Abstract
In the present study, we standardized a TaqMan locked nucleic acid (LNA) real-time polymerase chain reaction (PCR) probe for the accurate quantification and detection of hepatitis B virus (HBV) DNA in serum (plasma), and evaluated its methodology. LNA probe technology had a much better detection performance in HBV DNA than the common TaqMan probe. The assay based on the LNA probe had a wider linear detection range, higher sensitivity, stability and amplification efficiency, and a lower concentration of probes than the TaqMan probe. Among the 15 cases with chronic hepatitis B surface antigen (HBsAg) (+) alone, only 4 cases that were detected by TaqMan real-time PCR were negative; however, the same samples were positive by LNA real-time PCR (p<0.05). A positive correlation between viral load measurements for the 35 samples with HBV-positive DNA was detected in both LNA and TaqMan real-time PCR.
Keywords: human hepatitis B virus, real-time polymerase chain reaction, probe, locked nucleic acid, TaqMan
Introduction
Measurements of hepatitis B virus (HBV) DNA levels are routinely used to identify infectious, chronic carriers and to predict and monitor efficacies of antiviral treatment regimens (1–4). In addition, when serological testing could be inconclusive for the diagnosis of a HBV infection (e.g., due to the presence of genetic variants of HBV), the detection of HBV DNA may help to resolve these uncertainties (5). In recent years, real-time polymerase chain reaction (PCR) has been widely accepted as the gold standard for quantification of viral nucleic acids due to its improved speed, sensitivity and reproducibility, and its reduced risk of contamination (6,7). In the present study, we developed and evaluated a sensitive locked nucleic acid (LNA) real-time PCR probe for the accurate detection of HBV in plasma or serum samples, and compared it with the TaqMan probe.
Materials and methods
Samples
For the present study, a total of 39 human serum samples were obtained from chronic hepatitis B patients, which were previously assayed by enzyme-linked immunosor-bent assay (ELISA) (Kehua, Shanghai, China) at the Clinical Laboratory Qingdao Medical School Affiliated Hospital (China). The clinical serum samples were divided into 3 groups: group I contained 15 cases with hepatitis B surface antigen (HBsAg) (+) alone; group II contained 10 cases with HBsAg (+), HBeAg (+) and HBc (+); and group III contained 14 cases with HBsAg (+), HBeAb (+) and HBc (+). A further 19 cases of normal serum were obtained from volunteers. All samples were stored at −20°C until use.
Human cytomegalovirus-, herpes simplex virus-, hepatitis A virus-, hepatitis C virus- and human immunodeficiency virus type 1 (HIV-1)-positive specimens, as well as human genomic DNA were used to determine the specificity provided by the Shandong Provincial Key Laboratory of Molecular Virology (China).
Primers and probes
The primers (P1, P2) and probes were selected from the X gene of the HBV genome (GenBank ID: NC-00962), which generated a product of 114 bp. The primer and probe sequences are provided for LNA and TaqMan real-time PCR in Table I.
Table I.
Primers and probes for the detection of HBV by real-time PCR.
| Primers/probes | Sequence | Bases | Tm (°C) | Manufacturer |
|---|---|---|---|---|
| P1 | GACCACCAAATGCCCCTAT | 19 | 55 | Shanghai Biotech Inc. |
| P2 | CCRAGAYYGAGATCTTCTGCGAC | 23 | 53 | Shanghai Biotech Inc. |
| LNAa | FAM-TCGTCTAACAACAGT-BHQ1 | 15 | 66 | Takara |
| TaqMan | FAM-TCGTCTAACAACAGT(TAMMRA) | Shanghai Biotech Inc. | ||
| AGTTTCCGGAAGTGT-P | 30 | 64 |
The underlined nucleotides indicate a LNA monomer substitution. Tm, temperature; LNA, locked nucleic acid; HBV, hepatitis B virus; PCR, polymerase chain reaction.
