Abstract
The Hippo pathway controls organ size by multiple mechanisms that ultimately regulate the transcriptional co-activator Yorkie (Yki). Downregulation of Hippo signalling leads to tissue overgrowths due to Yki-mediated activation of target genes, whereas overexpression of the pathway triggers apoptosis in developing tissues. However, the mechanism underlying cell death induced by Hippo (Hpo)-activation is not understood. We found that overexpression of Hpo leads to induction of Dronc (Drosophila Caspase-9 homologue) expression and downregulation of dronc can suppress/block Hpo-mediated apoptosis. Furthermore, upregulation of Dronc activity strongly suppressed the overgrowth caused by Yki overexpression thereby suggesting that Hippo signalling restricts Dronc activity. Hippo-mediated cell death requires the activity of the initiator caspase Dronc. Loss-of-function of dronc in genetic mosaics leads to cell survival and increased cell proliferation in imaginal discs. dronc is transcriptionally suppressed in Yki overexpressing cells or cells mutant for other Hippo pathway components like warts (wts). We propose that Dronc is a transcriptional target of the Hippo signalling pathway. The Hippo–Dronc connection has implications in control of overall organ size and other growth regulatory mechanisms like compensatory proliferation and cell competition.
Keywords: caspases, Hippo pathway, organ size, dronc, Drosophila, imaginal discs
Hippo signalling pathway has emerged as a key size regulatory pathway that restricts cell proliferation and promotes apoptosis simultaneously to regulate organ size in flies and vertebrates.1, 2 At the centre of the Hippo signalling pathway is a core kinase cascade composed of two Serine/Threonine-specific kinases Hippo (Hpo)3, 4, 5, 6, 7 and Warts (Wts),8, 9 and their adaptor proteins Salvador10, 11 and Mob as tumour suppressor (Mats).12 The kinase cascade inactivates the oncoprotein Yorkie (Yki),13 which acts as a transcriptional co-activator of the Hippo pathway, and responds to multiple upstream regulators like the FERM-domain containing proteins Expanded (Ex) and Merlin (Mer),14 the WW- and C2-domain containing protein Kibra,15, 16, 17 and the single-pass trans-membrane proteins Crumbs (Crb)18, 19, 20 and Fat.21, 22, 23, 24 Yki partners with at least three transcription factors, Scalloped, Homothorax and Teashirt,1 to regulate transcription of genes regulating cellular growth (myc, bantam), cell cycle progression (cyclinB, cyclinD, cyclinE, e2F1) and inhibition of apoptosis (diap1). Hippo signalling also transcriptionally regulates several upstream components by feedback regulation, for example, mer, ex, four-jointed, kibra and crb.1 With its multiple inputs and targets, Hippo signalling has emerged as a pleiotropic pathway that acts in a context-dependent and tissue-specific manner to regulate organ size.
According to current models of Hippo signalling, activated Hpo can bind Sav, and phosphorylate Wts and its co-factor Mats, promoting Mats–Wts complex formation and increased Wts kinase activity. Activated Wts/Mats phosphorylates and inhibits Yki.1 Alternatively, Hpo, Wts and Ex can directly interact with Yki in a phosphorylation-independent manner leading to cytoplasmic sequestration and inhibition of Yki activity.1 Loss of Hippo signalling causes overgrowth in organs as mutant cells proliferate faster than wild-type cells and are resistant to developmentally regulated proapoptotic signals, which normally eliminate extra cells in a growing organ.2 Hyperactivation of Hippo signalling in flies, by overexpression of Hpo, Sav, Mats and Wts has been shown to induce cell death and form smaller organs.6, 7, 25 The proapoptotic gene hid and the bantam microRNA are induced in response to overexpression of Hippo pathway components; however, the molecular mechanism of Hippo-mediated cell death remains unclear.2 In this study, we investigated genes in the intrinsic cell death pathway to identify the mechanism of Hpo-mediated cell death. Our studies demonstrate that the Hippo pathway regulates Dronc (Drosophila Nedd-2-like caspase orthologous to human Caspase 9) levels to control both cell proliferation and cell survival, to regulate organ size.
Dronc encodes a caspase (cysteine protease) that forms the core of the apoptotic cell death machinery.26, 27 Caspases are synthesised as inactive zymogens that are activated by proteolytic cleavage regulated by the inhibitor of apoptosis (IAP) protein family. In flies, the Drosophila IAP (DIAP1) ensures cell viability by directly binding to caspases and regulating their activities in an ubiquitin-dependent manner.28 In response to apoptotic stimuli, Reaper, Hid and Grim (RHG) proteins bind the BIR domain of DIAP1 resulting in ubiquitination and proteosomal degradation of DIAP1, and activation of the caspase cascade in a Dark (Drosophila Apaf-1-related killer a.k.a dAPAF-1, Drosophila apoptosis promoting factor-1-like adaptor protein) -dependent manner. This involves the initiator caspase Dronc and the effector caspases Drice and DCP1.28, 29
Both DIAP1 and Dronc are expressed ubiquitously, but they have opposite roles in apoptosis. Loss of diap1 triggers caspase-mediated cell death,28 whereas loss of dronc blocks most apoptosis in flies.26, 27, 30 DIAP1 is activated by multiple mechanisms like auto-activation, post-transcriptional regulation by RHG proteins and transcriptional regulation by the Hippo pathway.6, 7 Dronc is activated by an autocatalytic mechanism31 and post-transcriptionally by Dark.32 Dronc activity is regulated by degradative (DIAP1-dependent) and non-degradative (DIAP1-independent) mechanisms.28 Overexpression of DIAP1 does not reduce Dronc levels or affect the catalytic activity of Dronc or target Dronc for degradation in living cells. Downstream of DIAP1, the overexpression of p35 (the baculovirus caspase inhibitor) inhibits Dronc-dependent cell death, but it does not inhibit Dronc. Here, we present evidences, suggesting that Hippo pathway regulates dronc transcription, and hyperactivation of Hippo signalling induces Dronc that contributes to apoptosis. Increased Dronc activity (via binding its adaptor Dark) can suppress the overgrowths caused by downregulation of Hippo signalling. Dronc loss of function causes increased proliferation and decreased apoptosis. In summary, our results suggest that Hippo signalling components need to control Dronc activity levels to control cell proliferation and cell survival.
Results
Overexpression of Hippo signalling components induces Caspase-dependent cell death
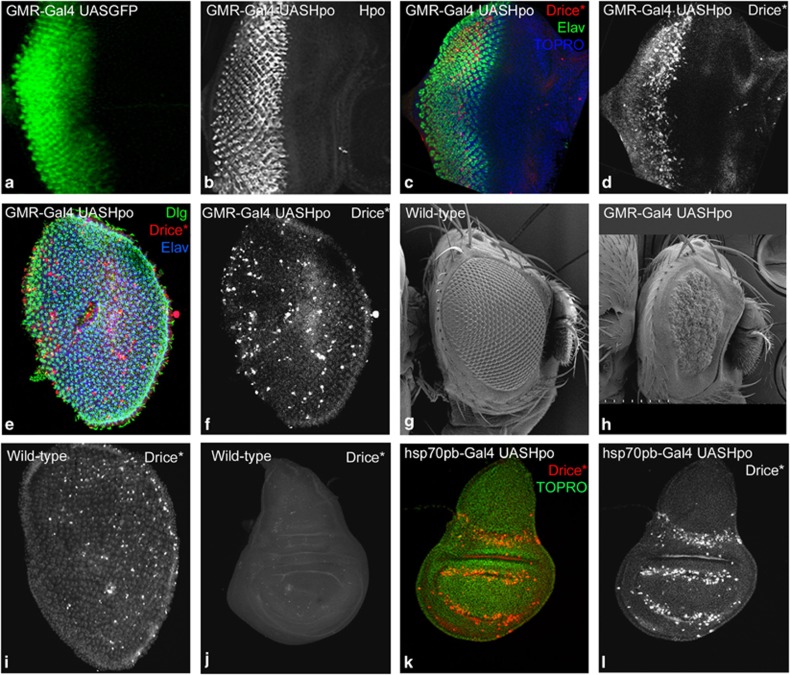
We overexpressed Hpo in the developing eyeantennal discs using the GMR-Gal4 driver (Figure 1a). Overexpression of Hpo (GMR-Gal4 UASHpo, Figure 1b) in the GMR domain induced massive cell death (activated Drice, Figure 1c and d) without disrupting cell differentiation or patterning (ELAV Figure 1c) in the eyeantennal discs. In mid-pupal retina (∼24 h APF), both photo-receptor and pigment cells continued to die precociously (Figure 1e and f). In comparison, in wild-type pupal retinae cell death occurs prominently in the pigment cells, and around the retinal boundary (Figure 1i). Compared with wild-type adult eyes (Figure 1g), Hpo overexpressing adult flies are arrested as pharates at RT, and at 18 °C show small rough eyes (Figure 1h). Hpo overexpression caused apoptosis in all epithelial tissues, for example, in the wing discs short pulses of Hpo overexpression using heat shock (hsp70)-Gal4 caused apoptosis (Figure 1i). In comparison, very few cells undergo apoptosis in wild-type wing discs (Figure 1j). Thus, consistent with previous findings, overexpression of Hippo leads to the activation of cell death in all growing organs. Co-expression of Yki and Hpo in eyeantennal discs (GMR-Gal4 UASYki, UASHpo) completely rescued the small eye phenotype of Hpo activation (Supplementary Figure S1). This suggests that Hpo exerts its apoptotic effects via the classical pathway involving the Yki co-activator. Genetic epistasis experiments revealed that Hpo acts upstream of the pro-apoptotic genes as in cells mutant for Df(3L)H99 (that uncovers the region of the RHG genes), overexpression of Hpo fails to induce apoptosis (Supplementary Figure S2). Thus, Hpo requires pro-apoptotic genes (RHG) to induce apoptosis (Supplementary Figure S2).
