Abstract
Stroke is one of the leading causes of death and disability worldwide. In past decades, researchers have studied the physiopathology and biochemistry of stroke, but knowledge of the molecular mechanisms underlying this disease remains at an early stage. To date, only recombinant tissue plasminogen activator (rtPA) has been approved by the USA FDA for acute ischemic stroke. However, as the limiting therapy time window is 4.5 h after stroke onset and patients must meet the applicable conditions, a small number of patients benefit from this therapy. Therefore, the research and development of new drugs for stroke are a big challenge for scientists. MicroRNAs (miRNAs) are short (20–23 nucleotides) single-stranded non-coding RNAs. The seed sequences at positions 2–7 from the 5′ end which are partially or complementary to one or more mRNAs inhibit or degrade target mRNAs, thus playing an important role in the post-transcriptional regulation of gene expression. Disregulated miRNAs have revealed their complex role in pathophysiological processes, and have also shown their potential role in disease diagnosis, and use as drug targets in neurodegenerative diseases and cancer. Recently, studies have found aberrantly expressed miRNAs in stroke; however, the implication of deregulated miRNA expression in stroke remains largely unknown. This review briefly summarizes recent studies concerning miRNA expression in stroke in vivo and in vitro, focuses on aberrant miRNA expression, as well as discusses their potential therapeutic role for stroke.
Keywords: bioinformatics, microRNA, neuropathology, stroke
Contents
Introduction
Biogenesis of miRNAs and mode of action
miRNA expression in the brain
Aberrant miRNA expression in stroke brain
miRNA targets and their potential therapeutic role in stroke
Conclusion
1. Introduction
Stroke is a complex disorder precipitated by a variety of genetic and environmental factors and is the leading cause of long-term disability and mortality in China. Its incidence is more than 4–5 times higher than that in developed countries (Chen: Report of The Third National Mortality Retrospective Sampling Survey, 2008). Ischemic stroke accounts for 85% of all strokes, while primary hemorrhagic stroke accounts for the remainder (1). Cerebral ischemia triggers a cascade of pathological events that ultimately cause irreversible neuronal injury in stroke-affected brain tissue within minutes of stroke onset. The pathophysiological disorders include excitotoxicity within minutes, a robust inflammatory response within hours, and programmed cell death (apoptosis) within hours and days of stroke onset. For the past two decades, the search for drugs for acute ischemic stroke has been ongoing. Many early successes in pre-clinical studies have been transferred to clinical studies; however, the results are frustrating. At present, only recombinant tissue plasminogen activator (rtPA) has been approved by the US FDA for treatment of acute ischemic stroke. However, as the therapy time window is 4.5 h after stroke onset and patients must meet the applicable conditions, the number of patients who can benefit from this therapy is small. Therefore, the need to develop an effective treatment for stroke is urgent.
MicroRNAs (miRNAs) constitute a class of endogenous small non-coding RNAs conserved through evolution. They mediate post-transcriptional regulation of protein-coding genes by binding to the 3′-untranslated region (3′-UTR) of target mRNAs, leading to translational inhibition or mRNA degradation, depending on the degree of sequence complementarity. Increased evidence indicates that a set of miRNAs, expressed in a spatially and temporally controlled manner in the brain, are involved in neuronal development, differentiation and synaptic plasticity. The strict spatial and temporal expression of miRNAs in a controlled manner is also known to play important roles in many physiological and pathological processes, including tumorigenesis (2), proliferation (3), hematopoiesis (4), metabolism (5), immune function (6), epigenetics and neurodegenerative diseases (7). miRNAs have also been found to be of benefit in identifying the etiology of lymphoma (8) and progression of certain neurological diseases (9). During the past few years, miRNA expression profiling has been examined in stroke in vivo and in vitro. The studies indicate that miRNAs have emerged as key mediators in both pathogenic and pathological aspects of ischemic stroke biology. In this review, we briefly summarize current research regarding the involvement of miRNAs in stroke with specific focus on aberrant expression of miRNAs, as well as discuss their potential therapeutic role in stoke.