In-house HBV DNA standard
We constructed the recombinant plasmid pUCm-T-HBV containing the HBV X gene region. The HBV plasmid DNA was isolated using the Takara MiniBEST Plasmid Purification Kit (Takara, Dalian, China). The extracted DNA was examined with ultraviolet (UV) spectrometry, and DNA samples with a UV reading A260:A280 ratio in the range of 1.6 to 1.8 were retained for use. The purified recombinant plasmid pUCm-T-HBV concentration was tested in triplicate (in parallel) with HBV DNA reference standards (3.83x103, 2.69x104, 1.33x105, 3.53x106 and 1.33x107 IU/ml; Beijing Control & Standards Biotechnology Co., Ltd; China). The standards were tested in duplicate in order to generate the standard curve. Following quantification, the HBV plasmid was serially diluted 10-fold from 40 to 4x107 IU/ml, and 2 μl of each diluted HBV plasmid were used as PCR templates.
HBV DNA extraction
The DNA was extracted using the polyethylene glycol (PEG) precipitation method. A total of 100 μl of serum or plasma was mixed with 100 μl of extraction I followed by centrifugation of the mixture at 13,000 x g for 10 min. The supernatant was removed and discarded. A total of 25 ml of extraction II was added to the pellet, mixed well and boiled for 10 min. The boiled samples were then spun down at 13,000 x g for 10 min and 2 μl supernatant was used as the PCR template.
LNA real-time PCR probe examination
Real-time PCR was performed with 40 μl PCR mixture [200 nM of each primer; 75 nM LNA probe; 200 μM of dGTP, dATP and dCTP; 400 μM dUTP; 3.5 mM MgCl2; 2 U of HotStarTaq DNA polymerase (Takara); 0.5 U uracil DNA glycosylase (UDG; MBI Fermentas, Burlington, ON, Canada) and 2 μl DNA sample]. The amplification was performed in the iCycler iQ5 (Bio-Rad, Hercules, CA, USA), and the protocol was as follows: initial activation of UDG at 37°C for 5 min; UDG inactivation; activation of HotStarTaq DNA polymerase and template denaturation at 95°C for 3 min; 40 cycles in two steps: 95°C for 5 sec, 60°C for 30 sec. For each cycle, standard curves were created in the range of 40-4x107 IU/ml by 1:10 serial dilutions of the pUCm-T-HBV standard, and all samples were run in duplicate. Standard curves were created by using the threshold cycle (Ct) as the Y axis and the log of HBV DNA concentration as the X axis. The potential sample was examined using Ct values corresponding to HBV DNA.
TaqMan real-time PCR probe examination
The PCR protocol was the same as the LNA probe real-time PCR examination; however, a 200 nM TaqMan probe was used instead of a 75 nM LNA probe.
Data analysis and statistics
The Ct values and amplification data were analyzed using the Bio-Rad iQ5 optical system software version 2.0 (Bio-Rad). Data were collected at the extension step of every cycle, and Ct values were determined with the PCR baseline subtracted curve fit analysis method using fluorescein-normalized data. A standard curve was created automatically with the Bio-Rad software for each run by plotting the Ct value threshold cycle numbers against virus loadings (IU/ml) for each standard, and quantification of HBV DNA for unknown samples was inferred from the regression line.
Pearson's correlation coefficient was used to assess the strength of linear correlations between the log transformed values of the two data sets using SPSS software (version 16.0). p<0.05 was considered to indicate a statistically significant difference.
Results
Calibration of in-house standards against reference standards for HBV DNA
The concentration of HBV plasmid DNA assessed against reference standards was 4.0x107 IU/ml. With optimized LNA real-time PCR probe conditions, the recombinant HBV plasmid was serially diluted 1:10, 6 times and 7 standard products (40-4.0x107 IU/ml) were obtained in order to create a standard curve. A linear relationship was obtained between the Ct values and the log10 concentrations of HBV DNA, which displayed a positive correlation in the region of 40 to 4.0x107 IU/ml per reaction. A typical standard curve amplification plot and linear regression analysis of the data are shown in Figs. 1 and 2, respectively. The regression analysis yielded a correlation co-efficient of 0.999 and a Y-intercept value of 45.72. The slope of −3.425 closely approximates the theoretical maximum amplification efficiency of 95.9%. This standard curve over a dynamic range of 40 to 4.0x107 IU/ml was used for the quantification of HBV DNA in the test samples. The minimum cut-off for quantification of HBV DNA was 40 IU/ml. The linear detection range for the TaqMan probe was typically from 4x102 to 4x107 DNA IU/ml per reaction. The minimum cut-off for the quantification of HBV DNA for the TaqMan probe was 4x102 IU/ml.
