Abstract
The accumulation of single nucleotide polymorphisms (SNPs) in the displacement loop (D-loop) of mitochondrial DNA (mtDNA) has been associated with various types of cancer. The association of SNPs with cancer risk and disease outcome has been exhaustively studied. In this study, we investigated the association of age-at-onset and SNPs in the mitochondrial D-loop using a population-based series of hepatocellular carcinoma (HCC) patients. Haplogroup M (489C) and allele 235G were identified for their association with the late onset of HCC by the log-rank test. In an overall multivariate analysis, haplogroup M (489C) was identified as an independent predictive factor for the age-at-onset of HCC at borderline significant levels [relative risk, 1.736; 95% confidence interval (CI), 0.967–3.115; p=0.065]. Genetic polymorphisms in the D-loop are predictive markers for age-at-onset in HCC patients. Accordingly, the analysis of genetic polymorphisms in the mitochondrial D-loop may help to identify HCC patient subgroups at high risk of early onset of the disease.
Keywords: hepatocellular carcinoma, displacement loop, single nucleotide polymorphism, age-at-onset
Introduction
Hepatocellular carcinoma (HCC) is the fifth most frequent type of cancer and the third leading cause of cancer mortality worldwide, with over half a million cases of mortality every year (1). HCC is also common in China. According to the annual cancer incidence and mortality report, the incidence and mortality rates of HCC in China over the last decade were 300,000 and 306,000 cases, respectively (2,3). This disease is strongly associated with several risk factors, including chronic hepatitis B virus (HBV) and chronic hepatitis C virus (HCV) infection, as well as alcohol abuse (4). Certain epidemic factors have been identified as risk factors or outcome predictors for HCC (5–7); however, the true mechanism of this cancer remains unknown. To date, few studies have focused on the genetic factors associated with age-at-onset of this cancer, although they have demonstrated the genetic prevalence of this disease (8).
Hepatitis virus infection and alcohol abuse are associated with increased oxidative stress in liver cells, resulting in DNA changes including mitochondrial DNA (mtDNA) instability (9,10). The human mitochondrial genome is 16 kb in length and is a closed-circular duplex molecule that contains 37 genes, including two ribosomal RNAs and complete sets of 22 transfer RNAs (tRNAs) (11). mtDNA is believed to be more susceptible to DNA damage and acquires mutations at a higher rate than nuclear DNA owing to high levels of reactive oxygen species (ROS), lack of protective histones and limited capacity for DNA repair in the mitochondria (12–14). Thus, somatic mtDNA mutations occur in a wide variety of degenerative diseases and cancers (15,16) and may be homoplasmic by clonal expansion (17,18) or heteroplasmic in tumor tissues (19,20). In a number of cancers, including hepatitis virus-related HCC, somatic mutations are frequently located in the mtDNA non-coding region, termed the displacement loop (D-loop) (21,22). This region is important for regulating the replication and expression of the mitochondrial genome since it contains the leading-strand origin of replication and is the main promoter for transcription (23).
We sequenced the D-loop that contains a length of 1,122 bps (nucleotides 16024-16569 and 1–576; www.mitomap.org) in the blood from HCC patients and identified 92 single nucleotide polymorphims (SNPs) in the D-loop. We also identified cancer risk and outcome associated SNPs (24,25). In the present study, we assess the correlation between germline SNPs of the D-loop and age-at-onset in HCC patients.
Materials and methods
Tissue specimens and DNA extraction
Blood samples were collected at the Fourth Hospital of Hebei Medical University (China) from 60 HCC patients who underwent HCC resection in the Department of Hepatobiliary Surgery between 2007 and 2008. All patients originated from the Hebei Province of China, a high-risk area for HCC. Whole blood was obtained from corresponding HCC patients. Mitochondria isolation and mtDNA extraction were carried out using the Blood Mitochondrial DNA Extraction Kit (Genmed Scientific Inc., Shanghai, China). The study was approved by the Human Tissue Research Committee of the Fourth Hospital of Hebei Medical University. All patients provided written informed consent for the collection of samples and subsequent analysis.
