Abstract
The aim of the present study was to screen for possible serum biomarkers for gastric adenocarcinoma. Surface-enhanced laser desorption ionization time of flight mass spectrometry (SELDI-TOF-MS) was used to screen serum samples from 109 cases of gastric adenocarcinoma and 106 control subjects (60 healthy subjects, 30 patients with chronic superficial gastritis and 16 cases of chronic atrophic gastritis). The differentially expressed protein peaks were selected and isolated using high performance liquid chromatography (HPLC) and processed with enzyme prior to liquid chromatography-mass spectrometry tandem mass spectrometry (LC-MS/MS) analysis and data mining with software XCalibur program components BioWorks 3.2. Among the gastric cancer cases, three differentially expressed protein peaks were selected as potential serum biomarkers: the m/z peaks at 5,906.5 showed increased expression (8.53±4.33 in the cancer group, and 0.88±0.31 in the control group); the m/z peaks at 6,635.7 and 8,716.3 showed decreased expression (6.54±2.44 and 0.93±0.29, respectively, in the cancer group and 17.56±4.43 and 2.16±0.98, respectively, in the control group) (P<0.01). The m/z peaks at 5,906.5, 6,635.7 and 8,716.3, were identified as fibrinogen α-chain, apolipo-protein A-II and apolipoprotein C-I. The combined use of the three biomarkers distinguished the cancer group patients from the control group samples at a sensitivity of 93.85% (61/65) and a specificity of 94.34% (50/53). In conclusion, fibrinogen α-chain, apolipoprotein A-II and apolipoprotein C-I were identified as potential markers for gastric cancer and appear to have diagnostic value for clinical applications.
Keywords: serum, proteome, surface-enhanced laser desorption ionization time of flight mass spectrometry, biomarker, gastric cancer, fibrinogen α-chain, apolipoprotein A-II, apolipoprotein C-I
Introduction
Gastric adenocarcinoma is a common gastrointestinal malignant tumor, accounting for 23.2% of cancer-related deaths in China (approximately 16 million individuals) (1,2). At present, the clinical diagnosis of gastric adenocarcinoma mainly relies on physical and histological examinations, which are accurate only for middle- or late-stage cases. Therefore, many patients are diagnosed at a late stage of the disease. Biochemical markers such as carcinoembryonic antigen, carbohydrate antigen 19-9 and carbohydrate antigen 125 are used as markers for diagnosis; however they are non-specific and lack adequate sensitivity (3–6). The present study aimed to design a new diagnostic system with high sensitivity and specificity for early-stage gastric cancer detection. We employed a newly emerging technique, surface-enhanced laser desorption ionization time-of-flight mass spectrometry (SELDI-TOF-MS) (7–12), to analyze the serum proteome from healthy volunteers, gastric cancer patients and gastritis patients, and to screen for specific protein biomarkers for gastric cancer (13–15).
Materials and methods
Clinical data
One hundred and nine gastric cancer patients (males 41, females 68, age range 32–89 years) (25 cases Dukes’ A, 22 cases Dukes’ B, 28 cases Dukes’ C and 34 cases of Dukes’ D), and 106 cases of controls (males 50, females 56, age range 26–85 years) (60 healthy volunteers, 16 cases of chronic atrophic gastritis patients, and 30 cases of chronic superficial gastritis) were recruited for this study at the First Affiliated Hospital of Zhejiang University and Zhejiang Taizhou Municipal Hospital. The subjects were assigned into an experimental group and a verification group according to Table I. The peripheral blood samples were collected in the morning after overnight fasting. The blood samples were then maintained at 4°C for 1–2 h prior to centrifugation at 3,000 rpm at 4°C for 10 min to separate out the serum. The serum samples were frozen at −80°C in a freezer for storage. All protocols and experiments were approved by the Taizhou Medical College Ethics Committee for clinical experiments and use of human samples; written informed consent was obtained from all subjects participating in this study. The study complied with the World Medical Association Declaration of Helsinki regarding ethical conduct of research involving human subjects.
Table I.