Figure 1.
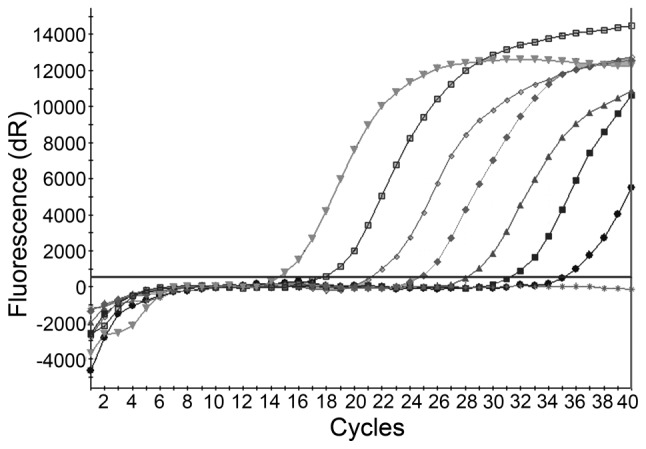
Amplification plots of in-house HBV standards using the TaqMan and LNA real-time PCR probes. The X axis denotes the number of cycles, and the Y axis denotes fluorescent intensity values. The horizontal line across the graph denotes the threshold line. The plots from left to right are 4x107, 4x106, 4x105, 4x104, 4x103, 4x102, 40 IU/ml and water. HBV, hepatitis B virus; PCR, polymerase chain reaction; LNA, locked nucleic acid; dR, double ratio.
Figure 2.
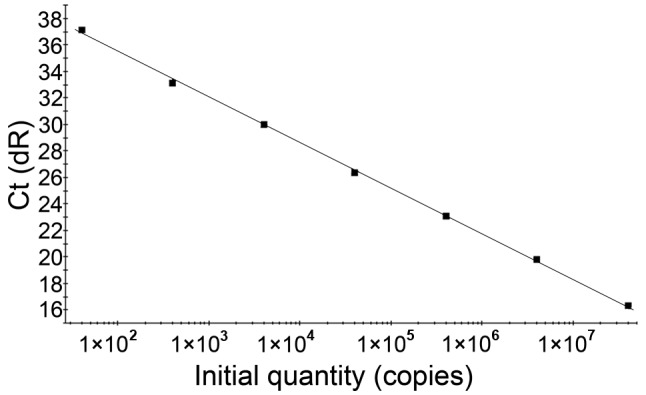
Standard curve of HBV DNA obtained using the LNA real-time PCR probe. The data show a positive correlation in the region of 40 to 4.0x107 IU/ml. The equation is Y=−3.425 log (X) + 45.72, with a correlation co-efficient of 0.999. HBV, hepatitis B virus; PCR, polymerase chain reaction; LNA, locked nucleic acid; dR, double ratio; Ct, threshold cycle.
Sensitivity (the lowest examination limit)
To evaluate the sensitivity of the LNA real-time PCR probe, serial dilutions of HBV plasmid DNA ranging from 4 to 640 (4, 40, 80, 160, 320 and 640) IU/ml were tested in 20 replicates. Following 40 amplification cycles, when the lowest plasmid amplification S-shaped plots and concentrations were able to be consistently differentiated from the double distilled water (ddH2O)-negative controls, DNA concentration was set as the lowest limit and used to evaluate the sensitivity of this method. At times, amplification performance was capable of quantifying HBV DNA with a sensitivity as low as 4 IU/ml using the LNA and TaqMan PCR probes; however, this concentration was not detected consistently. The number of positive cases at 4 IU/ml was 12 in the LNA PCR probe and 1 in the TaqMan PCR probe (Table II). The lowest concentration of HBV DNA quantified at a frequency of 100% was 40 IU/ml. The lowest concentration of HBV-positive DNA quantified at a frequency of 100% was 40 IU/ml, using the LNA PCR probe and 80 IU/ml using the TaqMan PCR probe.