Figure 1.
Hpo overexpression induces cell death. (a) GMR-Gal4 UASGFP eyeantennal imaginal disc showing the GFP reporter expression in GMR domain in the eye disc. (b–d) GMR-Gal4 UASHpo eye-antennal imaginal discs stained for (b) Hpo, (c) activated Drice (Drice*, red), Elav (green) and TOPRO (blue). Drice* marks dying cells in the eyeantennal discs (d, greyscale). (e and f) Mid-pupal retina (∼24APF) from GMR-Gal4 UASHpo animals showing ectopic cell death marked by expression of Drice* (e red, f greyscale), photoreceptor cells marked by Elav (e, blue), and cell shape marked by Discs large (Dlg) (e, green). (f) Ectopic cell death in the retina causes disruption of the retinal lattice. (g and h) Compared to wild type (g), the phenotype of Hpo over expressing retina (h) becomes progressively worse and results in the development of small rough eyes in the adult. (i) Wild-type mid-pupal retina comparable to retina in f marked by Drice*shows developmental apoptosis. (j–l) Wild-type wing imaginal discs (j) from third instar larvae show very little Drice* expression, whereas wing imaginal discs from hsp70-Gal4 UASHpo animals (k and l) show robust induction of cell death as seen by activated Drice* expression (i, red) a few hours after Hpo activation. The orientation of images in a-i is anterior to the right and dorsal up, and the orientation of the wing imaginal discs(j and l) is anterior to left
Diap1 levels remain unchanged in Hpo overexpressing cells
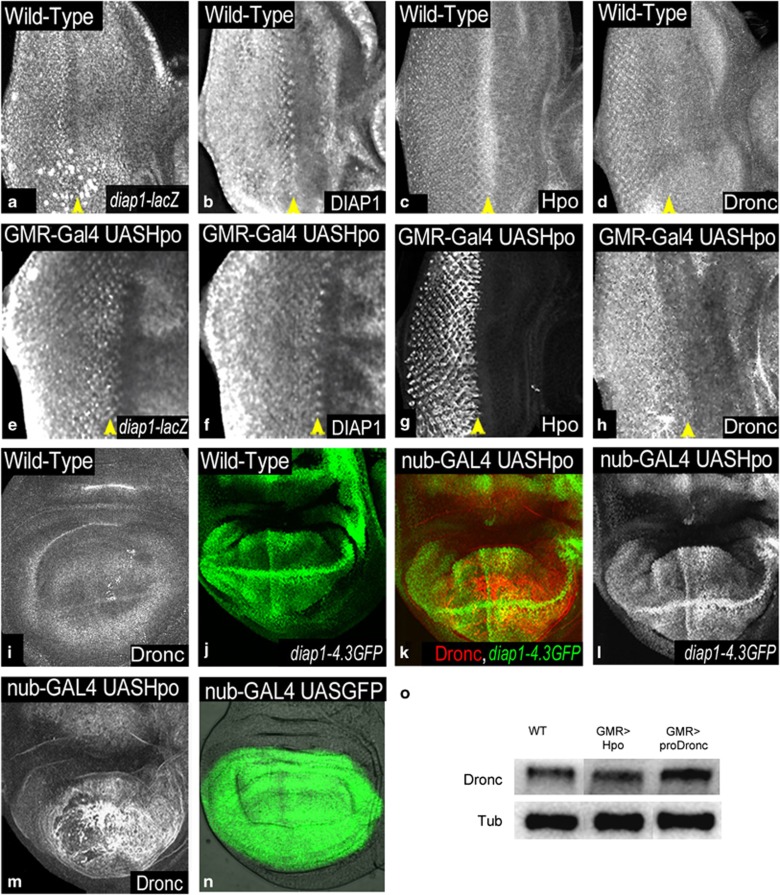
Given that Diap1 is a transcriptional target of the Hippo signalling pathway, and the pro-apoptotic genes act upstream of Diap1, we tested if overexpression of Hpo leads to downregulation of diap1 levels. We found that in GMR-Gal4 UASHpo eyeantennal discs (Figure 2e–h), the levels of diap1-lacZ (Figure 2e) and DIAP1 protein (Figure 2f) remain similar to wild type (Figure 2a and b). Similar results were observed by overexpression of Sav, Wts, or co-expression of pair-wise combination of Sav Wts (Supplementary Figure S3), Hpo Sav (data not shown), or Hpo Wts (data not shown). Next, we tested the Hpo response element (diap1-4.3-GFP) that recapitulates the expression of diap1 under Hpo regulation in the eyeantennal and the wing discs (Figure 2j). Overexpression of Hpo under nub-Gal4 (nub-Gal4 UASHpo) (Figure 2k–m) causes extensive cell death in the wing discs, and results in the development of a small-wing phenotype.6, 7 nub-Gal4 drives expression of transgenes in the wing pouch (nub-Gal4 UASGFP, Figure 2n). diap1-4.3-GFP expression remains unchanged in nub-Gal4 UASHpo wing discs (Figure 2k and l). This result was intriguing as cell death occurred in Hpo-overexpressing cells despite normal DIAP1 expression. DIAP1 overexpression is known to block apoptosis in several apoptotic contexts (e.g., overexpression of Hid).28 As Hpo overexpressing cells induced high levels of cleaved Caspase 3 (Casp3*), a ‘functional read-out' of Dronc activity,33 we tested Dronc expression in cells overexpressing Hpo.
Figure 2.