Polymerase chain reaction (PCR) amplification and sequence analysis
The forward primer, 5′-CCCCATGCTTACAAGCAA GT-3′ (nucleotide 16190–16209); and reverse primer, 5′-GCTTT GAGGAGGTAAGCTAC-3′ (nucleotide 602-583) were used for the amplification of a 982-bp product from the mtDNA D-loop region as previously described (15). PCR was performed according to the protocol of the PCR Master Mix Kit (Promega, Madison, WI, USA) and purified prior to sequencing. Cycle sequencing was carried out using the Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA) and the products were then separated on the ABI PRISM Genetic Analyzer 3100 (Applied Biosystems). Polymorphisms were confirmed by repeated analyses from the two strands. SNPs were identified directly from blood mito-chondria.
Statistical analysis
The age-at-onset curve of the HCC patients was calculated using the Kaplan-Meier method at each SNP site, and compared using the log-rank test. Multivariate survival analysis was performed using a Cox proportional hazards model. The statistical analyses were carried out using the SPSS 11.5 software package (SPSS Company, Chicago, IL, USA). A p-value of <0.05 was considered to indicate statistically significant differences.
Results
A total of 60 patients, including 49 HBV-associated and 11 alcohol-associated HCC patients, were enrolled in this study. The age-at-onset distribution of HCC patients is listed in Table I. Those analyzed included 6 patients aged <40 years, 12 patients aged 40–50, 30 patients aged 51–60 and 12 patients aged >60. None of these patients had received any adjuvant chemotherapy or radiation therapy following HCC resection. The age-at-onset and clinical characteristics of the HCC patients were analyzed using the Kaplan-Meier method and were compared using the log-rank test. Gender, portal vein thrombosis, child classification and tumor quantity were not associated with age-at-onset according to the results of the log-rank test. However, TNM classification and tumor size correlated with age-at-onset at statistically significant levels (Table II).
Table I.
Age-at-onset distribution in ESCC patients.
| Age (years) | No. of cases
|
|
|---|---|---|
| Male | Female | |
| ≤40 | 4 | 2 |
| 41–50 | 11 | 1 |
| 51–60 | 28 | 2 |
| >60 | 10 | 2 |
ESCC, esophageal squamous cell carcinoma.
Table II.
Clinical characteristics and their association with age-at-onset in HCC patients.
| Characteristics | No. of cases | p-value |
|---|---|---|
| Gender | 0.370 | |
| Male | 53 | |
| Female | 7 | |
| TNM classification | 0.062 | |
| I | 15 | |
| II | 45 | |
| Portal vein thrombosis | 0.405 | |
| Yes | 10 | |
| No | 50 | |
| Child classification | 0.122 | |
| A | 57 | |
| B | 3 | |
| Tumor size (diameter) | 0.024 | |
| <5 cm | 10 | |
| ≥5 cm | 50 | |
| Tumor quantity | 0.271 | |
| Single | 50 | |
| Multiple | 10 |
HCC, hepatocellular carcinoma.
Subsequently, the correlation between mtDNA genotype and age-at-onset was compared. We exluded the SNPs with a minor allele frequency of <5% and obtained 21 SNPs for further analysis. The HCC patients were divided into two groups on the basis of their genotype at each SNP site, and the age-at-onset curve was plotted using the Kaplan-Meier method for all HCC patients at these sites. A dramatic difference in age-at-onset appeared at sites 489 and 235 as shown by the log-rank test (Fig. 1). The 489C genotype, known as mtDNA haplogroup M, was significantly associated with a late age-at-onset of HCC when compared with halpogroup N (p=0.015) (Fig. 1). Another age-at-onset-associated SNP was identified at site 235 with frequency allele 235G and was linked with the late onset of HCC (p=0.009).
Figure 1.
