Abstract
Organelle genomes show remarkable variation in architecture and coding content, yet their nucleotide composition is relatively unvarying across the eukaryotic domain, with most having a high adenine and thymine (AT) content. Recent studies, however, have uncovered guanine and cytosine (GC)-rich mitochondrial and plastid genomes. These sequences come from a small but eclectic list of species, including certain green plants and animals. Here, I review GC-rich organelle DNAs and the insights they have provided into the evolution of nucleotide landscape. I emphasize that GC-biased mitochondrial and plastid DNAs are more widespread than once thought, sometimes occurring together in the same species, and suggest that the forces biasing their nucleotide content can differ both among and within lineages, and may be associated with specific genome architectural features and life history traits.
Keywords: Coccomyxa, GC content, mitochondrial DNA, plastid DNA, Polytomella, Selaginella, RNA editing
Introduction
Mitochondria and plastids are the products of ancient endosymbiotic events, involving a proteobacterium and a cyanobacterium, respectively (Lang et al., 1999; Palmer, 2003). Mitochondria arrived early and probably existed in the common ancestor of all eukaryotes (Gray et al., 1999). Plastids came later, first arising in the Archaeplastida (Plantae), and then being passed on laterally to diverse lineages through eukaryote–eukaryote endosymbioses (Archibald, 2009; Keeling, 2010). The genomes within contemporary mitochondria and plastids have been fashioned through coexistence and coevolution with their eukaryotic hosts, and in many instances have acquired bizarre and complex architectures (Palmer, 1985; Gray et al., 2004; Green, 2011).
Organelle DNAs boast an impressive, and often puzzling, array of sizes (<10 to >1000 kb), conformations (circular or linear), chromosome numbers (monomeric to highly fragmented), compactnesses (<10 to >90% non-coding DNA), and gene repertoires (<5 to >250 genes). Moreover, many organelle genomes use a non-standard genetic code (Jukes and Osawa, 1993), and some employ complicated editing systems that alter the sequences of RNA transcripts (Covello and Gray, 1989; Simpson and Thiemann, 1995). One feature of organelle DNA that has proven to be relatively constant across lineages is its nucleotide composition. Almost all completely sequenced mitochondrial and plastid DNAs (mtDNAs and ptDNAs) have a high adenine and thymine (AT) content (Kusumi and Tachida, 2005; Min and Hickey, 2007). Various hypotheses have tried to explain this AT bias, but the topic remains poorly understood.
Recently, it was shown that guanine and cytosine (GC)-rich organelle DNAs do exist (Tsuji et al., 2007; Smith and Lee, 2008; Hecht et al., 2011). These genomes come from a small but diverse group of species, including various green plants and animals, and sometimes have linear conformations or undergo large amounts of post-transcriptional editing. Unraveling the mechanism responsible for their GC enrichment may help explain the near-ubiquity of AT-rich mitochondrial and plastid genomes throughout the eukaryotic domain, and could give insights into other aspects organelle genome architecture, such as the origins of RNA editing. The existence of GC-rich organelle genomes, however, is poorly chronicled in the scientific literature, even though these sequences could impact how we use organelle DNA for studying molecular evolution (Foster and Hickey, 1999).
This review showcases GC-biased organelle genomes and the species in which they are found. GC enrichment is discussed in context to mutation, recombination, population genetics, and genome architecture. It is emphasized that GC-rich mtDNAs and ptDNAs are more common than once thought – occasionally occurring together in the same species – and that the processes promoting GC enrichment can differ within and among lineages.
The Near-Ubiquity of AT-Rich Organelle Genomes
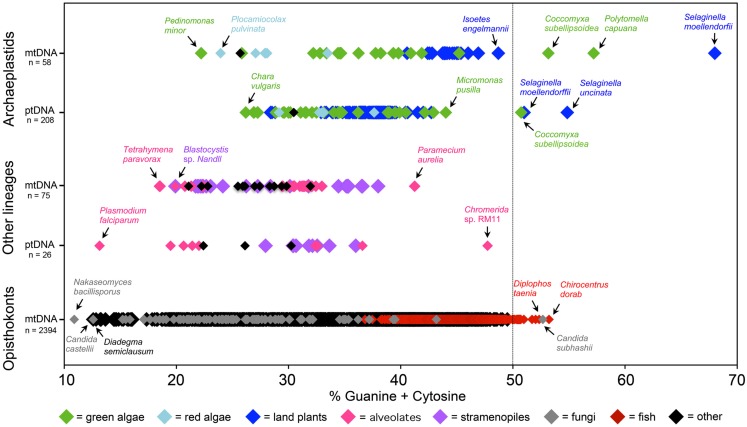
The sequencing of large numbers of organelle genomes from diverse lineages has revealed an almost universal AT bias in mtDNAs and ptDNAs across the eukaryotic domain (Figure 1). Of the ∼2,500 mitochondrial and plastid genomes that have been sequenced, as of January 1, 2012, most have an AT content above 50% (average ≈65%; Figure 1). Convergent evolution to AT richness is found in other types of organelle-located DNAs, such as mitochondrial plasmids (Handa, 2008), nucleomorph genomes (Moore and Archibald, 2009), and the genomes of mitochondrial viruses (Wu et al., 2010). Moreover, the genomes of bacterial and eukaryotic endosymbionts and intracellular parasites tend to have higher AT compositions than those of their free-living close relatives (Pallen and Wren, 2007; Nowack et al., 2008; McCutcheon and Moran, 2010).