Hpo overexpression induces Dronc. (a–d) Wild-type expression of (a) diap1-lacZ, (b) DIAP1, (c) Hpo and (d) Dronc in third instar eyeantennal imaginal discs is shown. (e–h) GMR-Gal4 UASHpo eyeantennal imaginal discs showing expression of (e) diap1-lacZ, (f) DIAP1, (g) Hpo and (h) Dronc, respectively. (i and j) Wild-type wing imaginal discs showing normal levels of expression of (i) Dronc and (j) Hpo-response element diap1-4.3GFP. nub-GAL4 UASHpo wing discs show no change in the expression of diap1-4.3GFP (green k, greyscale l), and show induction of Dronc (red k, greyscale m). (n) Wild-type third instar wing imaginal discs showing the domain of expression of nub-Gal4 marked by UAS GFP. (o) Semi-quantitative western blot to assess levels of expression of Dronc. Western blot analysis was performed on protein extracts from wild type (WT), GMR-Gal4 UASHpo and GMR-Gal4 UASproDronc eyeantennal imaginal discs. α-Tubulin is the loading control. Orientation of images is identical in a–h (anterior to the right, dorsal up), and in i–m (anterior left). Yellow arrowheads mark the morphogenetic furrow that coincides with the boundary of the GMR-Gal4 expression domain
Hippo induces Dronc, and requires elevated Dronc activity levels to induce apoptosis
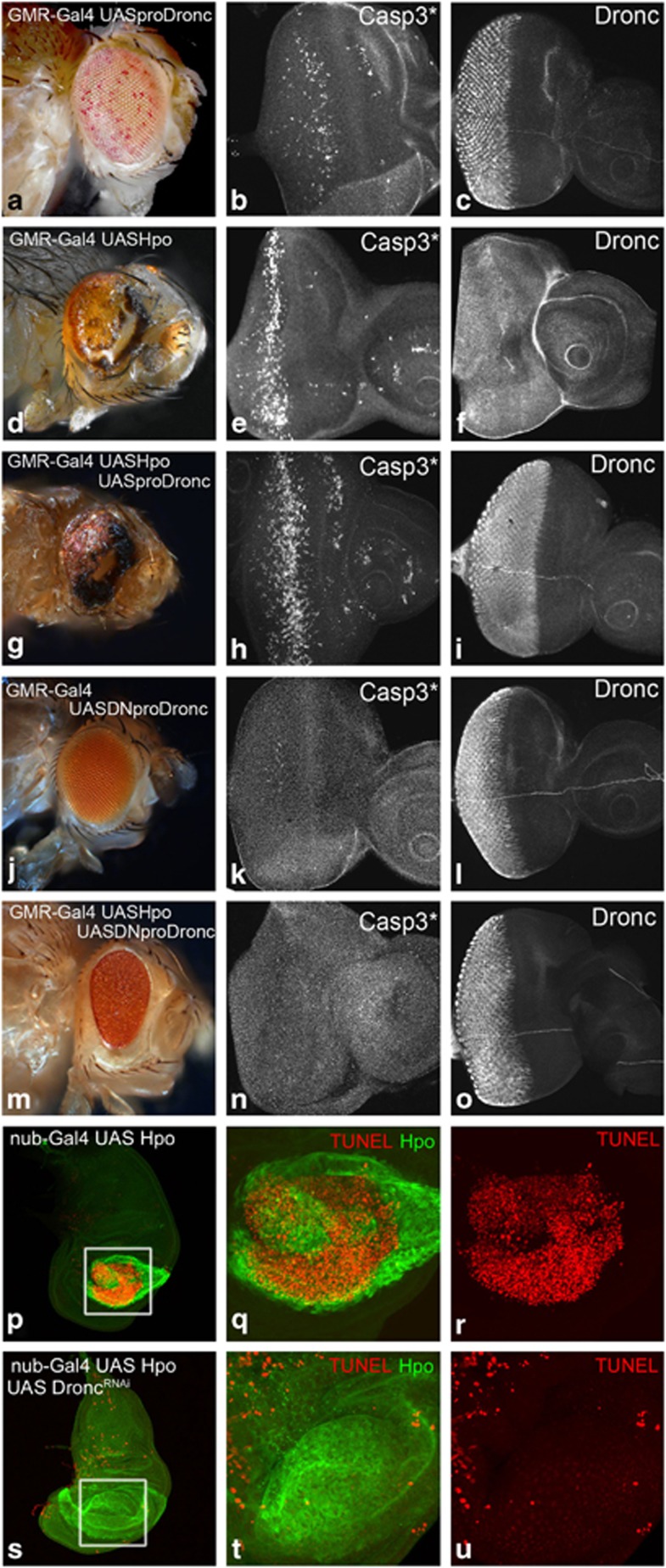
Compared to wild type (Figure 2d and i), overexpression of Hpo induces upregulation of Dronc expression in the eyeantennal (GMR-Gal4 UASHpo, Figure 2h), and the wing discs (nub-Gal4 UASHpo, Figure 2m). Assessment of Dronc protein levels by semi-quantitative western blots further supports this observation (Figure 2o). To understand the Hpo Dronc interaction, we tested the effects of Dronc overexpression (Figure 3, Supplementary Figure S4). In the eyeantennal discs, overexpression of pro-Dronc (GMR-Gal4 UASproDronc, Figure 3c) induces cell death (Casp3*, Figure 3b), without affecting the levels and localisation of Hpo (Supplementary Figure S4) or Yki (Supplementary Figure S4). Consistent with earlier data,27 the adult eyes appear normal sized, and show a ‘spotty-pigmented-eye' phenotype (Figure 3a). Overexpression of Hpo (GMR-Gal4 UASHpo) results in a ‘small-eye' phenotype in the adult (Figure 3d) caused by induction of cell death (Figure 3e) and Dronc (Figure 3f) in eye discs. Co-expression of pro-Dronc with Hpo (GMR-Gal4 UASHpo, UASproDronc, Figure 3g–i) causes increased cell death (Figure 3h) compared to Dronc (Figure 3b) or Hpo alone (Figure 3e); accumulation of Dronc (Figure 3i), and nuclear Yki localisation (Supplementary Figure S4). GMR-Gal4 UASHpo, UASproDronc larvae develop into pharate adults with small eyes and show increased cell death (Figure 3g) compared with eyes from GMR-Gal4 UASproDronc (Figure 3a) or GMR-Gal4 UASHpo (Figure 3d) animals. Thus, hpo genetically interacts with dronc.
Figure 3.
Hpo genetically interacts with Dronc to regulate cell death. Phenotypes of overexpression of GMR-Gal4 UASproDronc, (d–f) GMR-Gal4 UASHpo, (g–i) GMR-Gal4 UASHpo UASproDronc, (j–l) GMR-Gal4 UASDNproDronc and (m–o) GMR-Gal4 UASHpo UAS DNproDronc are shown. A comparison of cell death induced by overexpression of each transgene is shown by expression of activated caspase 3 (Casp3*) antibody in third instar eyeantennal imaginal discs. (a, d and g) The resulting adult phenotype of caspase induction shows mild effects for overexpresssion of UASprodronc (a and b) that are enhanced by co-expression of UASHpo (g and h). Cell death induced by overexpression of UASHpo alone is shown in (d and e) for comparison. (j–o) Overexpression of UASDNproDronc does not induce activated caspase (k) expression resulting in an almost wild-type adult eye phenotype (j). (m–o) Co-expression of UASHpo and UASDNproDronc is unable to induce activated caspase (n) during third instar larval development resulting in a complete rescue of UASHpo-mediated apoptosis in the adult eye (compare m with d). (c, f, i, l and o) These panels show eyeantennal imaginal discs of the indicated genotypes stained for expression of Dronc to confirm overexpression of Dronc transgenes. Note that Dronc levels are upregulated by over expression of UASHpo (f). (p–u) Wing imaginal discs from (p–r) nub-Gal4 UASHpo, and (s–u) nub-Gal4 UASHpo UASDroncRNAi stained for Hpo (green) and apoptosis (TUNEL, red). nub-Gal4 UASHpo discs were identified by the upregulated levels of Hpo expression (green) in the nub-Gal4 domain using an antibody against Hpo. Note that the extensive cell death caused by Hpo over expression (q and r) is suppressed by downregulation of Dronc levels (t and u). However, downregulation of Dronc does not affect Hpo expression (compare q with t). The areas magnified in q, r, t, u are boxed in white in p and s. Orientation of images is identical in all panels
The catalytically inactive form of Dronc acts as dominant negative, and is known to suppress cell death in the Hid/Reaper pathway.27 Adult eyes of flies overexpressing dominant-negative Dronc (GMR-Gal4 UASDNproDronc) appear phenotypically normal (Figure 3j), and precocious cell death is not induced in these eyeantennal discs (as measured by expression of Casp3*) (Figure 3k). To test if apoptosis induced by Hpo requires dronc function, we co-expressed dominant-negative Dronc with Hpo (GMR-Gal4 UASHpo, DNproDronc) (Figure 3m–o). Cell death induced by Hpo overexpression is completely suppressed by the expression of dominant negative proDronc as seen by dramatically reduced levels of Casp3* (Figure 3n). This results in significant rescue of the eye in the adult (Figure 3m) compared to GMR-Gal4 UASHpo alone (Figure 3d), suggesting that apoptosis induced by Hpo requires increased levels of Dronc activity or normal dronc function. In other developing organs, like in the wing discs, overexpression of Hpo with nub-Gal4 (nub-Gal4 UASHpo, Figure 3p–r) results in robust cell death in the wing pouch (Figure 3r) and small wings in the adult. To test if overexpression of Hpo can induce cell death when dronc is downregulated, Hpo was co-expressed with droncRNAi (nub-Gal4 UASHpo, UASdroncRNAi, Figure 3s–u). Cell death was dramatically reduced in these wing discs (Figure 3u) within the Hpo overexpressing cells. Quantification of nub-Gal4 domains in wing discs of larvae overexpressing Hpo (nub-Gal4 UAS-Hpo) or co-expressing Hpo with droncRNAi (nub-Gal4 UASHpo, UASdroncRNAi) showed significant rescue of the nub-Gal4 domain, and the adult wing size is restored (P<0.05, n=4, data not shown). Thus, apoptosis induced by Hpo overexpression requires Dronc.