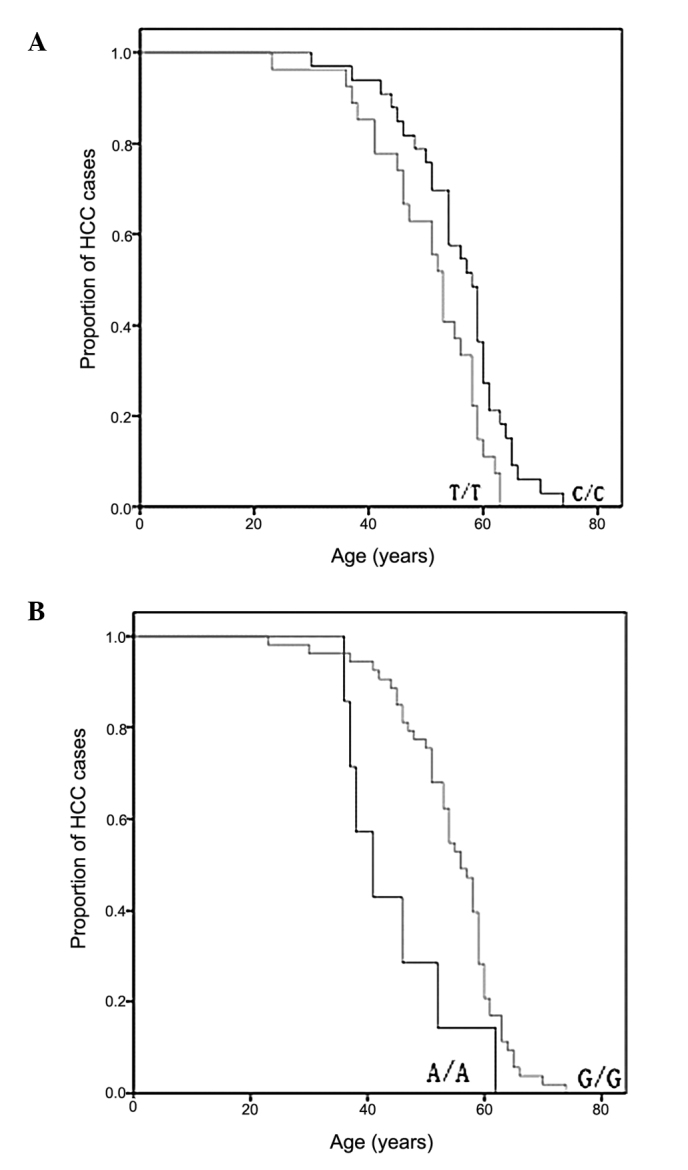
Comparison of the assocation between the age-at-onset of HCC and the (A) 489 and (B) 235 sites in the D-loop. D-loop, displacement loop; HCC, hepatocellular carcinoma.
Using the Cox proportional hazards model, we performed multivariate analysis of HCC age-at-onset predictors including TNM classification, tumor size and the two SNPs. As shown in Table III, haplogroup M (489C) was identified as an independent predictive factor for age-at-onset of HCC at borderline significant levels [relative risk, 1.736; 95% confidence interval (CI), 0.967–3.115; p=0.065].
Table III.
Multivariate analysis of predictive factors associated with the age-at-onset of HCC.
| Factors | Relative risk | 95% CI | p-value |
|---|---|---|---|
| Tumor size | 2.023 | 0.831–4.926 | 0.121 |
| TNM classification | 1.179 | 0.573–2.427 | 0.654 |
| 235 (G/A) | 0.611 | 0.254–1.470 | 0.271 |
| 489 (C/T) | 1.736 | 0.967–3.115 | 0.065 |
HCC, hepatocellular carcinoma; CI, confidence interval.
Discussion
Selected SNPs in the D-loop region have previously been examined for their ability to predict cancer risk and outcome in many types of cancer (26–28). The present study has extended these analyses to determine the relationships between age-at-onset and germline SNPs in a continuous sequence of mtDNA between nucleotides 16190 and 583 in HCC patients. The SNPsm, 489C/T and 235G/A, were identified for their association with age-at-onset at statistically significant levels by the log-rank test. In an overall multivariate analysis, haplogroup M (489C) was identified as an independent predictive factor for age-at-onset of HCC at borderline significant levels. The results from the log-rank test revealed that disease advancement with a larger tumor size and a more serious clinical stage was more prevalent in younger patients. However, this was not shown in the multivariate analysis.