Table II.
Number of replicate PCR analyses performed and number of positives cases found with LNA and TaqMan probes at various DNA concentrations.
| HBV plasmid DNA (IU/ml) | Total no. of replicates | No. of positives
|
|
|---|---|---|---|
| LNA probe | TaqMan probe | ||
| 4 | 20 | 12 | 1 |
| 40 | 20 | 20 | 10 |
| 80 | 20 | 20 | 20 |
| 160 | 20 | 20 | 20 |
| 320 | 20 | 20 | 20 |
| 640 | 20 | 20 | 20 |
LNA, locked nucleic acid; HBV, hepatitis B virus; PCR, polymerase chain reaction.
Specificity
The extractions from HBV HBeAg (+) patients, cytomegalovirus, herpes simplex virus, hepatitis A virus, hepatitis C virus (genotype 1), HIV-1 positive samples and human genomic DNA were examined in order to determine the specificity using the LNA real-time PCR probe system. Our method was able to detect HBeAg (+) serum samples. It revealed negative results with cytomegalovirus, herpes simplex, hepatitis A and hepatitis C viruses (genotype 1), as well as HIV-1-positive samples and human genomic DNA.
Reproducibility
High, middle and low concentration samples, including A01, A02 and A03 (HBV DNA virus load of 104, 105 and 107 IU/ml), were examined in order to determine reproducibility between the same and different batch samples. Every sample was examined 5 times per day for 3 days.
The intra-assay co-efficient of variation (CV) for the LNA PCR probe ranged from 0.6 to 2.82% (Table III). The inter-assay CV for the LNA PCR probe ranged from 1.09 to 2.54% (Table IV).
Table III.
Intra-assay variation of HBV viral DNA load in LNA real-time PCR (Ct values).
| Assay no. (day) | Sample ID | Replicates
|
Mean Ct | SD | CV (%) | ||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |||||
| 1 | A01 | 31.33 | 31.42 | 31.69 | - | - | 31.48 | 0.187 | 0.6 |
| A02 | 26.86 | 26.69 | 25.87 | 26.47 | - | 26.34 | 0.432 | 1.63 | |
| A03 | 18.47 | 19.54 | 18.95 | - | - | 19.25 | 0.457 | 2.82 | |
| 2 | A01 | 31.28 | 31.77 | 30.72 | 31.27 | 31.57 | 31.32 | 0.355 | 1.27 |
| A02 | 25.76 | 25.89 | 25.99 | 26.38 | 26.17 | 26.04 | 0.217 | 0.93 | |
| A03 | 17.88 | 18.70 | 18.38 | 18.55 | 18.08 | 18.32 | 0.301 | 1.84 | |
| 3 | A01 | 31.66 | 31.94 | 32.02 | 32.29 | 31.99 | 31.98 | 0.201 | 0.70 |
| A02 | 27.19 | 27.99 | 27.16 | 27.12 | 27.05 | 27.30 | 0.347 | 1.42 | |
| A03 | 18.73 | 19.42 | 19.14 | 19.10 | 18.59 | 18.99 | 0.299 | 1.76 | |
LNA, locked nucleic acid; HBV, hepatitis B virus; PCR, polymerase chain reaction; SD, standard deviation; CV, co-efficient of variation; Ct, threshold cycle.
Table IV.