Clinical subjects involved in the study.
| Groups | Experimental | Verification | Total |
|---|---|---|---|
| Gastric adenocarcinoma | |||
| Dukes’ A | 15 | 10 | 25 |
| Dukes’ B | 12 | 10 | 22 |
| Dukes’ C | 20 | 8 | 28 |
| Dukes’ D | 18 | 16 | 34 |
| Control | |||
| Chronic superficial gastritis | 15 | 15 | 30 |
| Chronic atrophic gastritis | 8 | 8 | 16 |
| Healthy controls | 30 | 30 | 60 |
| Total | 118 | 97 | 215 |
Protein chip analysis
After thawing and 10 min of centrifugation (10,000 rpm), a 20-μl serum sample without fraction treatment (which does not affect the protein mining efficiency as shown by the authors) was added to 30 μl 0.5% U9 (9 mol/l urea, 2% CHAPS (3[(3-cholamidopropyl)dimethylammonio]-l-propanesulfonate), 1% DTT (DL-dithiothreitol)) in a 96-well plate and incubated for 20 min at 4°C with 600 rpm vigorous agitation. The ProteinChip array cassette was put into a 96-well bioprocessor and 100 μl U1 buffer (50 mmol/l Tris-HCL diluted 10% U9 buffer) was added into each well, and incubated for 10 min at 4°C with 600 rpm vigorous agitation. Q10 buffer (200 μl) (100 mM Tris-HCl buffer pH 9.0) was then added and a 5-min incubation was carried out 2 times with agitation. All experimental reagents were obtained from Shanghai Shenggong Company, Shanghai, China.
Fifty microliters of the protein-denatured serum samples were removed to a new tube, and 200 μl Q10 buffer was added to dilute the samples before being applied onto the Q10 chip Bioprocessor (Ciphergen) for 60 min. Then each plate of the Q10 chip was added together with 200 μl Q10 buffer, incubation was carried out for 5 min two times with agitation, and finally 20 mm/l HEPES (pH 7.4) buffer was added for washing before drying. SPA (0.5 μl) was added 2 times into each plate with drying between each addition. Mass spectrometry was set as laser intensity 185, sensitivity 8, 2,000–20,000 m/z. Inter-chip CV was <10%. All-in-one control chip was used to adjust the system with a systemic error <0.1%. All of the data were processed with ProteinChip 3.0 software, then the Biomarker Wizard software 3.1 and Biomarker Wizard software 4.0.1. P<0.01 was determined to indicate statistically significant differences in the comparison between two protein peaks.
Purification and identification of specific protein peaks
Serum samples (100 μl) with 300 μl water and 700 μl acetonitrile were mixed and maintained at −20°C in a freezer for 30 min prior to centrifugation at 3000 rpm for 10 min. The supernatant was freeze-dried for 20 min before collection for HPLC. The purified solutions were collected at different time periods, freeze-dried to obtain a 20-μl volume solution. Solution (0.5 μl) was mixed with 1.5 μl matrix solution (10 mg/ ml CHCA) to be placed on chip points, with crystallization for MALDI-TOF MS detection. The conditions included pulsed nitrogen laser (337 nm), accelerating voltage 20 KV, linear analysis mode, mass range 3,000–20,000 m/z. The samples corresponding to the specific protein peaks in SELDI-TOF MS were subsequently identified.
LC-MS/MS analysis
Purified target protein of 20 μl was mixed with 60 μl 8M urea (final concentration of urea 6 M) and was agitated at room temperature for 20 min. Then 0.8 μl 1 M DTT (final concentration 10 mM) was added and mixed at room temperature for 1 h, and 3.2 μl 1 M iodine acetyl amine (final concentration 40 mM) was added and maintained for 45 min in the dark. DTT (3.2 μl 1 M) (final concentration 40 mM) was added for 20 min, and then 400 μl 50 mM NH4HCO3 was added to dilute the solution, with urea concentration at 1 M and pH 8.0. Subsequently, 0.1 μg protease in a 37°C water bath for 1 h, and formic acid was adjusted to a pH <3 to terminate the reaction. The hydrolysates were subjected to LC-MS/MS analysis. Sample solutions were put in a self-made C18 capillary column for liquid chromatography: inner diameter 100 μm, filled part 100 mm, filled particles with diameter 5 μm. The flow phase A was water and 0.1% formic acid; the flow phase B was acetonitrile and 0.1% formic acid. The washout followed the sequences below: 100% A (0 min) - 100% A (5 min) - 5% B (5.1 min) - 65% B (60 min) - 100% B (75 min) - 100% B (85 min). The flow speed was 200–800 nl/min. The data-dependent mode was used; the scanning ranges were from 400 to 2,000 m/z; the five strongest signal peaks of each full scan were selected for secondary MS (MS2) analysis.