Hippo pathway downregulates Dronc activity levels to control tissue size
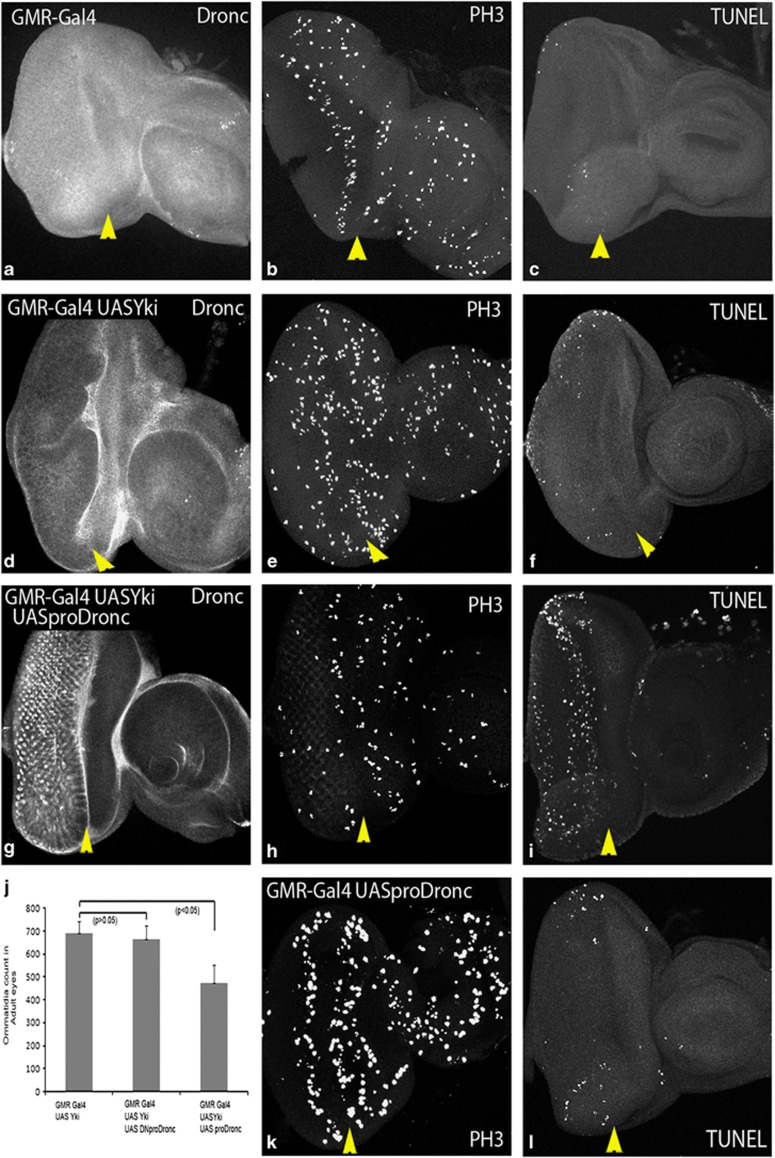
Next, we asked if altered levels of Dronc activity specifically affect Hippo functions connected to the regulation of apoptosis, or it also affects other Hippo functions like restriction of cell proliferation. The transcriptional co-activator Yki is a major target inhibited by the Hippo pathway. Compared to wild type (Figure 4a–c), overexpression of Yki (GMR-Gal4 UASYki; Figure 4d-f) results in overgrowth of adult eyes (Figure 4j) due to downregulation of Hippo signalling. GMR-Gal4 UASYki eyeantennal discs show downregulation of Dronc in the GMR expression domain (Figure 4d) compared with the wild type (Figure 4a). Upregulation of Dronc activity by co-expression of proDronc with Yki (GMR-Gal4 UASYki, UASproDronc; Figure 4g–i) caused a significant reduction in the adult eye-size (Figure 4j). Quantification of adult eye size substantiated our observation that increased Dronc activity significantly reduces Yki-induced overgrowth (Figure 4j, n=5, P<0.05). This data suggests that dronc levels are reduced in cells where Hippo signalling is downregulated, and increased Dronc activity can suppress the effects of downregulation of Hippo signalling.
Figure 4.
Hpo genetically interacts with Dronc to regulate organ size. A comparison of Dronc levels, mitotic figures and TUNEL-profiles in the GMR-Gal4 domain of eyeantennal imaginal discs from wild type, (d–f) GMR-Gal4 UASYki, (g–i) GMR-Gal4 UASYki, UASproDronc and (k–l) GMR-Gal4 UASproDronc animals is shown. Compared to wild type (a), Dronc levels are downregulated in cells overexpressing Yki (d). (b and c) Show a typical proliferation and cell death profile of wild-type eyeantennal imaginal discs, where cells undergo mitotic arrest posterior to the SMW (second mitotic wave) (b). Apoptosis is not induced at this stage of development (c). (e and f) In GMR-Gal4 UASYki eyeantennal discs the numbers of mitotic figures (PH3, e) are significantly increased in Yki overexpressing cells posterior to the SMW (e), and apoptosis is not induced (f). Co-expression of UASproDronc with UASYki causes a reduction in the number of mitotic figures posterior to SMW (h) and induces apoptosis (i). Control discs showing overexpression of UASproDronc stained for PH3 (k) and apoptosis (l) are shown for comparison. (j) Shows quantification of adult eye size from flies overexpressing UASYki alone or together with UASDNproDronc (P>0.05) or UASproDronc (P<0.05). Adult eye size was measured by counting the total number of ommatidia (n=5) for each genotype. Magnification and orientation of images is identical in all panels. The yellow arrows indicate the position of morphogenetic furrow that marks the boundary of the GMR-Gal4 domain. All comparisons are made in the region posterior (i.e. left) of the morphogenetic furrow. The color reproduction of this figure is available at the Cell Death and Differentiation journal online
This effect could either be due to the pro-apoptotic effects of Dronc, or Yki-expressing cells may show reduced proliferation when Dronc is overexpressed. To distinguish between these possibilities, we marked cells entering M-phase of cell division with Phospho-Histone H3 (PH3) antibody, and cells undergoing apoptosis with TUNEL. In wild-type eyeantennal discs, the pattern of cell division is random anterior to the MF, whereas posterior to the MF, the cells undergo cell cycle arrest and divide once in the second mitotic wave (SMW, Figure 4b). In comparison, not many cells undergo apoptosis at this stage of development in wild-type eyeantennal discs (Figure 4c). Overexpression of Yki (GMR-Gal4 UASYki) resulted in high levels of PH3 expression posterior to the SMW in the GMR domain, indicating ectopic proliferation (Figure 4e), whereas little apoptosis was seen in the eyeantennal discs (Figure 4f). Overexpression of pro-Dronc alone (GMR-Gal4 UASproDronc) shows no effect on PH3 levels (Figure 4k) and mild affect on apoptosis (Figure 4l) compared to wild type (Figure 4b and c). Co-expression of proDronc in Yki-expressing cells (GMR-Gal4 UASYki, UASproDronc) showed fewer PH3-positive mitotic cells (Figure 4h), and a dramatic increase in TUNEL labelling (Figure 4i), compared to GMR-Gal4 UASYki (Figure 4e and f). Thus, the reduction in the size of GMR-Gal4 UASYki, proDronc eyes could be attributed to both decreased proliferation and increased apoptosis in cells where Hippo signalling is downregulated.
