformaldehyde in PBS. Coronal brain sections (20 μm) prepared by cryostat sectioning were permeabilized and incubated with diluted monoclonal anti-tyrosine hydroxylase antibody (TH; Millipore), anti-NeuN monoclonal antiserum (Chemicon, Temecula, CA, USA), monoclonal anti-α-synuclein antibody (BD Transduction Laboratories, San Diego, CA, USA), monoclonal FK2 antibody (Enzo Life Sciences, Farmingdale, NY, USA), or polyclonal anti-phospho-α-synucleinSer129 antibody (Abcam, Cambridge, MA, USA). Subsequently, sections were incubated with biotinylated horse anti-mouse or goat anti-rabbit IgG followed by incubation with avidin-biotin-horseradish peroxidase complex (Vector Laboratories, Burlingame, CA, USA). TH-positive dopaminergic neurons or NeuN-positive neurons were visualized and counted by a Leica DM2500 microscope equipped with an RET 2000R CCD camera (Qimaging, Surrey, BC, Canada) and a 3-axis computer-controlled MAC 600 motorized stage (Ludl Electronics, Hawthorne, NY, USA) with the aid of StereoInvestigator software (MBF Bioscience, Williston, VT, USA). The optical density of striatal TH staining was measured using ImageJ (freeware Java version; National Institutes of Health, Bethesda, MD, USA) Software. Each analysis included the processing of 15–20 brain sections per mouse.
To visualize active caspase-3 or caspase-9 in SN dopaminergic neurons, SN sections were incubated with monoclonal anti-TH antibody and anti-active caspase-9 or caspase-3 polyclonal antiserum. Subsequently, sections were incubated with Alexa Fluor 568- and FITC-conjugated secondary antibodies. Sections were then viewed under a Leica DM6000 microscope equipped with Leica TCS SP5 confocal spectral scanning system and a × 100 oil-immersion objective. Confocal fluorescence images were processed and analyzed by LAS AF software (Leica, Mannheim, Germany).
Animal image study with [99mTc]-TRODAT
Animal imaging study using a high-resolution dual modality system (NanoSPECT/CT, Bioscan, Washington, DC, USA) was performed at Molecular Imaging Center, Chang Gung Memorial Hospital. This system features helical scanning for both SPECT and computed tomography (CT) acquisitions using a translation stage with variable axial scanning ranged from 30 to 300 mm. According to our previous study,40 185 MBq (concentrated in a total volume of 0.2 ml) of DAT tracer 99mTc-TRODAT (INER, Taiwan) was injected into tail vein of mouse. SPECT acquisition protocol was started after 1.5 h update period, and total acquisition time was 30 min. A CT scan with the same bed position was performed immediately after the SPECT acquisition for anatomical reference. Final image was reconstructed using iterative method. Coregistered SPECT/CT images were displayed and analyzed using the PMOD image analysis software (PMOD Technologies, Zurich, Switzerland). The bilateral striatal or cerebellar volume of interest was manually drawn on the fused SPECT/CT images. The specific binding ratio of bilateral striatal was calculated as (striatal average counts/cerebellum average counts)–1. Animal imaging was performed under anesthesia (1.5% isoflurane in 100% oxygen).
Wild-type or (G2019S) LRRK2-mediated phosphorylation of MKK4 in HEK 293 cells
The cDNA of FLAG-tagged MKK4 was transiently transfected into 293 cells stably expressing HA-tagged wild-type or (G2019S) LRRK2. Three days after the transfection, FLAG-tagged MKK4 was purified using FLAG immnunoprecipitation kit (Sigma). Briefly, HEK 293 cells were solubilized with ice-cold lysis buffer provided in the kit. Then, FLAG-tagged MKK4 was immunoprecipitated using anti-FLAG-M2 affinity gel. Immunoprecipitated FLAG-tagged MKK4 was eluted from the resin by competition with FLAG peptide. Subsequently, immunoprecipitated protein samples was fractionated on 10% SDS-polyacrylamide and transferred onto PVDF membrane. To visualize wild-type or (G2019S) LRRK2-induced phosphorylation of FLAG-tagged MKK4 at Ser257, PVDF membrane was incubated with diluted rabbit monoclonal anti-phospho-MKK4Ser257 antiserum and then with donkey anti-rabbit horseradish peroxidase-linked secondary antibody (Amersham).