2. Biogenesis of miRNAs and mode of action
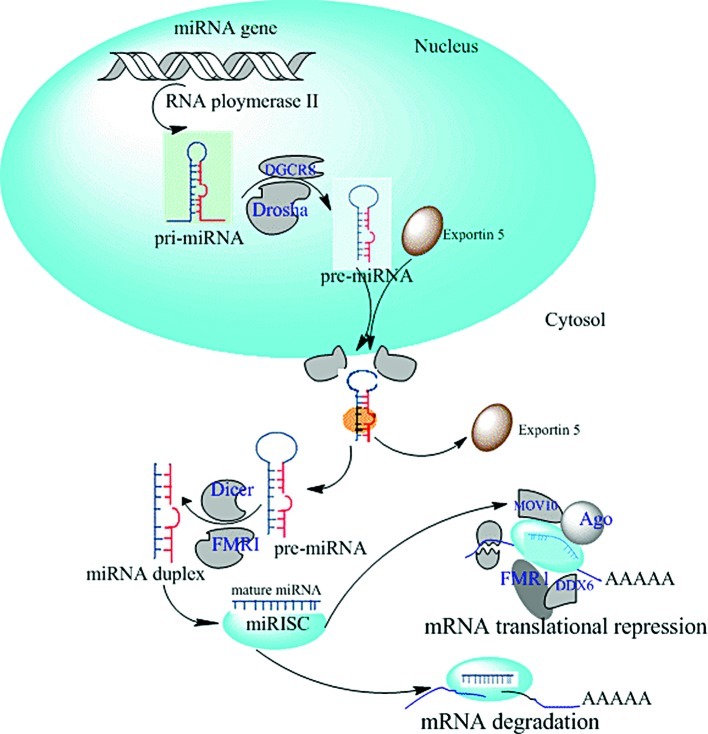
miRNAs are processed and matured through a complex biogenetic process involving multiple protein catalytic accessory proteins and macromolecular complexes following a coordinated series of events. miRNA genes transcribed by RNA polymerase II (Pol II), lead the generation of primary miRNA transcripts (pri-miRNAs), which are cleaved in the nucleus by the microprocessor complex (including the proteins Drosha and DgcR8) into precursor hairpin miRNAs (pre-miRNA). The pre-miRNA is exported to the cytoplasm by an exportin 5-dependent, then further processed by the RNase Dice to an intermediate miRNA duplex. Depending on the thermodynamic characteristics of the miRNA duplex, one strand (the leading strand, the sense strand or mature miRNA) is loaded into a multi-protein complex (miRNA-induced silencing complex; miRISC), whereas the other strand (the passenger strand, the antisense miRNA) is usually degraded. The miRNA-associated miRISC is guided to target mRNAs owing to extensive, but imperfect, pairing of the miRNA to target sequences that are preferentially located within the 3′-UTR of the mRNA. The partial or complementary interaction leads to translational repression and/or degradation of the target mRNAs. Although many mechanisms for RNA silencing have been proposed, the prevailing view in the field is still the mechanism by miRISC. Notably, the components of miRISC include Argonaute (Ago) and the helicases MOV10 (Armitage in Drosophila melanogaster) and DDX6 (also known as RCK and p54). In addition, accessory RNA-binding proteins (FMR1 and PuM2) are thought to modulate the activity of miRISC in neurons (10). At present, more than 1,733 human miRNAs have been identified (miRBase version 17; released on April 26, 2011).
3. miRNA expression in the brain
The effect of miRNAs on the neuronal function of the brain has been reported in the past few years. miRNA expression profiles in mouse and humans are phylogenetically conserved, organ-enriched, organ-specific, and with time and space are strictly expressed. One study that characterized miRNA expression profiles in mouse and human organs found that eight miRNAs are enriched in the brain. Moreover, eight brain miRNAs were found to have induced expression during neuronal differentiation of human and mouse EC cells after retinoic acid application, suggesting that there may be a conserved role in mammalian neuronal development (11). Several studies have indicated a relationship between miRNAs and neuronal development and differentiation. For example, miR-124a, the most studied brain-specific miRNA, was >100 times more highly expressed in mouse CNS than in other organs (12). It was found to be perfectly conserved at the nucleotide level from worms to humans and expressed throughout the embryonic and adult CNS (13). Studies have demonstrated that miR-124 plays an important role in neuronal differentiation and function, as the overexpression of miR-124 in differentiating mouse P19 cells promotes neurite outgrowth, while the blockade of miR-124 decreases neurite outgrowth and levels of acetylated α-tubulin (14). miR-134 is the prototypic dendritically localized miRNA that plays a crucial role in dendritic protein synthesis of hippocampal neurons (15), and has stage-specific effects on cortical progenitors, migratory neurons and differentiated neurons (16). In mouse cortical neurons, miR-132 expression was found to induce neurite outgrowth (17). One study found that the mechanism of miR-132 for enhancing dendrite morphogenesis in hippocampal neurons was through suppression of the expression of the GTPase-activating protein p250 GAP (18). miR-132 also mediates anti-inflammatory effects via the targeting of acetylcholinesterase, leading to an increase in the neurotransmitter acetylcholine to block brain inflammation (19).