Dronc activity levels affect Hippo function
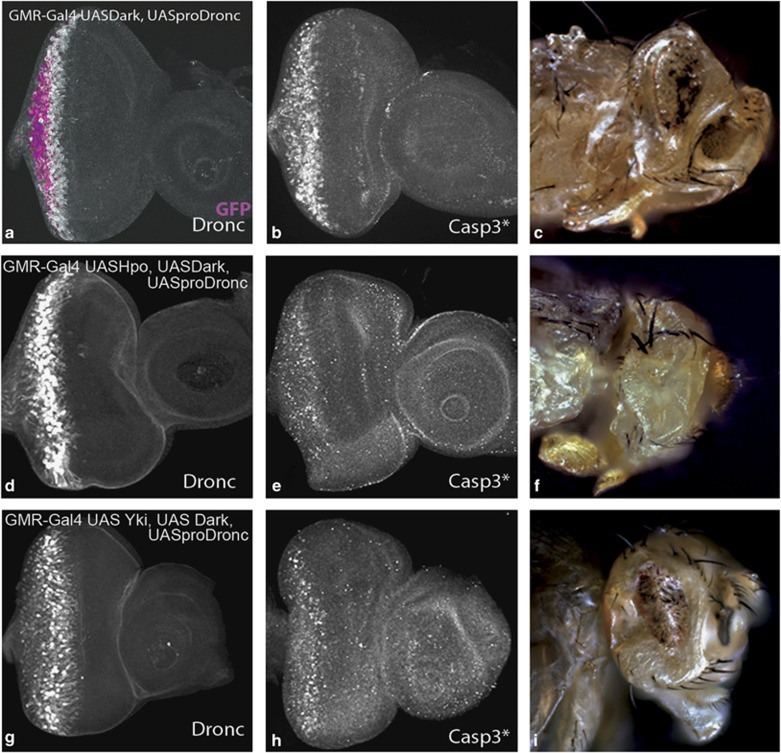
Dronc requires Dark to form the apoptosome holoenzyme, and they act in a feedback inhibitory loop to regulate caspase activation.32 To further investigate the effect of Dronc activity levels on the regulation of cell death, we co-expressed Dronc with its binding partner Dark. Overexpression of Dark in the developing eyeantennal discs (GMR-Gal4 UASDark) does not induce significant apoptosis.32 Co-expression of Dark and Dronc together (GMR-Gal4 UASDark, UASproDronc, UASGFP) induced massive apoptosis in the eyeantennal discs (Figure 5a and b) and adult eyes (Figure 5c).32 This phenotype is comparable to the GMR-Gal4 UASHpo phenotype (Figure 1h). Co-expression of Dark, Dronc and Hpo (GMR-Gal4 UASDark, UASproDronc, UASHpo) induced massive apoptosis in the eyeantennal disc (Figure 5d and e) and adult (Figure 5f). This phenotype (Figure 5d and f) appears stronger compared with the controls (Figure 5c) because both photoreceptor and pigment cells are eventually eliminated in animals co-expressing Hpo (Figure 5e). In all experiments where we co-expressed Dark and Dronc, we observed that Dronc is expressed strongly in a band (Figure 5a and d) that roughly corresponds with the band of Casp3* (Figure 5b and e), indicating the region in which Dronc activity is high. Thus, our data suggest that Hpo-mediated apoptosis is enhanced by increased Dronc activity. We next tested if Hippo-mediated regulation of cell proliferation is affected by increased Dronc activity (Figure 5g–i). Co-expression of Dark and Dronc with Yki (GMR-Gal4 UASDark, UASproDronc, UASYki) completely suppressed the overgrowth phenotype of Yki overexpression (Figure 5i), due to induction of apoptosis (Figure 5h) in response to increased Dronc activity (Figure 5g). In summary, these data further validate our hypothesis that Dronc acts genetically downstream of Hpo, and keeping Dronc activity levels under control are important for Hippo to regulate both cell proliferation and cell death.
Figure 5.
Hippo pathway regulates Dronc activity levels to control growth and apoptosis. Co-expression of Dark and proDronc with UASGFP (magenta in a) (GMR-Gal4 UASGFP, UASDark, UASproDronc) induces (a) Dronc (greyscale), and (b) activated Caspase 3 (Casp3*) in eyeantennal imaginal discs, resulting in development of small eyes in adult flies (c). The effect of co-expression of Dark and Dronc with Hpo (GMR-Gal4 UASHpo, UASDark, UASproDronc) (d–f) and Yki (GMR-Gal4 UASYki, UASDark, UASproDronc) (g–i) on the expression of Dronc (d and g), and Casp3* (e and h) in eyeantennal imaginal discs and in adult eyes (f and i) is shown. Magnification and orientation of images is identical in all panels. The color reproduction of this figure is available at the Cell Death and Differentiation journal online
Loss of dronc induces excess proliferation and cell survival
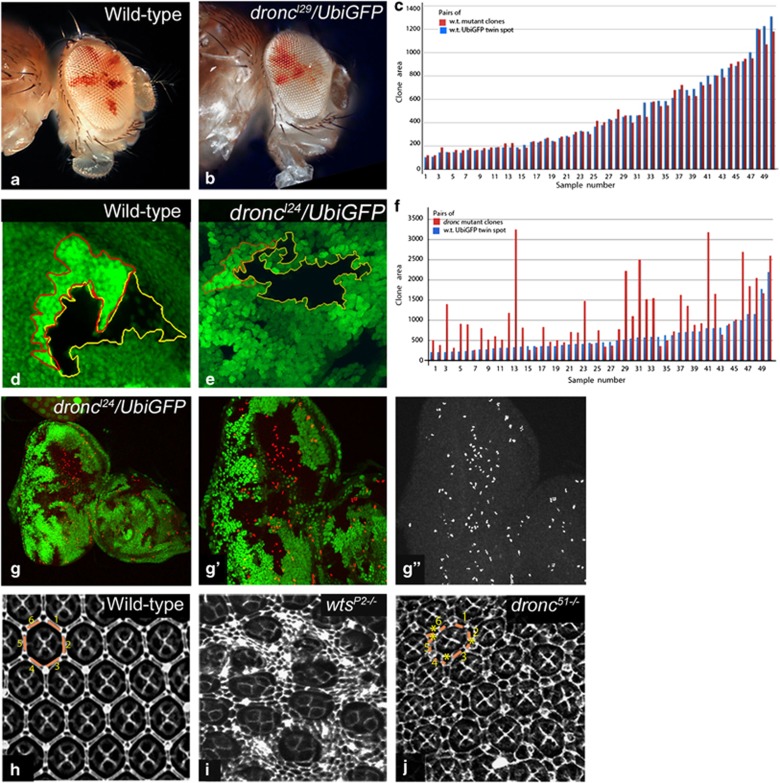
So far our data suggests that decreased Dronc levels coincide with increased cell proliferation (Figure 4d and e). To directly test if dronc affects cell proliferation, we compared growth rates of wild-type and dronc mutant cells in genetic mosaics (Figure 6a and b) generated using the eyFLP system.11 In wild-type controls (Figure 6a), the adult eye is comprised of roughly equal number of white and red cells, indicating that the growth rates for both cell populations are comparable. The adult eyes mosaic for dronc mutant cells are predominantly white (Figure 6b). This indicates that dronc mutant cells (Figure 6b, white) proliferate faster than wild-type cells (red) and out-compete them. Next, we compared the size of dronc mutant clones to their wild-type twin spots for three null alleles droncI24, droncI29 and dronc2 in wing discs to rule out the effects of background mutations.34 We also confirmed that droncI24, droncI29 and dronc2 failed to complement dronc51. Compared to wild-type clones (Figure 6c and d), the size of droncI24 mutant clones is much larger than their wild-type twin spots (Figure 6e and f), and the difference in cell numbers is significant when assessed using the t-test (P<0.003). The same experiment with droncI29 and dronc2 showed similar difference (data not shown). We also tested PH3 expression in dronc mutant clones (Figure 6g–g″) and found that loss of dronc induced ectopic proliferation (Figure 6g″). Compared with wild type (Figure 6h), Hippo pathway mutants show robust survival of extra pigment cells (Figure 6i) in the pupal retina at ∼20hAPF. dronc null mutants (for example, dronc51, droncI24, droncI29 or dronc2) also showed extra interommatidial pigment cells in the pupal retina (Figure 6j).34 Taken together, our data demonstrates that dronc affects cell proliferation and developmental cell death.