Inter-assay variation of HBV viral DNA load in LNA real-time PCR (Ct values).
| Sample ID | Assay no. (day)
|
Mean | SD | CV (%) | ||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | ||||
| A01 | 31.48 | 31.32 | 31.98 | 31.59 | 0.344 | 1.09 |
| A02 | 26.34 | 26.04 | 27.3 | 26.56 | 0.658 | 2.48 |
| A03 | 19.25 | 18.32 | 18.99 | 18.85 | 0.480 | 2.54 |
LNA, locked nucleic acid; HBV, hepatitis B virus; PCR, polymerase chain reaction; SD, standard deviation; CV, coefficient of variation; Ct, threshold cycle.
Stability
To evaluate the stability of the LNA and TaqMan probes, the PCR mixture including the probe was repeatedly frozen and thawed once a day for 5 days. A comparison of threshold Ct values of the 7 standard products prior to and following reagent freezing and thawing, displayed no significant difference (p>0.05) (Table V). There was a minor affect of on the standard curves (Fig. 3). Prior to freezing and thawing the reagents, background fluorescence of the LNA probe (16,000 to 34,000 fluorescence units) was higher than the TaqMan probe (12,000 to 20,000 fluorescence units) and, compared to the TaqMan probe (~36%), the LNA probe fluorescence slightly decreased (~1%) with freezing and thawing (5 times; Fig. 4).
Table V.
The changes in Ct values prior to and following reagent freezing and thawing.
| Standards | Day 1 | Day 2 | Day 3 | Day 4 | Day 5a |
|---|---|---|---|---|---|
| 4.00E+01 | 36.21 | 36.51 | 35.1 | 36.2 | 35.9 |
| 4.00E+02 | 31.57 | 32.63 | 31.35 | 31.5 | 32.1 |
| 4.00E+03 | 28.63 | 29.39 | 28.07 | 28.21 | 28.55 |
| 4.00E+04 | 25.49 | 25.92 | 24.82 | 23.99 | 25.36 |
| 4.00E+05 | 21.63 | 22.51 | 21.25 | 21.27 | 21.31 |
| 4.00E+06 | 18.06 | 19.26 | 17.82 | 18.04 | 17.71 |
| 4.00E+07 | 14.65 | 15.79 | 14.63 | 15.0 | 15.0 |
The Ct values displayed no significant difference (p>0.05) prior to and following reagent freezing. Ct, threshold cycle.
Figure 3.
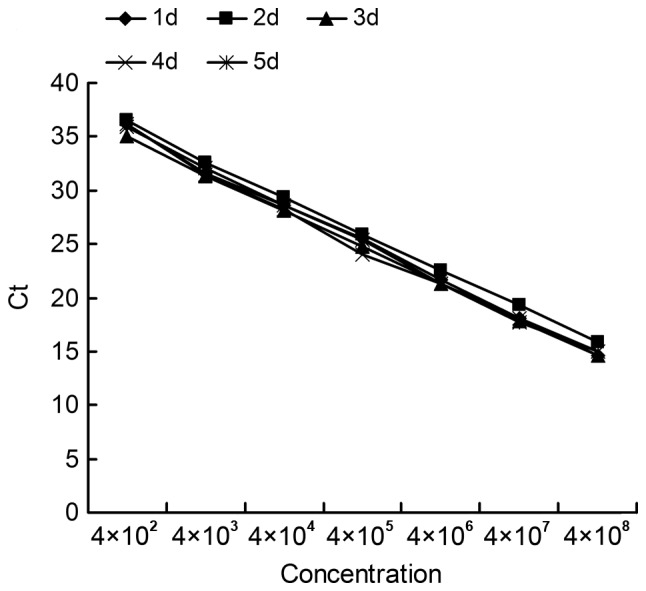
Effect of freezing and thawing on the standard curve. Ct, threshold cycle.
Figure 4.