Real-time quantitative RT-PCR assays of Bim or FasL
Total RNA was prepared from the SN using Trizol Reagent (Invitrogen, Carlsbad, CA, USA). Then, the first-strand cDNA was synthesized in a reaction mixture containing total RNA (1 μg), 8 ng/μl oligodT primer, 1 mM of each dNTP, 20 U of ribonuclease inhibitor and 100 U of SuperScript III RT (InVitrogen). Reaction was performed for 1 h at 50 °C and terminated by incubating for 15 min at 70 °C. Real-time PCR was carried out in the StepOne Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) using SYBR Green PCR Master Mix (Applied Biosystems). The reaction mixture (40 μl) consisted of cDNA aliquot, 400 nM of forward or reverse primer and 1 × SYBR Green PCR Master Mix containing AmpliTaq Gold DNA polymerase and SYBR Green 1 dye. PCR amplification of GAPDH mRNA was used as the normalization control. Relative change in mRNA expressions was determined by the equation: Fold change=2−[ΔΔCt], ΔΔCt=(CtBim/FasL–CtGAPDH)(G2019S)LRRK2–(CtBim/FasL– CtGAPDH)wild type. Ct value is the cycle number at which fluorescence signal crosses the threshold.
Statistics
All results are expressed as the mean±S.E. value of n experiments. Statistical significance among multiple experimental groups was determined by one-way ANOVA followed by Dunnett′s test. Unpaired Student′s t-test (two-tailed) was used to determine the significant difference between two groups of data. A P-value of <0.05 was considered significant.
Acknowledgments
This work was supported by the National Science Council of ROC (NSC97-2321-B-182–006, NSC 98-2314-B-182-034-MY2, NSC98-3112-B-182-003, and NSC99-3112-B-182-003), Chang Gung Medical Research Projects (CMRPD150153 and CMRPD170402), and Healthy Aging Research Center of Chang Gung University (EMRPD1A0831).
Glossary
- PARK8
familial type 8 of Parkinson′s disease
- LRRK2
leucine-rich repeat kinase 2
- SN
substantia nigra
- MKK4
MAPK kinase 4
- JNK
c-Jun N-terminal kinases
The authors declare no conflict of interest.
Footnotes
Edited by L Greene
References
- Hardy J, Lewis P, Revesz T, Lees A, Paisan-Ruiz C. The genetics of Parkinson's syndromes: a critical review. Curr Opin Genet Dev. 2009;19:254–265. doi: 10.1016/j.gde.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Lesage S, Brice A. Parkinson's disease: from monogenic forms to genetic susceptibility factors. Hum Mol Genet. 2009;18:48–59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- Funayama M, Hasegawa K, Kowa H, Saito M, Tsuji S, Obata F. A new locus for Parkinson's disease (PARK8) maps to chromosome 12p11.2-q13.1. Ann Neurol. 2002;51:296–301. doi: 10.1002/ana.10113. [DOI] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA. LRRK2 in Parkinson's disease: protein domains and functional insights. Trends Neurosci. 2006;29:286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson's disease. Nat Rev Neurosci. 2010;11:791–797. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biskup S, Moore DJ, Celsi F, Higashi S, West AB, Andrabi SA, et al. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann Neurol. 2006;60:557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- Westerlund M, Belin AC, Anvret A, Bickford P, Olson L, Galter D. Developmental regulation of leucine-rich repeat kinase 1 and 2 expression in the brain and other rodent and human organs: implications for Parkinson's disease. Neuroscience. 2008;152:429–436. doi: 10.1016/j.neuroscience.2007.10.062. [DOI] [PubMed] [Google Scholar]