Figure 6.
dronc mutants show cell survival. (a and b) Shows comparison of wild-type and droncI29 adult mosaic eyes made using the cl w+ technique11 to compare proliferation rate of wild-type (red) and dronc mutant (white cells). Note that dronc mosaic eyes show fewer red cells, indicating that dronc mutant cells proliferate faster than wild-type cells. (c–f) Analysis of clone sizes between GFP negative clones and their wild-type twin spots (GFP positive) for wild-type (c and d) and droncI24 mutants (e and f) are shown. Clone sizes were measured from wing imaginal discs dissected from mid third instar larvae. Representative twin clones from wild-type (d) and droncI24 (e) wing imaginal discs are shown for reference. (g-g″) droncI29 mutant clones (GFP negative) induce ectopic cell proliferation (PH3, red) posterior to the second mitotic wave in the eyeantennal disc. (g′ and g″) Higher magnification of the area boxed in g is shown in g′ to show extra cell proliferation in mutant clones. The PH3 expression is shown in greyscale in g″. (h–j) Mid-pupal retinae from wild-type (h) wtsP2−/− (i) and dronc51−/− (j) mutants are shown. Cell outlines in mid-pupal retinae are marked using Dlg antibody. dronc mutant cells show survival of extra inter-ommatidial cells as marked by asterisks (see Figure 6j) The pigment cells for one wild-type (h) and one dronc mutant (j) ommatidium are numbered and marked by yellow lines. Magnification is identical for adult flies (a and b), imaginal discs (d, e, g′–g″) and pupal retinae in h–j. Imaginal discs in panels g is at a lower magnification (20 × )
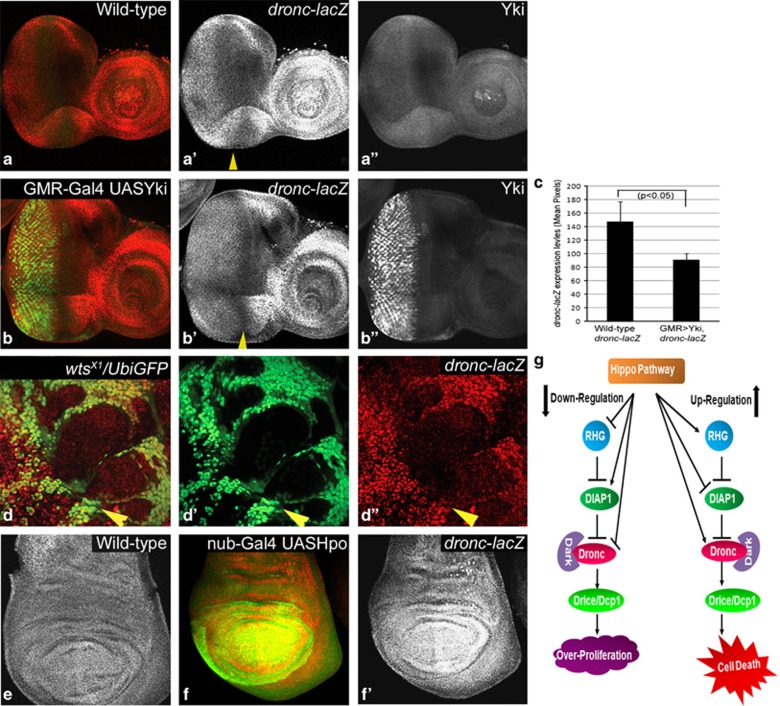
Our data suggests that dronc may be directly controlled by Hippo signalling. To investigate this, we analysed the expression of a dronc-transcriptional reporter (dronc1.7 kb-lacZ) where 1.7 kb of dronc genomic sequence drives lacZ expression.35 dronc1.7 kb-lacZ is ubiquitously expressed in wild-type eyeantennal (Figure 7a–a″) and wing (Figure 7e) discs. The expression of dronc1.7 kb-lacZ is significantly suppressed by overexpression of Yki (Figure 7c, n=5, P<0.033) in the GMR expression domain (GMR-Gal4 UASYki, Figure 7b–b″). Compared to the strong suppression of Dronc in cells overexpressing Yki (Figure 4d), the expression of dronc1.7 kb-lacZ appears to be not as dramatically suppressed, perhaps due to perdurance of the β-galactosidase. However, in wts mutant cells the expression of dronc1.7 kb-lacZ is strongly suppressed throughout the eyeantennal discs (Figure 7d–d″). Overexpression of Hpo in the eyeantennal (GMR-Gal4 UASHpo, data not shown) or wing disc (nub-Gal4 UASHpo, Figure 7f′) results in robust induction of dronc1.7 kb-lacZ (Figure 7f′) expression. Thus, dronc is a transcriptional target of the Hippo signalling pathway.
Figure 7.
Hippo regulates dronc transcription. Third instar eyeantennal imaginal discs from wild type (a-a″), GMR-Gal4 UASYki/dronc1.7 kb-lacZ (b-b″) and eyFLP; dronc1.7 kb-lacZ/+ FRT82 wtsX1/FRT82B UbiGFP (d-d″), larvae stained with α-β-gal to detect dronc1.7 kb-lacZ expression levels. (c) Quantification of dronc1.7 kb-lacZ levels measured by pixel intensity values of m β-gal expression in the GMR-Gal4 domain from eyeantennal imaginal discs of wild-type and GMR-Gal4 UAS Yki larvae (n=5, P<0.05). Third instar wing imaginal discs from wild-type (e) and nub-Gal4 UASHpo/ dronc1.7 kb−lacZ (f and f′) larvae stained with α β-gal to detect dronc1.7 kb−-lacZ expression levels. (g) Model for Hippo pathway mediated regulation of genes within the intrinsic cell death pathway. During homoeostasis, Diap1 restricts activation of caspases via proteosomal degradation. However, in many cells low levels of active apoptosome complexes are constantly generated in vivo by association of Dronc and Dark. Loss of function of Hippo pathway restricts Hid and Dronc expression and positively regulates Diap1 in living cells, which allows cells to proliferate and evade apoptosis. In naturally occurring cell death pathways that require Hippo activity, such as the killing of interommatidial cells, a death-inducing signal may activate Hpo, which in turn may induce expression and activation of Hid and Dronc, as well as downregulation of DIAP1 to trigger apoptosis. A similar scenario may be envisaged for other situations like stress (induced by overexpression of Hpo) that induce apoptotic response. The color reproduction of this figure is available at the Cell Death and Differentiation journal online
Discussion
During organogenesis, optimum levels of Hippo signalling are crucial for maintaining tissue homoeostasis. Our analysis of the regulation of cell death by overexpression of Hpo (Figure 1)3, 4, 6, 7 revealed that Hpo requires its own signalling pathway involving the transcriptional co-activator Yki (Supplementary Figure S1), and pro-apoptotic genes of the intrinsic cell death pathway to induce cell death (Supplementary Figure S2). We found that overexpression of Sav, Wts or Hpo does not accompany changes in Diap1 levels (Figure 2, Supplementary Figure S3). Thus, it is possible that in Hpo overexpressing cells diap1 levels may not be detectably altered or that more sensitive assay may be required to resolve the effects of Hpo overexpression on regulation of Diap1 expression. As overexpression of Sav, Wts or Hpo caused robust induction of activated Caspase 3 (Drosophila Drice/Casp3*), we decided to test if the initiator caspase Dronc that controls Casp3* activation is regulated by the Hippo pathway. Here, we show that dronc, the Drosophila homologue of mammalian Caspase9 and a member of the intrinsic cell death pathway is a novel target of the Hippo pathway. We show that Hippo signalling regulates dronc levels to control tissue size, and Dronc genetically acts downstream of Hpo. dronc mutants show increased proliferation and decreased apoptosis, a phenotype shared by several tumour suppressor genes within the Hippo pathway (e.g., hpo, sav, wts).
We present several lines of evidence indicating that Dronc may be important for Hippo functions. First, the levels of Dronc expression and activity correlate with increased or reduced Hippo activity levels. Dronc is induced when Hpo is overexpressed (Figure 2h and m), and suppressed when Hippo signalling is downregulated (Figure 4d). Dronc activity was monitored by the expression of activated Casp3*, which is induced (Figure 3b) in cells showing high levels of active Dronc (e.g., in GMR-Gal4 UASproDronc) and suppressed (Figure 3k) in cells where Dronc activity is reduced (e.g., in GMR-Gal4 UASDNproDronc). When Dronc activity is high co-expression of Hpo (GMR-Gal4 UASproDronc, UASHpo) leads to increased cell death (Figure 3h), however, when Dronc activity is reduced by overexpression of UASDNproDronc co-expression of Hpo (GMR-Gal4 UASDNproDronc, UASHpo) is unable to induce cell death (Figure 3n). Second, dronc genetically interacts with members of the Hippo pathway (Figures 3 and 4). Genetic epistasis experiments (Figure 3j–u) show that Hpo requires dronc to induce cell death, and Hpo-mediated apoptosis can be suppressed when dronc is downregulated (Figure 3s–u). Thus, dronc acts downstream of Hpo. Third, increased Dronc activity suppresses the overgrowth phenotypes of loss of Hippo signalling (Figure 4g–i). Co-expression of Yki and proDronc showed that increased Dronc levels led to reduced cell proliferation and increased cell death (Figure 4h and i), suggesting that dronc levels need to be regulated in order for Hippo pathway to control organ size. Co-expression of Dark and Dronc can suppress the overgrowth phenotype of Hippo pathway components as well as enhance the cell death induced by Hpo overexpression (Figure 5). The Dronc Dark interactions show that Hpo-induced cell death is mediated through Dronc activity as cell death (Casp3*, Figure 5e and h) is seen only in the cells where Dronc is upregulated (Figure 5d and g). Taken together, our data suggests the Hippo signalling pathway components control Dronc activity levels to control cell proliferation and cell survival.