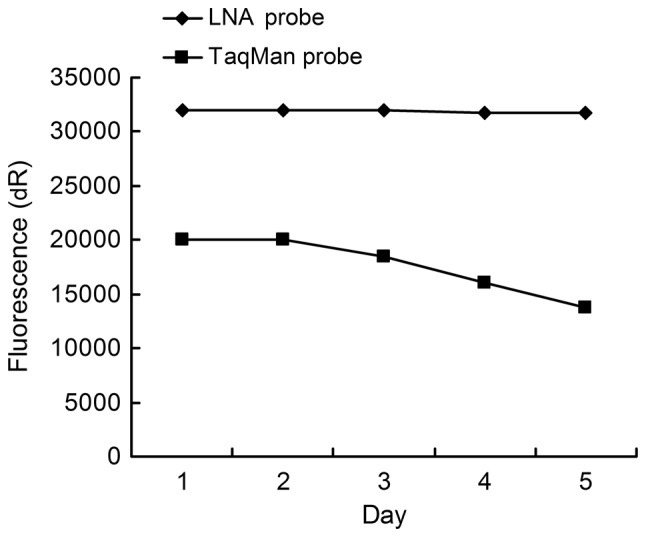
Comparative study on the change in background fluorescence with freezing and thawing (5 times). Background fluorescence of the LNA probe (16,000 to 34,000 fluorescence units) was higher than the TaqMan probe (12,000 to 20,000), and the LNA probe fluorescence slightly decreased with freezing and thawing (~1%) as compared to the TaqMan probe (~36%) (5 times). LNA, locked nucleic acid; dR, double ratio.
Comparative evaluation of LNA and TaqMan real-time PCR assays
Among the 39 cases of known chronic hepatitis B, 100% of the cases (39/39) were shown to be positive by LNA real-time PCR and 76.9% of cases (35/39) were shown to be positive by TaqMan real-time PCR. Out of the 15 cases of chronic hepatitis B HBsAg (+), 4 were shown to be negative by TaqMan real-time PCR and positive by LNA real-time PCR (p<0.05). A positive correlation was observed between the viral load measurements for 35 samples, where both LNA and TaqMan real-time PCR detected HBV-positive DNA. HBV-positive DNA within the linearity ranges was observed in both the LNA and TaqMan real-time PCR assays (Fig. 5).
Figure 5.
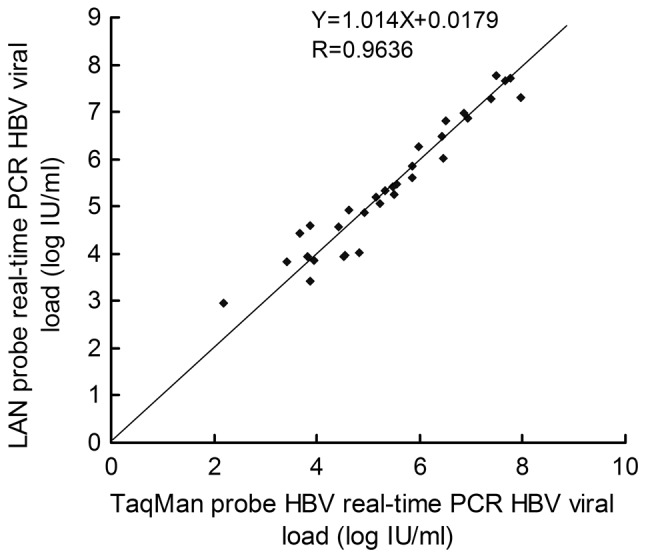
Correlation between viral load measurements for 35 HBV-positive DNA serum samples within the linear range for both LNA and TaqMan real-time PCR. LNA, locked nucleic acid; HBV, hepatitis B virus; PCR, polymerase chain reaction.
The results of 19 cases with normal serum from the two methods were below the minimum cut-off value (TaqMan: 80 IU/ml and LNA: 40 IU/ml).