4. Aberrant miRNA expression in stroke brain
The expression profiles of miRNAs in vivo or in vitro in stroke brains have revealed alterations in many individual miRNAs. These studies indicate the main involvement of deregulated miRNA expression in the pathogenesis and the process of stroke (Table I). The first miRNA screening study of ischemic rats subjected to transient focal ischemia by middle cerebral ischemia artery occlusion (MCAO) reported alterations in several miRNAs in the brain and blood at 24 and 48 h after reperfusion (20). The authors identified the expression of 114 miRNAs in ischemic brain samples by microRNA array. Among them, 106 and 82 transcripts were detected in the 24- and 48-h reperfusion brain samples, respectively. Their data also show that miRNAs are actively regulated and that their expression pattern changed with reperfusion time, thus indicating temporal expression during ischemic injury. The let-7 family, which has been implicated in neural cell specification of miRNAs (let-7a, b, c and e), was up-regulated in the 48-h-reperfused MCAO brain samples along with miR-150 and -125a (20).
Table I.
Stroke-induced miRNAs.
| Authors, year (ref.) | Sample source/type of stroke | Regulation | miRNAs aberrantly expressed | Notes |
|---|---|---|---|---|
| Jeyaseelan et al, 2008 (20) | Male SD rat blood and brain, transient focal ischemia by MCAO | Up | (24 h) miR-16, -23a, -103, -107, -150, -185, -191, -292-5p, -320, -451, -494 and let-7 (a, d, f and i) | Screening of blood and brain samples at 24 and 48 h after reperfusion. |
| (48 h) miR-26a, -26b, -103, -107, -140*, -150, -185, -195, -191, -214, -320, -328, -352, -494 and let-7 (a, c and i) | miRNAs whose expression changed by >1-fold are presented. | |||
| Down | (both 24 and 48 h) miR-7, -27a, -29b,c, -30e,- 98, -101a, -137, -148b, -204, -218, -301, -338, -335, -365-5p, -376b,b* | |||
| Dharap et al, 2009 (21) | Adult male spontaneously hypertensive rat brain, transient focal ischemia by MCAO | Up | miR-140,-145,-331 | MCAO 1 h |
| Down | miR-376b-5p, -153, -29c, -98, -204 | Screening of blood samples at 3, 6, 12, 24 and 72 h. | ||
| The miRNAs presented had fold changes at all time points. | ||||
| Tan et al, 2009 (22) | Human peripheral whole blood, ischemic stroke | Up | Has-let-7e, miR-1184, -1246, -1261, 1275, -1285, -1290, -181a, -25*, -513a-5p, -550, 602, -665, -891, -933,-923 | Ischemic patients between 18-49 years of age of all subtypes (LA, SA, CEmb and UND). miRNAs whose expression changed by >1-fold are presented. |
| Down | Has-let-7f, miR-126, -1259, -142-3p, -15b, -186, -519e, -768-5p | |||
| Liu et al, 2001 (23) | Male SD rat brain and blood, three different injuries | Up | (brain) miR-542-3p | Focal ischemia, intracerebral hemorrhage and Kainate seizures. |
| (blood) miR-96, -152, 298, -333, -505 | ||||
| Down | (brain) miR-155, 362-3p, -122, -450a-5p | miRNAs in all three conditions were regulated by >2-fold. | ||
| (blood) miR-125a-5p, -130b, -142-3p, -330, -342-5p, -685, -347 | ||||
| Lee et al, 2001 (26) | Male C57BL/6 mice brain, ischemic preconditioning | Up | (3 h) miR-200a,b,c, miR-141, -429, -182, -183, 96 | Ischemic preconditioning for 15 min. Screening of miRNA expression after reperfusion of 3 and 24 h. |
| (24 h) miR-681, 197 | ||||
| Down | (3 h) no down-regulated miRNAs | |||
| (24 h) miR-468 | miRNAs whose expression was changed by >1-fold are presented. | |||
| Dharap and Vemuganti, 2010 (27) | Adult male spontaneously hypertensive rat brain, ischemic preconditioning | Up | miR-374, 98, -340, -5p, -21, -352, -37*, -335, -181b, -26b,- 181b, -15b, -146a, let-7a | Preconditioning for 10 min, reperfusion at 6, 24 and 72 h. |
| Down | miR-466c, -292-5p, -328, -873, -494, -345-5p, -92b, -30c-2*, 322*, let-7d* | miRNAs whose expression was changed by >1.5-fold at all of the three time points. |
SD, Sprague-Dawley rat; MCAO, middle cerebral ischemia artery occlusion; LA, large artery stroke; SA, small artery stroke; CEmb, cardioembolic stroke; UND, undetermined cause.