- Kumari K, Tan EK. LRRK2 in Parkinson's disease: genetic and clinical studies from patients. FEBS J. 2009;361:310–321. doi: 10.1111/j.1742-4658.2009.07344.x. [DOI] [PubMed] [Google Scholar]
- Gilks WP, Abou-Sleiman PM, Gandhi S, Jain S, Singleton A, Lees AJ, et al. A common LRRK2 mutation in idiopathic Parkinson's disease. Lancet. 2005;365:415–416. doi: 10.1016/S0140-6736(05)17830-1. [DOI] [PubMed] [Google Scholar]
- Giasson BI, Covy JP, Bonini NM, Hurtig HI, Farrer MJ, Trojanowsk JQ, et al. Biochemical and pathological characterization of Lrrk2. Ann Neurol. 2006;59:315–322. doi: 10.1002/ana.20791. [DOI] [PubMed] [Google Scholar]
- Rajput A, Dickson DW, Robinson CA, Ross OA, Dachsel JC, Lincoln SJ, et al. Parkinsonism, Lrrk2 G2019S, and tau neuopathology. Neurology. 2006;67:1506–1508. doi: 10.1212/01.wnl.0000240220.33950.0c. [DOI] [PubMed] [Google Scholar]
- Wider C, Dickson DW, Wszolek ZK. Leucine-rich repeat kinase 2 gene-associated disease: redefining genotype-phenotype correlation. Neurodegener Dis. 2010;7:175–179. doi: 10.1159/000289232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, et al. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- Wang LH, Besirli CG, Johnson EM. Mixed lineage kinases: a target for the prevention of neurodegeneration. Annu Rev Pharmacol Toxicol. 2004;44:451–474. doi: 10.1146/annurev.pharmtox.44.101802.121840. [DOI] [PubMed] [Google Scholar]
- Burke RE. Inhibition of mitogen-activated protein kinase and stimulation of Akt kinase signaling pathways: two approaches with therapeutic potential in the treatment of neurodegenerative disease. Pharmacol Ther. 2007;114:261–277. doi: 10.1016/j.pharmthera.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht S, Kirchhof R, Chromik A, Willesen M, Nicolaus T, Raivich G, et al. Specific pathophysiological functions of JNK isoforms in the brain. Eur J Neurosci. 2005;21:363–377. doi: 10.1111/j.1460-9568.2005.03857.x. [DOI] [PubMed] [Google Scholar]
- Ries V, Silva RM, Oo TF, Cheng HC, Rzhetskaya M, Kholodilov N, et al. JNK2 and JNK3 combined are essential for apoptosis in dopamine neurons of the substantia nigra, but are not required for axon degeneration. J Neurochem. 2008;107:1578–1588. doi: 10.1111/j.1471-4159.2008.05713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah E, Rockenstein E, Veinbergs I, Mallory M, Hashimoto M, Takeda A, et al. Dopaminergic loss and inclusion body formation in alpha-synuclein mice: implications for neurodegenerative disorders. Science. 2000;287:1265–1269. doi: 10.1126/science.287.5456.1265. [DOI] [PubMed] [Google Scholar]
- Chou AH, Chen CY, Chen SY, Chen WJ, Chen YL, Weng YS, et al. Polyglutamine-expanded ataxin-7 causes cerebellar dysfunction by inducing transcriptional dysregulation. Neurochem Int. 2010;56:329–339. doi: 10.1016/j.neuint.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Wang HL, Chou AH, Lin AC, Chen SY, Weng YH, Yeh TH. Polyglutamine-expanded ataxin-7 upregulates Bax expression by activating p53 in cerebellar and inferior olivary neurons. Exp Neurol. 2010;224:486–494. doi: 10.1016/j.expneurol.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Wang CY, Guo HY, Lim TM, Ng YK, Neo HP, Hwang PYK, et al. Improved neuronal transgene expression from an AAV-2 vector with a hybrid CMV enhancer/PDGF-β promoter. J Gene Med. 2005;7:945–955. doi: 10.1002/jgm.742. [DOI] [PubMed] [Google Scholar]
- Dickson DW, Braak H, Duda JE, Duyckaerts C, Gasser T, Halliday GM, et al. Neuropathological assessment of Parkinson's disease: refining the diagnostic criteria. Lancet Neurol. 2009;8:1150–1157. doi: 10.1016/S1474-4422(09)70238-8. [DOI] [PubMed] [Google Scholar]