Discussion
LNA is a recently developed technology. It is a nucleotide chemical modification technology (also known as 3rd generation reverse oligonucleotide) (8). The modification occurs through a 2-methyl sugar link between O2 and C4. With LNA modification, the stability and affinity for DNA molecules has been increased in PCR reactions. It has been reported that the oligonucleotide melting temperature increased by 9.6°C following each extra nucleotide insertion (9). Mutation modification has enabled the significant amplification of test environments and the recognition of single base mismatches. In addition, it has allowed for more complicated experiments to be carried out in a single test tube. Reported advantages of these short probes include improved sensitivity towards single base mismatches (specificity), ease of design and improved signal-to-noise ratio, partly due to reduced fluorescence from spurious binding and the close proximity of the quencher and reporter dye (10). This opinion has been in accordance with the findings of Costa et al, who demonstrated LNA probes to be more sensitive than conventional DNA probes when testing fetal DNA in maternal serum samples. This result was attributed to improved PCR efficiency when using shorter probes as a result of less amplification interference (11). A comparative study on sensitivity (12), indicated an improved performance of LNA probes compared to TaqMan probes when detecting Salmonella in a range of sample matrices. The LNA probe reached higher fluorescence plateaus and produced lower Ct values and standard deviations. Josefsen et al published the same conclusion, that the LNA real-time PCR probes were the most sensitive assay of 4 real-time PCR probes (LNA, MGB, Scorpion and TaqMan) (13). Nevertheless, the reported sensitivity of the LNA probes was not significantly better compared to other probes, when comparing 9 different real-time PCR chemistries for qualitative and quantitative applications (14).
In the present study, we presented a quantitative assay based on the LNA real-time PCR probe for the detection of HBV DNA. Due to the advantages mentioned previously, LNA probe technology should have greater detection performance for HBV DNA than common probes, and we have demonstrated this result through our research. The assay detection limit of LNA probes was capable of reaching 40 IU/ml DNA per reaction, which could not occur with TaqMan probes. In addition, the assay based on the LNA probe had a wider linear detection range. The linear standard curve was obtained between 40 and 4x107 DNA IU/ml per reaction (r=0.999), while the 1inear detection range based on the TaqMan probe was typically from 4x102 to 4x107 DNA IU/ml per reaction. Furthermore, the advantages of LNA real-time PCR probes were higher sensitivity, stability, amplification efficiency and lower concentration of probes (Table VI).
Table VI.
Comparative studies on sensitivity, stability, background Ct values and amplification efficiency between LNA and TaqMan.
| Probe | Sensitivity (IU/ml) | Stability (fluorescence decrease) | Optimal probe concentration | Ct values of sample A02 | Amplification efficiency |
|---|---|---|---|---|---|
| LNA | 40 | 1% | 75 | 26.56a | 95.9 |
| TaqMan | 80 | 36% | 200 | 28.67 | 88.2 |
p<0.05 of the Ct values denotes statistically significant differences vs. the use of the LNA probe (analysis of variance). LNA, locked nucleic acid; Ct, threshold cycle.
In 35 samples, HBV-positive DNA was detected by both LNA and TaqMan real-time PCR. According to the related reference standard (15–18), the LNA real-time PCR viral load in 35 HBV DNA-positive samples compared well with the TaqMan real-time PCR assay viral load (r=0.9636, p<0.05) (Fig. 5). Due to the lower virus quantities in the serum of patients from group I than the other 2 groups, the quantification of HBV DNA in group I patients required higher sensitivity real-time PCR. Out of the 15 HBsAg (+) cases, 4 were shown to be negative by TaqMan real-time PCR and positive by LNA real-time PCR (p<0.05), which further demonstrates that LNA real-time PCR probes have greater sensitivity than TaqMan probes.
Collectively, our data demonstrate that the LNA real-time PCR probe for the quantitative detection of HBV DNA exhibits greater advantages, including a high degree of precision, wide range of linearity, clinically relevant detection limit, ease of design and an improved signal-to-noise ratio, compared to TaqMan real-time PCR.