We have previously performed a number of studies on D-loop SNPs as predictive factors for digestive tract cancers (24,25,29). In the present study, for the first time, we suggest that, other than as predictors for cancer risk and outcome, SNPs in the D-loop are also predictors for age-at-onset in HCC patients. All non-African lineages belong to two founder clusters, namely haplogroups M and N; 489C, defined as haplogroup M, and 489T, defined as haplogroup N. According to the ‘out of Africa’ theory, these are both derived from the L3 mtDNA African lineage (30). The functional significance of the mtDNA haplo-groups and their association with human behaviors requires further study, although certain haplogroups have been identified as markers for special diseases (31,32). The results from this study require validation in the form of larger population sizes and laboratory-based functional studies.
The D-loop region of mtDNA is crucial for the regulation of mitochondrial genome replication and expression. SNPs in this region might affect mtDNA replication and lead to the alteration of the electron transport chain, which is responsible for the release of highly reactive oxygen species (ROS) and could contribute to nuclear genome damage as well as cancer initiation and promotion (33–35). These two SNPs may alter the transcription of the mitochondrial genome, and this may subsequently enhance the production of ROS when the mitochondrial transcription is altered (36). These ROS-mediated mechanisms may thereby accelerate tumor development.
In conclusion, SNPs in the D-loop have been found to be biomarkers for the age-at-onset of HCC. The analysis of genetic polymorphisms in the D-loop might help to identify patient subgroups at high risk for an early onset, thereby helping to refine therapeutic decisions in HCC cancers.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 30801384). The research was supported in part by the Natural Science Foundation of Hebei Province (No. C2008000958).
References
- 1.Gomaa AI, Khan SA, Toledano MB, Waked I, Taylor-Robinson SD. hepatocellular carcinoma: Epidemiology, risk factors and pathogenesis. World J Gastroenterol. 2008;14:4300–4308. doi: 10.3748/wjg.14.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun Z, Ming L, Zhu X, Lu J. Prevetion and control of hepatitis B in China. J Med Virol. 2002;67:447–450. doi: 10.1002/jmv.10094. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Bray F, Pisani P, Parkin DM, editors. Globocan 2000: Cancer Incidence, Mortality and Prevalence Worldwide Version 1.0. IARC Press; Lyon: p. 2001. [Google Scholar]
- 4.Caldwell S, Park SH. The epidemiology of hepatocellular cancer: from the perspectives of public health problem to tumor biology. J Gastroenterol. 2009;44:96–101. doi: 10.1007/s00535-008-2258-6. [DOI] [PubMed] [Google Scholar]
- 5.Maki A, Kono H, Gupta M, Asakawa M, Suzuki T, Matsuda M, Fujii H, Rusyn I. Predictive power of biomarkers of oxidative stress and inflammation in patients with hepatitis C virus-associated hepatocellular carcinoma. Anal Surg Oncol. 2007;14:1182–1190. doi: 10.1245/s10434-006-9049-1. [DOI] [PubMed] [Google Scholar]
- 6.Okada S, Shimada K, Yamamoto J, Takayama T, Kosuge T, Yamasaki S, Sakamoto M, Hirohashi S. Predictive factors for postoperative recurrence of hepatocellular carcinoma. Gastroenterology. 1994;106:1618–1624. doi: 10.1016/0016-5085(94)90419-7. [DOI] [PubMed] [Google Scholar]