Analysis of dronc loss of function reveals that dronc mutant cells proliferate faster than their wild-type neighbours (Figure 6a–f). dronc mutant cells show ectopic cell division posterior to the SMW (Figure 6g–g″), indicating that dronc mutant cells continue to divide when wild-type cells have stopped dividing. In addition, dronc mutant cells show extra cell survival as seen in mid-pupal retina that shows extra interommatidial cells (Figure 6h–j). Thus, dronc may be required to regulate both cell proliferation and apoptosis. It is interesting to note that all alleles of dronc are semi-lethal, and several of them cause homozygous larvae to enter an extended larval period (of up to 12days) during which the larvae develop hyperplasia of the brain lobes, imaginal discs and circulating hemocytes, melanotic tumours and increased larval body size.34, 36, 37, 38 These observations strongly support our findings that dronc mutants have defects in the control of cell proliferation, and in addition, there is decreased apoptosis that contributes to their mutant phenotype.
Other evidence shows that mutants of Drosophila APAF-1 (or ark or hac-1) have a growth advantage, as strong ark alleles have been identified in screens involving mitotic recombination where the mutant clones (ark−/−) were larger than the wild-type twin spots.39 These ark mutants (point mutations and deletions) were recovered from the same screen that yielded two hpo mutants.4 Furthermore, these strong ark mutants show similar pupariation defects as dronc mutants; and hyperplasia of the brain lobes and imaginal discs.39 These findings further support the idea that genes involved in the regulation of apoptosis may have growth-regulatory functions.
It is interesting to note that like most Hippo pathway components Dronc is expressed ubiquitously. Our findings show that dronc is transcriptionally regulated by the Hippo pathway – loss of Hippo signalling leads to transcriptional suppression of dronc (Figure 7b and d), whereas Hpo overexpression causes transcriptional induction of dronc (Figure 7f). Thus, it is likely that loss of function of Hippo pathway causes dronc suppression that protects mutant cells from activation of caspases, and contributes to their overgrowth phenotype. Overexpression of Hippo pathway components, on the other hand, upregulates dronc expression and activation of the downstream effector caspases (Drice/Casp3*) that results in cell death. In summary, we show that dronc is a novel transcriptional target of Hippo signalling.
We have developed a model (Figure 7) to reconcile our findings with the current models of Hippo-mediated regulation of genes in the intrinsic cell death pathway. Hippo pathway is known to regulate cell death by transcriptional regulation of Hid, Diap13, 6, 7 and the bantam miRNA.1 In normal cells there is a balance between diap1 expression and basal levels of activated caspases, and Hippo signalling maintains tissue homoeostasis by negatively regulating hid and dronc, and positively regulating diap1 (Figure 7). When cells experience stress (e.g., due to overexpression of Hpo), they activate the proapoptotic gene hid and the initiator caspase dronc downstream of the Hippo pathway to trigger apoptosis (Figure 7).
Both in flies and mammals, hyperactivation of Hippo or MST1/2 signalling is known to induce apoptosis.1, 40 MST1/2 are known targets of caspases. Furthermore, YAP1/2 are known to interact with p73 via a PDZ domain in YAP, and induce apoptotic target genes.40 However, these mechanisms of regulating apoptosis may not be conserved in flies because the site for caspase cleavage is not conserved in Drosophila Hpo,6 and Drosophila Yki does not have the conserved PDZ domain. Thus, Hpo overexpression in flies induces apoptosis through an alternate mechanism that does not involve caspase cleavage or p73. Many cell–cell interactions are important for normal development or are initiated in response to tissue damage, for example, compensatory proliferation, cell competition and regeneration. The Hippo pathway has emerged as an important regulator of all these cell–cell interactions and Dronc is linked to the compensatory proliferation response. Our studies now link Dronc to the Hippo pathway and provide a possible mechanism by which Hippo signalling regulates cellular responses in Drosophila. Thus, in the future, it will be interesting to investigate the molecular mechanism underlying Hpo-mediated Dronc regulation, and if this interaction is conserved in higher vertebrates.
Materials and Methods
Fly stocks
The following fly stocks were used for the experiments: Df(3L)H99 FRT80B/TM6B, dronc2 FRT80, droncI24 FRT80, droncI29 FRT80, dronc1.7 kb-lacZ, dronc2.8 kb-lacZ (A Bergmann), UASHpo, GMR-Gal4, nub-Gal4, hsp70pb-Gal4, FRT82B wtsP2/TM6B, UASDroncRNAi, diap1j5c8/TM6B, yw eyFLP; cl w+ FRT80/TM6B (G Halder), GMR-Dronc (B Hay), UASDiap1-4.3-GFP (J Jiang), UASHpoRNAi (N Tapon), UASYki (DJ Pan), UASproDronc, UASDNproDronc(C-A), UASDark (E Arama), dronc51 FRT79/TM6B (J Abrams), GMR-Sav, GMR-Wts (I Hariharan), and yw eyFLP; P[w+]Ubi-GFPnls FRT80B/TM6B (Bloomington). All crosses were performed at room temperature (22 °C) unless otherwise mentioned.
Overexpression using GAL4 UAS system
We have used GMR-GAL4 that drives expression of UAS-linked transgenes posterior to the morphogenetic furrow in the differentiating cells, and in post-mitotic cells of the developing eye (Figure 1a). nub-GAL4 drives expression of UAs-linked transgenes in the wing pouch of the developing wing disc (Figure 2n). Hsp70pb-GAL4 drives expression of transgenes following heat-shock induction. Hsp70pb-Gal4 UAS-Hpo larvae were heat shocked for 20 min by incubating at 37 °C in a water bath (Figure 1I and j).
Mosaic analysis
Mutant clones were induced in eyeantennal or wing discs using the FLP/FRT mitotic recombination system. We used three null alleles of dronc that were of independent origin; dronc2 was obtained in a different mutagenesis screen a few months after the mutagenesis that resulted in the isolation of droncI24 and droncI29 (A Bergmann, personal communication). To generate dronc mutant clones (Figure 6), dronc FRT80B/TM6B virgin females, carrying either y w eyFLP or y w UbxFLP on the X chromosome, were crossed to either y w; P[w+]Ubi-GFPnls FRT80B/TM6B or y w; cell lethal P[w+] FRT80B/ TM6B male flies. Twin-spot analysis and immunohistochemistry was performed on mutant clones from these crosses. In Supplementary Figure S2, Df(3L)H99 mutant clones were induced by out-crossing flies of the genotype y w; +: Df(3L)H99 FRT80B to y w eyFLP; + Ubi-GFP FRT80B. Similarly, the genetic epistasis experiment in Supplementary Figure S2 was done by crossing together y w eyFLP; GMR-Gal4; Ubi-GFP FRT80B and y w; UASHpo; Df(3L)H99 FRT80B flies.
Scanning electron microscopy (SEM), immunohistochemistry, TUNEL assay
Adult flies were processed for SEM by using the hexamethyldisilazane (HMDS) method with modifications.11 Flies were fixed for a day in 70% acetone, and washed twice in 100% acetone for 4 h each. Acetone was then exchanged with HMDS through two washes in 1 : 1 acetone:HMDS and two washes in 100% HMDS over 2 days. Dehydrated flies were mounted on electron microscopic stubs. Flies were coated with gold using a Denton vacuum sputter coater and analysed using a Hitachi (Schaumburg, IL, USA) S-4800 High Resolution Scanning Electron Microscope (HRSEM). Eyeantennal and wing discs were dissected from wandering third instar larvae in 1 × PBS and fixed in 4% paraformaldehyde (in PBS) for 20 min. They were washed three times with PBST (1 × PBS+2% Triton X-100) for 10 min each and blocked with 2% normal donkey serum for 1 h. The discs dissected from larvae were incubated overnight at 4 °C in primary antibody. The discs were washed in PBST twice for 10 min each. The disc samples were incubated for ∼2 h in secondary antibodies and washed in PBST for 10 min. The discs were washed in PBST (2 × 15 min), mounted in Vectashield mountant, and scanned on an Olympus (Center Valley, PA, USA) Fluoview 1000 confocal microscope. Pupal retinae dissected 24 h after puparium formation (APF) were processed essentially using the same protocol except that the sample was processed in a glass-well plate rather than a microfuge tube.