The data retrieval used the XCalibur program components BioWorks 3.2 (Thermo Finnigan) and the peptide sequences were searched for in the NCBI human protein database (human.ref) according to the mass spectrum. The following parameters were used: enzyme split site at random site; fixed modification at cysteine amine formylation modification; variable modification at methionine oxidation; retrieval parameters ΔCN >0.1; Sp >500; Rsp ≥5; Xcorr vs. Charge: Xcorr (+1) >1.9, Xcorr (+2)>2.5, Xcorr (+3) >3.75.
Results
The serum protein fingerprint spectrum
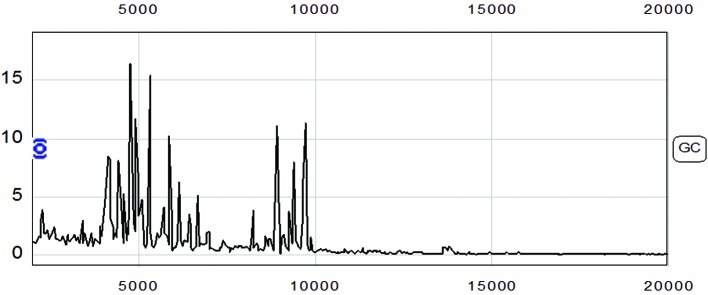
The protein fingerprint spectrum from 65 cases of gastric adenocarcinoma and 53 cases of control were normalized first, then analyzed using Biomarker wizard software. Two hundred and twenty-seven protein peaks were found in the m/z range from 2,000–50,000 (Fig. 1).
Figure 1.
Serum proteomic spectrum of the gastric adenocarcinoma patients.
Data analysis of the gastric adenocarcinoma protein fingerprint spectrum and the diagnostic model
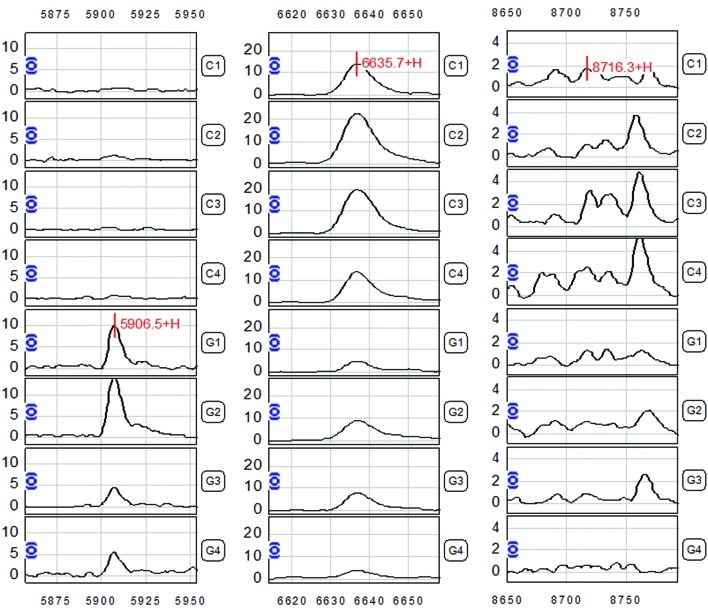
The Biomarker Wizard software analysis revealed three differentially expressed protein peaks in serum samples from gastric adenocarcinoma patients, with m/z at 5,906.5, 6,635.7 and 8,716.3 respectively (P<0.01) (Fig. 2 and Table II), which were considered as potential biomarkers for gastric adenocarcinoma. The 5,906.5 peak showed increased expression in the gastric adenocarcinoma cases; while the 6,635.7 and 8,716.3 peaks showed a decreased expression level in the gastric adenocarcinoma serum samples (Table II). The combined analysis with the three peaks as the basis for the diagnostic model showed a sensitivity of 93.85 (61/65) and a specificity of 94.34% (50/53) in analyzing the mass spectrometry data from the 65 gastric adenocarcinoma patients and 53 cases of control subjects (Table III).
Figure 2.