To understand the functional significance of miRNAs in ischemic pathophysiology, the profile of miRNAs in adult rat brain at different reperfusion times (3, 6, 12, 24 and 72 h) after MCAO was investigated. Of the 238 miRNAs evaluated, 8 showed increased and 12 showed decreased expression at a minimum of 4 out of 5 reperfusion time points studied between 3 h and 3 days compared to the sham group. Eight miRNAs showed a significant change (three increased and five decreased) at all time points (3 h to 3 days of reperfusion) after focal ischemia. miR-145 was one of these miRNAs. Prevention of miR-145 expression through continuously infused antagomir-145 into the lateral ventricles of cohorts of rats led to an increased protein expression of its downstream target superoxide dismutase-2 (SOD2) in the post-ischemic brain, and decreased the damage of the infarct. The researchers also found that the number of aberrant miRNAs gradually increased as the reperfusion time progressed. Investigation of the functions of these significantly induced miRNAs must be ongoing (21).
Induced miRNAs in the blood of cancer patients have been researched as a potential biomarker for cancer diagnosis. Likewise, the expression levels of miRNAs in the blood of stroke patients have also been found to be reproducible and indicative of the disease state (22). Use of blood samples obtained from young ischemic stroke patients (18–49 years) has shown that, in addition to the disease progression, the stroke subtype can also be identified via the miRNA profiles. By miRNA array chip, among the 157 miRNAs identified in stroke, expression levels of 138 were found to be up-regulated >1-fold, and the expression levels of 19 miRNAs were found to be down-regulated >1-fold. Among the 19 down-regulated miRNAs, 8 miRNAs (hsa-let-7f, miR-126, -1259, -142-3p, -15b, -186, -519e and -768-5p) were poorly expressed across the three subtypes of stroke (LA, large artery stroke; SA, small artery stroke; CEmb, cardioembolic stroke). Similarly, among the 138 highly expressed miRNAs, 17 miRNAs (hsa-let-7e, miR-1184, -1246, -1261, -1275, -1285, -1290, -181a, -25*, -513a-5p, -550, -602, -665, -891a, -933, -939 and -923) were identified as highly expressed in all subtypes of stroke. Another important finding showed that the miRNA expression patterns could distinguish the prognosis of patients. More miRNAs were down-regulated in stroke patients with a favorable outcome (mRS <2) compared to normal controls, irrespective of the subtype of stroke. These results showed the possible use of miRNAs in blood as biomarkers for specific types of brain injuries (22).
Different brain injuries (ischemia stroke, intracerebral hemorrhage and kainate seizures) lead to different miRNA expression profiles both in the blood and brain, but do share some miRNAs (23). Liu et al found that miR-542-3p was up-regulated and miR-155, -362-3p and -450a-5p were down-regulated in the brain after different injuries. miR-96, -152, -298, -333 and -505 were up-regulated and miR-125a-5p, -130b, -142-3p, -330, -243-5p and -685-347 were down-regulated in blood after different injuries (23).