- Fleming SM, Salcedo J, Fernagut PO, Rockenstein E, Masliah E, Levine MS, et al. Early and progressive sensorimotor anomalies in mice overexpressing wild-type human α-synuclein. J Neurosci. 2004;24:9434–9440. doi: 10.1523/JNEUROSCI.3080-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield J, Neame SJ, Paquet L, Bernard O, Ham J. Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron. 2001;29:629–643. doi: 10.1016/s0896-6273(01)00239-2. [DOI] [PubMed] [Google Scholar]
- Dunn C, Wiltshirea C, MacLarena A, Gillespiea D.A.F. Molecular mechanism and biological functions of c-Jun N-terminal kinase signalling via the c-Jun transcription factor. Cell Signal. 2002;14:585–593. doi: 10.1016/s0898-6568(01)00275-3. [DOI] [PubMed] [Google Scholar]
- Perier C, Bove J, Wu DC, Dehay B, Choi DK, Jackson-Lewis V, et al. Two molecular pathways initiate mitochondria-dependent dopaminergic neurodegeneration in experimental Parkinson's disease. Proc Natl Acad Sci USA. 2007;104:8161–8166. doi: 10.1073/pnas.0609874104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis SN, Adams JM. Life in the balance: how BH3-only proteins induce apoptosis. Curr Opin Cell Biol. 2005;17:617–625. doi: 10.1016/j.ceb.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carboni S, Antonsson B, Gaillard P, Gotteland JP, Gillon JY, Vitte PA. Control of death receptor and mitochondrial-dependent apoptosis by c-Jun N-terminal kinase in hippocampal CA1 neurones following global transient ischaemia. J Neurochem. 2005;92:1054–1060. doi: 10.1111/j.1471-4159.2004.02925.x. [DOI] [PubMed] [Google Scholar]
- Gaig C, Marti MJ, Ezquerra M, Rey MJ, Cardozo A, Tolosa E. G2019S LRRK2 mutation causing Parkinson's disease without Lewy bodies. J Neurol Neurosurg Psychiatry. 2007;78:626–628. doi: 10.1136/jnnp.2006.107904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YP, Liu WC, Oo TF, Wang L, Tang Y, Jackson-Lewis V, et al. Mutant LRRK2R1441G BAC transgenic mice recapitulate cardinal features of Parkinson's disease. Nat Neurosci. 2009;12:826–828. doi: 10.1038/nn.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloeckner CJ, Schumacher A, Boldt K, Ueffing M. The Parkinson disease-associated protein kinase LRRK2 exhibits MAPKKK activity and phosphorylates MKK3/ 6 and MKK4/ 7, in vitro. J Neurochem. 2009;109:959–968. doi: 10.1111/j.1471-4159.2009.06024.x. [DOI] [PubMed] [Google Scholar]
- Hsu CH, Chan D, Greggio E, Saha S, Guillily MD, Ferree A, et al. MKK6 binds and regulates expression of Parkinson's disease-related protein LRRK2. J Neurochem. 2009;112:1593–1604. doi: 10.1111/j.1471-4159.2010.06568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Iaccarino C, Crosio C, Vitale C, Sanna G, Carri MT, Barone P. Apoptotic mechanisms in mutant LRRK2-mediated cell death. Hum Mol Genet. 2007;16:1319–1326. doi: 10.1093/hmg/ddm080. [DOI] [PubMed] [Google Scholar]
- Ho CC, Rideout HJ, Ribe E, Troy CM, Dauer WT. The Parkinson disease protein leucine-rich repeat kinase 2 transduces death signals via Fas-associated protein with death domain and caspase-8 in a cellular model of neurodegeneration. J Neurosci. 2009;29:1011–1016. doi: 10.1523/JNEUROSCI.5175-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KJ, Liu HL, Hsu PH, Chung YH, Huang WC, Chen JC, et al. Quantitative micro-SPECT/CT for detecting focused ultrasound-induced blood-brain barrier opening in the rat. Nucl Med Biol. 2009;36:853–861. doi: 10.1016/j.nucmedbio.2009.04.011. [DOI] [PubMed] [Google Scholar]