Protein fingerprint of the three protein peaks (m/z 5,906.5, 6,635.7, 8,716.3) for the diagnostic model of the gastric adenocarcinoma cases.
Table II.
Intensity of the three differential peaks in gastric adenocarcinoma and control groups.
| (m/z) | Gastric adenocarcinoma | Control | P-values |
|---|---|---|---|
| 5,907.5 | 8.53±4.33 | 0.88±0.31 | 2.8×10−7 |
| 6,636.7 | 6.54±2.44 | 17.56±4.43 | 4.5×10−6 |
| 8,716.3 | 0.93±0.29 | 2.16±0.98 | 8.4×10−4 |
Table III.
Characteristics of the gastric adenocarcinoma diagnostic model.
| Set | Groups | Case number | Correct cases | Diagnosis rate (%) |
|---|---|---|---|---|
| Experimental | Gastric adenocarcinoma | 65 | 61 | 93.85 |
| Control | 53 | 50 | 94.34 | |
| Verification | Gastric adenocarcinoma | 44 | 40 | 90.91 |
| Control | 53 | 48 | 90.57 |
Purification of the protein peaks and the MS analysis
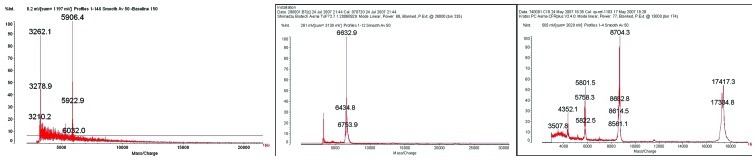
The protein peaks of the three biomarkers (m/z 5,906.5, 6,635.7 and 8,716.3) were isolated and purified with HPLC, and then collected into PCR tubes for MALDI-TOF-MS examination. The results showed that the three peaks were proteins with molecular weights of 5,906.4, 6,632.9 and 8,704.3, respectively (Fig. 3).
Figure 3.
MALDI-TOF image of the three differential protein peaks (m/z 5,906.5, 6,637.6, 8,716.3).
Following LC-MS/MS measurement of the digested proteins and the data screening from NCBI human protein database, the 5,906.4 peak was found to correspond to the fibrinogen α chain (100% match), the 6,632.9 peak corresponded to the apolipoprotein A-II (96% match), and the 8,704.3 peak corresponded to the lipid-laden protein C-I (60% match) (Table IV).
Table IV.
Amino acid sequence of the differential protein peaks in the gastric adenocarcinoma cases.
| (m/z) | Protein | Molecular weight | Amino acid sequence | Match rate (%) |
|---|---|---|---|---|
| 5,906.5 | Fibrinogen α-chain | 5,904.0 | SSSYSKQFTS STSYNRGDST FESKSYKMAD | 100 |
| EAGSEADHEG THSTKRGHAK SRPV | ||||
| 6,635.7 | Apolipoprotein A-II | 6,630.0 | TPDY SSALDKLKEF GNTLEDKARE | 96 |
| LISRIKQSEL SAKMREWFSE TFQKVKEKLK IDS | ||||
| 8,716.3 | Apolipoprotein C-I | 8,707.8 | QAK EPCV ESLVSQYFQT VTDYGKDLME | 60 |
| KVKSPELQAE AKSYFEKSKE | ||||
| QLTPLIKKAG TELVNFLSYF VELGTQPATQ |
Discussion
Gastric cancer is a cancer without clear symptoms at onset, and metastasis and recurrence are common (16,17). The prognosis of this disease is poor as well. Early diagnosis of gastric cancer urgently requires novel techniques with high sensitivity and specificity. Since the onset and progression of cancer lead to characteristic changes in the serum proteome, it is possible to employ proteomic techniques to screen for potential biomarkers of gastric cancer (11,18,19). The present study utilized SELDI-TOF MS, and successfully identified a new diagnostic model for gastric cancer, including suitability for early-stage patients.
SELDI-TOF MS is an ideal platform for proteomic studies with several advantages. i) A small amount of sample is required. The scan is fast and suitable for clinical diagnosis and high throughput-screening analysis. ii) The technique can identify the specific spectrum including several biomarkers at the same time. iii) Crude samples without prior purification can be used. iv) The technique can be combined with many genomic techniques. v) The technique is of high reliability and can be reproduced in repeated tests. vi) The technique is applicable to proteins that are not suitable for 2D-PAGE analysis, such as those with extremely small molecular weights, or hydrophobic, transmembrane, as well as isoelectric point (8–10). The technique currently shows progressive results in biomarker screening of autoimmune diseases, inflammation disorders and many types of malignant cancers (9,10,20–24), providing the basis for diagnosis and treatment of these diseases clinically.