The following primary antibodies were used: rat α-Elav (1 : 50, DSHB), m-β-gal (1 : 300, DSHB) rb α-activated Drice (1 : 1000, B Hay), m α-DIAP1 (1 : 250, B Hay), rb α-Dronc (1 : 1000, B Hay), rb α-DLG (1 : 200, K Cho), m α-Cut (1 : 200, DSHB), gp α-Dronc (1 : 500, HD Ryoo), rb α-Yki (1 : 400, K Irvine), rt α-Hpo (1 : 250, G Halder), gp α-Hpo (1 : 2000, G Halder), m α-Phospho-Histone3 (1 : 100, Upstate, Millipore, Billerica, MA, USA), rb α-active Casp3* (1 : 250, Cell Signaling). Nuclei were marked by TOPRO (1 : 1500, Molecular Probes Inc.). The following secondary antibodies (Jackson Immunoresearch) were used: α-m Cy3 (1 : 1000), α-rb FITC (1 : 250) α-rb Cy3 (1 : 1000), α-gp Cy3 (1 : 1000), α-gp FITC (1 : 250), α-gp Cy5 (1 : 400), α-rt FITC (1 : 250), α-rat Cy3 (1 : 1000) and α-gp Cy5 (1 : 250).
TUNEL labelling was performed on the imaginal discs and pupal retinae using an in-situ cell death detection kit (Roche, Indianapolis, IN, USA) following the manufacturer's protocol.
Western blot
Eyeantennal discs were dissected in cold PBS, and lysed on SDS sample buffer. Western blots were performed according to standard protocols. The antibodies used were m-αDIAP1 (1 : 6000), gp-αDronc (1 : 6000), and m α-alpha Tubulin (Sigma, 1 : 10 000). Images were captured using the BioSpectrum 500 Imaging System (UVP, Upland, CA, USA).
Statistical analysis
For counts presented in Figure 4j, total number of ommatidia in the adult eyes were counted for each genotype (n=5) and error bars indicate S.D. of mean. For twin-spot analysis in Figure 6c and f, clone sizes were determined by measuring mean pixel values using the Histogram function of Adobe Photoshop 5.5 (n=50). P-values were calculated using t-test (one-tailed) in Microsoft Office Excel 2007.
Acknowledgments
We would like to thank J Abrams, E Arama, A Bergmann, G Halder, I Hariharan, B Hay, KD Irvine, J Jiang, S Kumar, DJ Pan, HD Ryoo, A Singh, Bloomington Drosophila Stock Centre and the Developmental Studies Hybridoma Bank for fly stocks and antibodies; the NEST facility at the University of Dayton for access to the SEM and the Olympus Fluoview 1000 Confocal microscope, and members from Singh and Kango-Singh labs for many helpful discussions and comments on the manuscript. MKS and SV would like to thank Daniel McCorry, Katelin Hanes, Mackenzie Sullivan, Nanditha Ranganathan and Lauren Sciarappa for their assistance with sample preparation for immuno-histochemistry and SEM; and Dr. Amit Singh, Dr. Janice Fischer, Dr. Georg Halder and Dr. Tin Tin Su for comments on the manuscript. SV was supported in part by the Graduate Student Summer Fellowship from the University of Dayton. This publication was made possible by grants from the Knights Templar Eye Foundation and the University of Dayton start-up support to MKS.
Glossary
- Yki
Yorkie
- Hpo
Hippo
- Wts
Warts
- Mats
Mob as tumour suppressor
- Ex
Expanded
- Mer
Merlin
- Crb
Crumbs
- Sd
Scalloped
- Hth
Homothorax
- Tsh
Teashirt
- DIAP1
Drosophila inhibitor of apoptosis protein-1
- IAP
inhibitor of apoptosis protein
- Sav
Salvador
- HID
head involution defective
- Dronc
Drosophila Nedd-2-like caspase
- RHG
Reaper, Hid Grim
- Dark
Drosophila Apaf-1 related killer
- Casp3*
Caspase 3
- PH3
Phospho-Histone H3
- MF
morphogenetic furrow
- SMW
second mitotic wave
- APF
after puparium formation
- YAP
Yes-associated protein
- MST
Mammalian Ste 20-like kinase
- DLG
disc large tumour suppressor
- PDZ
PSD95, Drosophila (Dlg-1), ZO-1 protein
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labelling
- Drice
Drosophila interleukin converting enzyme
- UAS
upstream activator sequence
- GFP
green fluorescent protein
- FERM
4.1 protein, ezrin, radixin, moesin
- BIR
Baculovirus IAP Repeat
- GMR
glass multimeric repeat
- RT
room temperature
- Nub
nubbin
- DN
dominant negative
- DCP-1
Death Caspase-1
- ELAV
embryonic lethal, abnormal vision
- hsp
heat shock protein
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by H Steller
Supplementary Material
References
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kango-Singh M, Singh A. Regulation of organ size: insights from the Drosophila Hippo signaling pathway. Dev Dyn. 2009;238:1627–1637. doi: 10.1002/dvdy.21996. [DOI] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, Hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey K, Bell D, Wahrer D, Schiripo T, Haber D, et al. Salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, et al. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, et al. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, et al. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci USA. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, et al. The fat cadherin acts through the Hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the Hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, et al. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Quinn LM, Dorstyn L, Mills K, Colussi PA, Chen P, Coombe M, et al. An essential role for the caspase dronc in developmentally programmed cell death in Drosophila. J Biol Chem. 2000;275:40416–40424. doi: 10.1074/jbc.M002935200. [DOI] [PubMed] [Google Scholar]
- Meier P, Silke J, Leevers SJ, Evan GI. The Drosophila caspase DRONC is regulated by DIAP1. EMBO J. 2000;19:598–611. doi: 10.1093/emboj/19.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme M, Meier P. Inhibitor of apoptosis proteins in Drosophila: gatekeepers of death. Apoptosis. 2009;14:950–960. doi: 10.1007/s10495-009-0358-2. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams JM. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat Cell Biol. 1999;1:272–279. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- Hawkins CJ, Yoo SJ, Peterson EP, Wang SL, Vernooy SY, Hay BA. The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J Biol Chem. 2000;275:27084–27093. doi: 10.1074/jbc.M000869200. [DOI] [PubMed] [Google Scholar]
- Muro I, Monser K, Clem RJ. Mechanism of Dronc activation in Drosophila cells. J Cell Sci. 2004;117 (Pt 21:5035–5041. doi: 10.1242/jcs.01376. [DOI] [PubMed] [Google Scholar]
- Shapiro PJ, Hsu HH, Jung H, Robbins ES, Ryoo HD. Regulation of the Drosophila apoptosome through feedback inhibition. Nat Cell Biol. 2008;10:1440–1446. doi: 10.1038/ncb1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell Death Differ. 2010;17:534–539. doi: 10.1038/cdd.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Li Y, Arcaro M, Lackey M, Bergmann A. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development. 2005;132:2125–2134. doi: 10.1242/dev.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daish TJ, Cakouros D, Kumar S. Distinct promoter regions regulate spatial and temporal expression of the Drosophila caspase dronc. Cell Death Differ. 2003;10:1348–1356. doi: 10.1038/sj.cdd.4401312. [DOI] [PubMed] [Google Scholar]
- Waldhuber M, Emoto K, Petritsch C. The Drosophila caspase DRONC is required for metamorphosis and cell death in response to irradiation and developmental signals. Mech Dev. 2005;122:914–927. doi: 10.1016/j.mod.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Daish TJ, Mills K, Kumar S. Drosophila caspase DRONC is required for specific developmental cell death pathways and stress-induced apoptosis. Dev Cell. 2004;7:909–915. doi: 10.1016/j.devcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Chew SK, Akdemir F, Chen P, Lu WJ, Mills K, Daish T, et al. The apical caspase dronc governs programmed and unprogrammed cell death in Drosophila. Dev Cell. 2004;7:897–907. doi: 10.1016/j.devcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Mills K, Daish T, Harvey KF, Pfleger CM, Hariharan IK, Kumar S. The Drosophila melanogaster Apaf-1 homologue ARK is required for most, but not all, programmed cell death. J Cell Biol. 2006;172:809–815. doi: 10.1083/jcb.200512126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini E, Oka T, Sudol M, Strano S, Blandino G. YAP: at the crossroad between transformation and tumor suppression. Cell Cycle. 2009;8:49–57. doi: 10.4161/cc.8.1.7259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.