Ischemic preconditioning (IPC) describes the ischemic stimulus that triggers an endogenous, neuroprotective response that protects the brain during a subsequent severe ischemic injury, a phenomenon known as ‘tolerance’. Comparison and analysis of miRNA expression by miRNA array in mouse cortex in response to preconditioning, ischemic injury and tolerance was used to understand the mechanism of the protective effects of IPC. The study revealed that, after 24 h of reperfusion in the different groups, expression of 153 miRNAs was increased and expression of 4 was decreased and uniquely regulated in the preconditioned cortex, expression of 2 miRNAs was increased and expression of 24 was decreased in the ischemic cortex, and expression of 1 was increased and expression of 2 was decreased in tolerant cortex. The results suggested that preconditioning was the foremost regulator of miRNAs (24).
The gold relief time of stroke is within 3 h after stroke. A study by Kim et al found that the maximum down-regulation of gene expression occurs 3 h after ischemia (25). The change in miRNAs in response to ischemia are quite prompt, and indentifying miRNA changes during this period is very important. A recent study investigated the miRNA profiles at 3 and 24 h of reperfusion. After 15 min, after IPC, 8 miRNAs in the 3-h reperfusion group were categorized into two miRNA families (miR-200 and miR-182) and were found to be selectively up-regulated. Transfection of miR-200b, -200c, -429 and -182 to Neuro-2a cells showed neuroprotective effects after oxygen glucose deprivation was applied (26).
5. miRNA targets and their potential therapeutic role in stroke
An miRNA requires only a few base-pair interactions to effectively silence a target mRNA. The most important region for interaction is the seed sequence, which at positions 2–7 from the 5′ end occurs with high frequency in the genome by chance alone, and therefore the prediction of functional miRNA target sites is challenging, even just within 3′-UTR of mRNAs.
According to the above studies, it is well known that ischemia does lead to changes in miRNA expression, either during ischemic preconditioning or post-stroke, at different times. However, it was notable that miRNAs transcriptome following focal cerebral ischemia was found to occur independently of the miRNAs synthesis machinery. In particular, focal ischemia produced no significant effects in the mRNA expression of important proteins involved in miRNA processing, such as RNAses Drosha and Dicer, Drosha cofactor Pasha, or the premiRNA transporter exportin-5 up to 24 h post-reperfusion (27).
miR-298 was the only miRNA that was up-regulated in both brain and blood after different injuries (ischemic stroke and intracerebral hemorrhage) (23). Results of another study were consistent with this finding. miR-298 has been found to be up-regulated in the brain after transient MCA occlusions (20). Although miR-298 has many functions, the function in stroke is not yet clear; it was shown to recognize specific binding sites in the 3′-UTR of β-amyloid precursor protein-converting enzyme-1 (BACE1) mRNA and to regulate BACE1 protein expression in cultured neuronal cells (28).
The neuroprotective mechanism of IPC has been researched in the past few years. The induced miRNAs during IPC exhibited an important role in the protective effect. Targets of miRNAs significantly regulated in preconditioned mouse cortex were predicted by bioinformatic software tool miRanda (version 2005). The most common target of the regulated miRNAs was found to be mRNA encoding methyl CpG binding protein 2 (MeCP2) (24). DNA microarray studies have shown that MeCP2 mRNA was not significantly regulated by preconditioning (29). Among these significantly decreased miRNAs, down-regulated miR-132 induced a rapid increase in MeCP2 protein, but not mRNA. This first suggested that the link between miR-132 and MeCP2 may serve as an effector of IPC (24). Another study regarding the protective effects of IPC confirmed that miR-132 regulates MeCP2 protein expression through lose- and gain-function of miR-132, and was decreased in preconditioned mouse cortex. Western blotting and q-PCR analysis of the levels of MeCP2 protein and MeCP2 mRNA showed that IPC increases MeCP2 protein, but has no effect on MeCP2 mRNA. The finding indicates that MeCP2 plays an important role in ischemic preconditioned protection. MeCP2 KO mice were found to have a high mortality rate in response to ischemic challenge administered 72 h after preconditioning. This suggests that KO mice have an increased susceptibility to ischemia. These studies support the concept that both miR-132 and MeCP2 serve as novel effectors of the molecular mechanisms underlying ischemic preconditioning-induced tolerance (24).