The present study described three potential biomarkers for gastric cancer with high sensitivity and specificity in both the experimental and verification set, as mentioned in Results. The three identified markers were fibrinogen α-chain, apolipoprotein A-II, and lipid-laden proteins C-I. They may play different roles in the onset and progression of gastric cancer. Fibrinogen participates in blood coagulation processes, and it may mediate the interaction of cancer cells and platelet, which occurs during cancer metastasis (25,26). Apolipoprotein participates in lipid transport and may be involved in cell proliferation/apoptosis regulation (27,28). C-I is mainly synthesized in the liver, and is less well-known in cancer biology. Several previous studies have shown a lower expression of C-I in serum from cancer patients (29,30), which was consistent with the present study. However the detailed mechanism requires future studies.
Taken together, the present study proved the efficiency of SELDI-TOF MS in screening for biomarkers of gastric cancer in a serum proteome-based manner. The three discovered biomarkers could be effectively used for gastric cancer diagnosis. Due to the limited number of patients, we did not perform a correlation analysis between the stage of cancer progression and the biomarker profiles. It is necessary to recruit more patients with early-stage disease to identify various biomarkers for the diagnosis of patients as early as possible.
Acknowledgments
The study was supported by the Zhejiang Medicine and Health Science and Technology Program grant 2010KYB127, and the Zhejiang Gongyi Applied Technology Research Program grant 2011C33045.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Tseng CW, Yang JC, Chen CN, et al. Identification of 14-3-3beta in human gastric cancer cells and its potency as a diagnostic and prognostic biomarker. Proteomics. 2011;11:2423–2439. doi: 10.1002/pmic.201000449. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi Y, Niwa Y, Tajika M, et al. Serum tumor antigen REG4 as a useful diagnostic biomarker in gastric cancer. Hepatogastroenterology. 2010;57:1631–1634. [PubMed] [Google Scholar]
- 5.Yang S, Chung HC. Novel biomarker candidates for gastric cancer. Oncol Rep. 2008;19:675–680. [PubMed] [Google Scholar]
- 6.Chan DC, Chen CJ, Chu HC, et al. Evaluation of serum amyloid A as a biomarker for gastric cancer. Ann Surg Oncol. 2007;14:84–93. doi: 10.1245/s10434-006-9091-z. [DOI] [PubMed] [Google Scholar]
- 7.Poon TC. Opportunities and limitations of SELDI-TOF-MS in biomedical research: practical advices. Expert Rev Proteomics. 2007;4:51–65. doi: 10.1586/14789450.4.1.51. [DOI] [PubMed] [Google Scholar]
- 8.Cho WC. Research progress in SELDI-TOF MS and its clinical applications. Sheng Wu Gong Cheng Xue Bao. 2006;22:871–876. doi: 10.1016/S1872-2075(06)60061-7. (In Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke CH, Buckley JA, Fung ET. SELDI-TOF-MS proteomics of breast cancer. Clin Chem Lab Med. 2005;43:1314–1320. doi: 10.1515/CCLM.2005.225. [DOI] [PubMed] [Google Scholar]
- 10.Liu C. The application of SELDI-TOF-MS in clinical diagnosis of cancers. J Biomed Biotechnol. 2011;2011:245821. doi: 10.1155/2011/245821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caffrey RE. A review of experimental design best practices for proteomics based biomarker discovery: focus on SELDI-TOF. Methods Mol Biol. 2010;641:167–183. doi: 10.1007/978-1-60761-711-2_10. [DOI] [PubMed] [Google Scholar]
- 12.Kristina G, Radomir P, Eva B, et al. When one chip is not enough: augmenting the validity of SELDI-TOF proteomic profiles of clinical specimens. Lab Chip. 2009;9:1014–1017. doi: 10.1039/b815503h. [DOI] [PubMed] [Google Scholar]