- 7.Minagawa M, Makuuchi M, Takayama T, Kokudo N. Selection criteria for repeat hepatectomy in patients with recurrent hepatocellular carcinoma. Ann Surg. 2003;238:703–710. doi: 10.1097/01.sla.0000094549.11754.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong N, Yeo W, Wong WL, Wong NL, Chan KY, Mo FK, Koh J, Chan SL, Chan AT, Lai PB, et al. TOP2A overexpression in hepatocellular carcinoma correlates with early age onset, shorter patients survival and chemoresistance. Int J Cancer. 2009;124:644–652. doi: 10.1002/ijc.23968. [DOI] [PubMed] [Google Scholar]
- 9.Schwarz KB. Oxidative stress during viral infection: a review. Free Radical Biol Med. 1996;21:641–649. doi: 10.1016/0891-5849(96)00131-1. [DOI] [PubMed] [Google Scholar]
- 10.Mansouri A, Fromenty B, Berson A, Robin MA, Grimbert S, Beaugrand M, Erlingr S, Pessayre D. Multiple hepatic mitochondrial DNA deletions suggest premature oxidative aging in alcoholic patients. J Hepatol. 1997;27:96–102. doi: 10.1016/s0168-8278(97)80286-3. [DOI] [PubMed] [Google Scholar]
- 11.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 12.DiMauro S, Schon EA. Mitochondrial DNA mutations in human disease. Am J Med Genet. 2001;106:18–26. doi: 10.1002/ajmg.1392. [DOI] [PubMed] [Google Scholar]
- 13.Beal MF. Mitochondia, free radicals, and neurodegeneration. Curr Opin Neurobiol. 1996;6:661–666. doi: 10.1016/s0959-4388(96)80100-0. [DOI] [PubMed] [Google Scholar]
- 14.Lightowlers RN, Chinnery PF, Turnbull DM, Howell N. Mammalian mitochondrial genetics: heredity, heteroplasmy and disease. Trends Genet. 1997;13:450–455. doi: 10.1016/s0168-9525(97)01266-3. [DOI] [PubMed] [Google Scholar]
- 15.Wallace DC. Mouse models for mitochondrial disease. Am J Med Genet. 2001;106:71–93. doi: 10.1002/ajmg.1393. [DOI] [PubMed] [Google Scholar]
- 16.Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, Jen J, Sidransky D. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 17.Nomoto S, Yamashita K, Koshikawa K, Nakao A, Sidransky D. Mitochondrial D-loop mutation as clonal markers in multicentric hepatocellular carcimona and plasma. Clin Cancer Res. 2002;8:481–487. [PubMed] [Google Scholar]
- 18.Mambo E, Gao X, Cohen Y, Guo Z, Talalay P, Sidransky D. Electrophile and oxidant damage of mitochondrial DNA leading to rapid evolution of homoplasmic mutations. Proc Natl Acad Sci USA. 2003;100:1838–1843. doi: 10.1073/pnas.0437910100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoneyama H, Hara T, Kato Y, Yamori T, Matsuura ET, Koike K. Nucleotide sequence variation is frequently in the mitochondrial DNA displacement loop region of individual human tumor cells. Mol Cancer Res. 2005;3:14–20. [PubMed] [Google Scholar]
- 20.Jakupciak JP, Maragh S, Markowitz ME, Greenberg AK, Hoque MO, Maitra A, Barker PE, Wagner PD, Rom WN, Srivastava S, Sidransky D, O’Connell CD. Performance of mitochondrial DNA mutations detecting early stage cancer. BMC Cancer. 2008;8:285. doi: 10.1186/1471-2407-8-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nashikawa M, Nishiguchi S, Shiomi S, Tamori A, Koh N, Takeda T, Kubo S, Hirohashi K, Kinoshita H, Sato E, Inoue M. Somatic mutation of mitochondrial DNA in cancerous and noncancerous liver tissue in individuals with hepatocellular carcinoma. Cancer Res. 2001;61:1843–1845. [PubMed] [Google Scholar]
- 22.Sanchez-Cespedes M, Parrella P, Nomoto S, Cohen D, Xiao Y, Esteller M, Jeronimo C, Jordan RC, Nicol T, Koch WM, Schoenberg M, Mazzarelli P, Fazio VM, Sidransky D. Identification of a mononucleotide repeat as a major target for mitochondrial DNA alterations in human tumors. Cancer Res. 2001;61:7015–7019. [PubMed] [Google Scholar]