References
- 1.Schalm SW. Treatment of chronic hepatitis B. Neth J Med. 1994;44:103–109. [PubMed] [Google Scholar]
- 2.Jardi R, Buti M, Rodriguez-Frias F, et al. The value of quantitative detection of HBV-DNA amplified by PCR in the study of hepatitis B infection. J Hepatol. 1996;24:680–685. doi: 10.1016/s0168-8278(96)80263-7. [DOI] [PubMed] [Google Scholar]
- 3.Berger A, Braner J, Doerr HW, Weber B. Quantification of viral load: clinical relevance for human immunodeficiency virus, hepatitis B virus and hepatitis C virus infection. Intervirology. 1998;41:24–34. doi: 10.1159/000024912. [DOI] [PubMed] [Google Scholar]
- 4.Kessler HH, Preininger S, Stelzl E, et al. Identification of different states of hepatitis B virus infection with a quantitative PCR assay. Clin Diagn Lab Immunol. 2000;7:298–300. doi: 10.1128/cdli.7.2.298-300.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Deursen FJ, Hino K, Wyatt D, et al. Use of PCR in resolving diagnostic difficulties potentially caused by genetic variation of hepatitis B virus. J Clin Pathol. 1998;51:149–153. doi: 10.1136/jcp.51.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackay IM, Arden KE, Nitsche A. Real-time PCR in virology. Nucleic Acids Res. 2002;30:1292–1305. doi: 10.1093/nar/30.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez JB, Marquez VE, Nicklaus MC, Mitsuya H, Barchi JJ., Jr Conformationally locked nucleoside analogues. Synthesis of dideoxycarbocyclic nucleoside analogues structurally related to neplanocin. C J Med Chem. 1994;37:3389–3399. doi: 10.1021/jm00046a024. [DOI] [PubMed] [Google Scholar]
- 9.Kurreck J, Wyszko E, Gillen C, Erdmann VA. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30:1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kutyavin IV, Afonina IA, Mills A, et al. 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000;28:655–661. doi: 10.1093/nar/28.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa JM, Ernault P, Olivi M, Gaillon T, Arar K. Chimeric LNA/DNA probes as a detection system for real-time PCR. Clin Biochem. 2004;37:930–932. doi: 10.1016/j.clinbiochem.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 12.Reynisson E, Josefsen MH, Krause M, Hoorfar J. Evaluation of probe chemistries and platforms to improve the detection limit of real-time PCR. J Microbiol Methods. 2006;66:206–216. doi: 10.1016/j.mimet.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Josefsen MH, Lofstrom C, Sommer HM, Hoorfar J. Diagnostic PCR: comparative sensitivity of four probe chemistries. Mol Cell Probes. 2009;23:201–203. doi: 10.1016/j.mcp.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Buh Gasparic M, Tengs T, La Paz JL, et al. Comparison of nine different real-time PCR chemistries for qualitative and quantitative applications in GMO detection. Anal Bioanal Chem. 2010;396:2023–2029. doi: 10.1007/s00216-009-3418-0. [DOI] [PubMed] [Google Scholar]
- 15.Pas SD, Fries E, De Man RA, Osterhaus AD, Niesters HG. Development of a quantitative real-time detection assay for hepatitis B virus DNA and comparison with two commercial assays. J Clin Microbiol. 2000;38:2897–2901. doi: 10.1128/jcm.38.8.2897-2901.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shyamala V, Arcangel P, Cottrell J, et al. Assessment of the target-capture PCR hepatitis B virus (HBV) DNA quantitative assay and comparison with commercial HBV DNA quantitative assays. J Clin Microbiol. 2004;42:5199–5204. doi: 10.1128/JCM.42.11.5199-5204.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ronsin C, Pillet A, Bali C, Denoyel GA. Evaluation of the COBAS AmpliPrep-total nucleic acid isolation-COBAS TaqMan hepatitis B virus (HBV) quantitative test and comparison to the VERSANT HBV DNA 3.0 assay. J Clin Microbiol. 2006;44:1390–1399. doi: 10.1128/JCM.44.4.1390-1399.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olioso D, Boaretti M, Ligozzi M, Lo Cascio G, Fontana R. Detection and quantification of hepatitis B virus DNA by SYBR green real-time polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 2007;26:43–50. doi: 10.1007/s10096-006-0223-y. [DOI] [PubMed] [Google Scholar]