- 13.Liu W, Gao X, Cai Q, et al. Identification of novel serum biomarkers for gastric cancer by magnetic bead. Front Biosci (Elite Ed) 2010;2:961–971. doi: 10.2741/e155. [DOI] [PubMed] [Google Scholar]
- 14.Huang Q, Chen W, Wang L, Lin W, Lin J, Lin X. Identification of transgelin as a potential novel biomarker for gastric adenocarcinoma based on proteomics technology. J Cancer Res Clin Oncol. 2008;134:1219–1227. doi: 10.1007/s00432-008-0398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Umemura H, Togawa A, Sogawa K, et al. Identification of a high molecular weight kininogen fragment as a marker for early gastric cancer by serum proteome analysis. J Gastroenterol. 2011;46:577–585. doi: 10.1007/s00535-010-0369-3. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Yu JC, Kang WM, Ma ZQ. Treatment strategy for early gastric cancer. Surg Oncol. 2011 Jan 21; doi: 10.1016/j.suronc.2010.12.004. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 17.Saka M, Morita S, Fukagawa T, Katai H. Present and future status of gastric cancer surgery. Jpn J Clin Oncol. 2011;41:307–313. doi: 10.1093/jjco/hyq240. [DOI] [PubMed] [Google Scholar]
- 18.Cho WC. Proteomics technologies and challenges. Genomics Proteomics Bioinformatics. 2007;5:77–85. doi: 10.1016/S1672-0229(07)60018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho WC. Contribution of oncoproteomics to cancer biomarker discovery. Mol Cancer. 2007;6:25. doi: 10.1186/1476-4598-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei L, Wang XJ, Zheng ZG, et al. Identification of serum protein markers for breast cancer relapse with SELDI-TOF MS. Anat Rec (Hoboken) 2011;294:941–944. doi: 10.1002/ar.21399. [DOI] [PubMed] [Google Scholar]
- 21.Felix K, Fakelman F, Hartmann D, et al. Identification of serum proteins involved in pancreatic cancer cachexia. Life Sci. 2011;88:218–225. doi: 10.1016/j.lfs.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Liu L, Liu J, Wang Y, et al. A combined biomarker pattern improves the discrimination of lung cancer. Biomarkers. 2011;16:20–30. doi: 10.3109/1354750X.2010.521257. [DOI] [PubMed] [Google Scholar]
- 23.Hogdall E, Fung ET, Christensen IJ, et al. Proteomic biomarkers for overall and progression-free survival in ovarian cancer patients. Proteomics Clin Appl. 2010;4:940–952. doi: 10.1002/prca.200900171. [DOI] [PubMed] [Google Scholar]
- 24.Gemoll T, Roblick UJ, Auer G, Jornvall H, Habermann JK. SELDI-TOF serum proteomics and colorectal cancer: a current overview. Arch Physiol Biochem. 2010;116:188–196. doi: 10.3109/13813455.2010.495130. [DOI] [PubMed] [Google Scholar]
- 25.Konstantopoulos K, Thomas SN. Cancer cells in transit: the vascular interactions of tumor cells. Annu Rev Biomed Eng. 2009;11:177–202. doi: 10.1146/annurev-bioeng-061008-124949. [DOI] [PubMed] [Google Scholar]
- 26.Costantini V, Zacharski LR. The role of fibrin in tumor metastasis. Cancer Metastasis Rev. 1992;11:283–290. doi: 10.1007/BF01307183. [DOI] [PubMed] [Google Scholar]
- 27.Vanhollebeke B, Pays E. The function of apolipoproteins L. Cell Mol Life Sci. 2006;63:1937–1944. doi: 10.1007/s00018-006-6091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashe PC, Berry MD. Apoptotic signaling cascades. Prog Neuropsychopharmacol Biol Psychiatry. 2003;27:199–214. doi: 10.1016/S0278-5846(03)00016-2. [DOI] [PubMed] [Google Scholar]
- 29.Engwegen JY, Helgason HH, Cats A, et al. Identification of serum proteins discriminating colorectal cancer patients and healthy controls using surface-enhanced laser desorption ionisation-time of flight mass spectrometry. World J Gastroenterol. 2006;12:1536–1544. doi: 10.3748/wjg.v12.i10.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan Y, Wang J, Yang Y, et al. Detection and identification of potential biomarkers of breast cancer. J Cancer Res Clin Oncol. 2010;136:1243–1254. doi: 10.1007/s00432-010-0775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]