Two miRNA familes, including miR-200 (miR-200a, miR-200b and miR-429) and miR-182 (miR-182, miR-183 and miR-96), were up-regulated early after ischemic preconditioning. Through bioinformatics technology, prediction of the targeted mRNAs showed that the neuroprotective effects, mainly by down-regulating prolyl hydroxylase 2 (PHD2), were a possible mechanism in neuroprotection (26).
miR-497 was found to be induced in mouse brain transient MCAO, and its function was further researched and validated in stroke (30). One study demonstrated that the miR-497 directly hybridizes to the predicted 3′-UTR target sites of bcl-2, leading to translation inhibition. Repression of miR-497 using antagomirs was found to lower miR-497 levels, reduce MCAO-induced infarct effectively and improve neurological deficits with a corresponding increase in bcl-2 protein, an anti-apoptotic protein (30). Cerebral vascular endothelial cell (CEC) degeneration significantly contributes to blood-brain barrier (BBB) breakdown and neuronal loss after cerebral ischemia. A recent study demonstrated that peroxisome proliferator-activated receptor δ (PPARδ) was significantly reduced and miR-15a was up-regulated in oxygen-glucose deprivation (OGD)-induced mouse CEC death. Intracerebrovascular infusion of a specific PPARδ agonist significantly reduced ischemia-induced miR-15a expression, increased bcl-2 protein levels without a change in bcl-2 mRNA levels, and attenuated caspase-3 activity. Gain or loss of miR-15a significantly increased or reduced OGD-induced CEC death, respectively. Exploring the molecular mechanisms, it was found that up-regulation of PPARδ reduced ischemic vascular injury through alleviation of miR-15a expression. Then, the translation inhibition of the anti-apoptotic gene bcl-2 was alleviated, BBB disruption was ultimately protected and cerebral infarction in mice after transient focal cerebral ischemia was reduced. Therefore, the miR-15a transcript was regulated by PPARδ (31).
6. Conclusion
Increasing evidence indicates that aberrant expression of various miRNAs, both up-regulated and down-regulated species, plays an active role in the pathological processes or neuroprotective role of stroke. A single miRNA has a great impact on cellular function by mostly suppressing and occasionally activating numerous downstream mRNA targets. Therefore, even small-scale turbulence occurring in the miRNA-mediated gene regulation affects various biological pathways involved in both neurodegeneration and neuroprotection in stroke brains. Deregulated miRNAs in stroke provide evidence that miRNA profiles may aid in identifying the subtype of stroke, and show potential use as therapeutic targets. A deeper understanding of miRNAs may also benefit new drug research and development, uncover the pharmacological mechanisms of new drugs at the post-translational level and provide a better evaluation of the drug, which can maximize the success rate from bench to bedside.
However, to date, none of the induced miRNAs and miRNAs targets have been adequately validated in the context of stroke. Furthermore, a standard definition of aberrant miRNAs and the technology used in different experimental platforms have not been standardized, thus much work is still required. We need to elucidate the entire concept of regulated miRNAs, mRNAs, proteins and transcription factors; and the complex relationship among them is most important. We believe that further understanding of aberrant miRNAs in stroke may benefit stroke treatment in the future in a multi-faceted manner, as they do play an important role in the process of stroke pathophysiology.
Figure 1.
Biogenesis of miRNAs and mode of action. pri-miRNAs are transcribed from the intra- and inter-genetic regions of the genome by RNA Pol II, followed by processing by the RNase III enzyme Drosha into premiRNAs. After nuclear export, they are cleaved by the RNase III enzyme Dicer into mature miRNAs consisting of ∼23 nucleotides. Finally, a single-stranded miRNA is loaded onto the miRISC, where the seed sequence is located at positions 2–7 from the 5′ end of the miRNA binding to the target mRNA thus inhibiting gene transcription.
Acknowledgments
This study was supported by the Joint Project of the Chinese Academy of Sciences and the Guangdong Province Guangdong Science and Technology Plan Project (Project no 2009B091300129); and the Science and Technology Development Project of Guangzhou (Project no 2010U1-E00531-7).