- 23.Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410:103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 24.Zhang R, Zhang F, Wang C, Wang S, Shiao Y-H, Guo Z. Identification of sequence polymorphism in the D-loop region of mitochondria DNA as a risk factor for hepatocellular carcinoma with distinct etiology. J Exp Clin Cancer Res. 2010;29:130. doi: 10.1186/1756-9966-29-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C, Zhang F, Fan H, Peng L, Zhang R, Liu S, Guo Z. Sequence polymorphisms of mitochondrial D-loop and hepatocellular carcinoma outcome. Biochem Biophys Res Commun. 2011;406:493–496. doi: 10.1016/j.bbrc.2011.02.088. [DOI] [PubMed] [Google Scholar]
- 26.Navaglia F, Basso D, Fogar P, Sperti C, Greco E, Zambon CF, Stranges A, Falda A, Pizzi S, Parenti A, Pedrazzoli S, Plebani M. Mitochondrial DNA D-loop in pancreatic cancer: somatic mutations are epiphenomena while the germline 16519 T variant worsens metabolism and outcome. Am J Clin Pathol. 2006;126:593–601. doi: 10.1309/GQFCCJMH5KHNVX73. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Bamlet WR, de Andrade M, Boardman LA, Cunningham JM, Thibodeau SN, Petersen GM. Mitochondrial genetic polymorphisms and pancreatic cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:1455–1459. doi: 10.1158/1055-9965.EPI-07-0119. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, McDonnell SK, Hebbring SJ, Cunningham JM, St Sauver J, Cerhan JR, Isaya G, Schaid DJ, Thibodeau SN. Polymorphisms in mitochondrial genes and prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:3558–3566. doi: 10.1158/1055-9965.EPI-08-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang R, Wang R, Zhang F, Wu C, Fan H, li Y, Wang C, Guo Z. Single nucleotide polymorphisms in the mitochondrial displacement loop and outcome of esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2010;29:155. doi: 10.1186/1756-9966-29-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.González AM, Larruga JM, Abu-Amero KK, Shi Y, Pestano J, Cabrera VM. Mitochondrial lineage M1 traces an early human backflow to Africa. BMC Genomics. 2007;8:223. doi: 10.1186/1471-2164-8-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carelli V, Achilli A, Valentino ML, Rengo C, Semino O, Pala M, Olivieri A, Mattiazzi M, Pallotti F, Carrara F, et al. Haplogroup effects and recombination of mitochondrial DNA: novel clues from the analysis of Leber hereditary optic neuropathy pedigrees. Am J Hum Genet. 2006;78:564–574. doi: 10.1086/501236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Udar N, Atilano SR, Memarzadeh M, Boyer DS, Chwa M, Lu S, Maguen B, Langberg J, Coskun P, Wallace DC, et al. Mitochondrial DNA haplogroups associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009;50:1966–2974. doi: 10.1167/iovs.08-2646. [DOI] [PubMed] [Google Scholar]
- 33.Bandy B, Davision AJ. Mitochondrial mutations may increase oxidative stress: implications for carcinogenesis and aging? Free Radic Biol Med. 1990;8:523–539. doi: 10.1016/0891-5849(90)90152-9. [DOI] [PubMed] [Google Scholar]
- 34.Gille JJ, Joenje H. Cell culture models for oxidative stress: superoxide and hydrogen peroxide versus normobaric heperoxia. Mutat Res. 1992;275:405–414. doi: 10.1016/0921-8734(92)90043-o. [DOI] [PubMed] [Google Scholar]
- 35.Shigenaga MK, Hagen TM, Ames BN. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dement GA, Maloney SC, Reeves R. Nuclear HMGA1 nonhistone chromatin proteins directly influence mitochondrial transcription, maintenance, and function. Exp Cell Res. 2007;313:77–87. doi: 10.1016/j.yexcr.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]