References
- 1.Beal CC. Gender and stroke symptoms: a review of the current literature. J Neurosci Nurs. 2010;42:80–87. [PubMed] [Google Scholar]
- 2.Mocellin S, Pasquali S, Pilati P. Oncomirs: from tumor biology to molecularly targeted anticancer strategies. Mini Rev Med Chem. 2009;9:70–80. doi: 10.2174/138955709787001802. [DOI] [PubMed] [Google Scholar]
- 3.Johnnidis JB, Harris MH, Wheeler RT, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 4.Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur J Haematol. 2008;81:304–310. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 5.Aumiller V, Förstemann K. Roles of microRNAs beyond development – metabolism and neural plasticity. Biochim Biophys Acta. 2008;1779:692–696. doi: 10.1016/j.bbagrm.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Carissimi C, Fulci V, Macino G. MicroRNAs: novel regulators of immunity. Autoimmun Rev. 2009;8:520–524. doi: 10.1016/j.autrev.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Bushati N, Cohen SM. MicroRNAs in neurodegeneration. Curr Opin Neurobiol. 2008;18:292–296. doi: 10.1016/j.conb.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Lawrie CH, Soneji S, Marafioti T, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–1161. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 9.Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- 11.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishima T, Mizuguchi Y, Kawahigashi Y, Takizawa T, Takizawa T. RT-PCR-based analysis of microRNA (miR-1 and -124) expression in mouse CNS. Brain Res. 2007;1131:37–43. doi: 10.1016/j.brainres.2006.11.035. [DOI] [PubMed] [Google Scholar]
- 13.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 14.Yu JY, Chung KH, Deo M, Thompson RC, Turner DL. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp Cell Res. 2008;314:2618–2633. doi: 10.1016/j.yexcr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 16.Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P. Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cereb Cortex. 2011;21:1857–1869. doi: 10.1093/cercor/bhq262. [DOI] [PubMed] [Google Scholar]
- 17.Vo N, Klein ME, Varlamova O, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wayman GA, Davare M, Ando H, et al. An activity-regulated microRNA controls dendritic plasticity by downregulating p250GAP. Proc Natl Acad Sci USA. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Neill LA. Boosting the brain’s ability to block inflammation via microRNA-132. Immunity. 2009;31:854–855. doi: 10.1016/j.immuni.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- 21.Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab. 2009;29:675–687. doi: 10.1038/jcbfm.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan KS, Armugam A, Sepramaniam S, et al. Expression profile of microRNAs in young stroke patients. PLoS One. 2009;4:e7689. doi: 10.1371/journal.pone.0007689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu DZ, Tian Y, Ander BP, et al. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab. 2010;30:92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lusardi TA, Farr CD, Faulkner CL, et al. Ischemic preconditioning regulates expression of microRNAs and a predicted target, MeCP2, in mouse cortex. J Cereb Blood Flow Metab. 2010;30:744–756. doi: 10.1038/jcbfm.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JB, Piao CS, Lee KW, et al. Delayed genomic responses to transient middle cerebral artery occlusion in the rat. J Neurochem. 2004;89:1271–1282. doi: 10.1111/j.1471-4159.2004.02429.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee ST, Chu K, Jung KH, et al. MicroRNAs induced during ischemic preconditioning. Stroke. 2010;41:1646–1651. doi: 10.1161/STROKEAHA.110.579649. [DOI] [PubMed] [Google Scholar]
- 27.Dharap A, Vemuganti R. Ischemic pre-conditioning alters cerebral microRNAs that are upstream to neuroprotective signaling pathways. J Neurochem. 2010;113:1685–1691. doi: 10.1111/j.1471-4159.2010.06735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem. 2009;284:1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stenzel-Poore MP, Stevens SL, Xiong Z, et al. Effect of ischaemic preconditioning on genomic response to cerebral ischaemia: similarity to neuroprotective strategies in hibernation and hypoxia-tolerant states. Lancet. 2003;362:1028–1037. doi: 10.1016/S0140-6736(03)14412-1. [DOI] [PubMed] [Google Scholar]
- 30.Yin KJ, Deng Z, Huang H, et al. miR-497 regulates neuronal death in mouse brain after transient focal cerebral ischemia. Neurobiol Dis. 2010;38:17–26. doi: 10.1016/j.nbd.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin KJ, Deng Z, Hamblin M, et al. Peroxisome proliferator-activated receptor delta regulation of miR-15a in ischemia-induced cerebral vascular endothelial injury. J Neurosci. 2010;30:6398–6408. doi: 10.1523/JNEUROSCI.0780-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]