Figure 1.
Nucleotide composition continuum of completely sequenced mitochondrial DNA (mtDNA) and plastid DNA (ptDNA) sequences. Most of the complete organelle genome sequences deposited in GenBank have a GC content below 50%, with the exception of those from certain green algae, lycophytes, fish, and fungi. The number of genome sequences (n) within each group is shown beside the y-axis. Mitochondrial and plastid genome sequences were downloaded from GenBank on January 1, 2012.
Many processes can influence nucleotide landscape, including mutation, recombination, random genetic drift, and selection (Lynch, 2007; Charlesworth and Charlesworth, 2010). The net effect of these processes ultimately determines the equilibrium nucleotide composition of a genome. The origins of AT richness within mtDNAs and ptDNAs are thought to reflect the endosymbiotic history of these genomes, their location within the cell, the unique population-genetic features that define organelles, and selection for metabolic and translational efficiency.
The massive shedding of genes that characterized early mtDNA and ptDNA evolution resulted, at least for some lineages, in the loss of key DNA repair proteins and, consequently, diminished nucleotide repair capacities within organelles (Kleine et al., 2009; Bendich, 2010, but see Liu and Demple, 2010). Organelle DNAs are typically uniparentally inherited, non-recombining, and can experience severe bottlenecks during transmission, which implies that they are inefficient at purging deleterious mutations from their populations (Muller, 1964; Rand, 2001; but see Piganeau et al., 2004). Organelle genomes undergo multiple rounds of replication per cell division (Birky, 2001), predisposing them to replication errors, and they are housed in energy-producing compartments where high concentrations of reactive oxygen species promote GC→AT mutations through the deamination of cytosine and the oxidative conversion of guanine to 8-oxo-guanine (Martin, 1995; Asada, 2006; Murphy, 2009; Shokolenko et al., 2009). Together, these points suggest that organelle DNAs inhabit a highly mutagenic environment, where DNA repair is inefficient, and the mutational spectrum is skewed toward AT. As one might expect, many species, including most metazoans, appear to have high organelle DNA mutation rates (Lynch et al., 2006). There are some species, however, for which the organelle DNA mutation rate is estimated to be low (e.g., most angiosperms), yet their organelle genomes are still AT-rich (Drouin et al., 2008).
In addition to a genome-wide AT bias, mtDNAs and ptDNAs can exhibit regional and strand-specific nucleotide biases (Gibson, 2005; Kusumi and Tachida, 2005). The mutational consequences of organelle genome replication can give rise to AC vs. GT inequities because the DNA strand that spends more time in the mutationally vulnerable single-stranded state is prone to C→T and A→G transitions (Ames et al., 1995; Frank and Lobry, 1999; Faith and Pollock, 2003); but this does not impact the overall AT composition as the G’s and T’s of one strand are complemented by A’s and C’s on the other strand.
Natural selection is thought to have contributed to the high AT content of mitochondrial and plastid genomes. Selection for translational efficiency and accuracy is believed to have shaped the nucleotide composition of codons in organelle genes, in some cases enriching the thymine content of synonymous sites (Morton, 1998). Others have argued that AT richness is an adaptation for metabolic efficiency, noting the increased energetic costs of producing C vs. T and G vs. A and the varying abundance of A/T vs. G/C nucleotides during organelle DNA synthesis (Jukes and Bhushan, 1986; Wolfe, 1991; Rocha and Danchin, 2002).
Thus, a multitude of forces have likely helped generate the near-universal AT bias of organelle DNAs. The discovery of organelle genomes with a high GC content has provided an important point of comparison from which to better understand these forces.
Taxa with GC-Rich Organelle Genomes
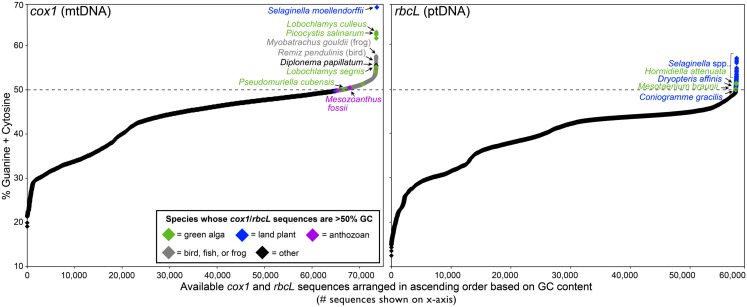
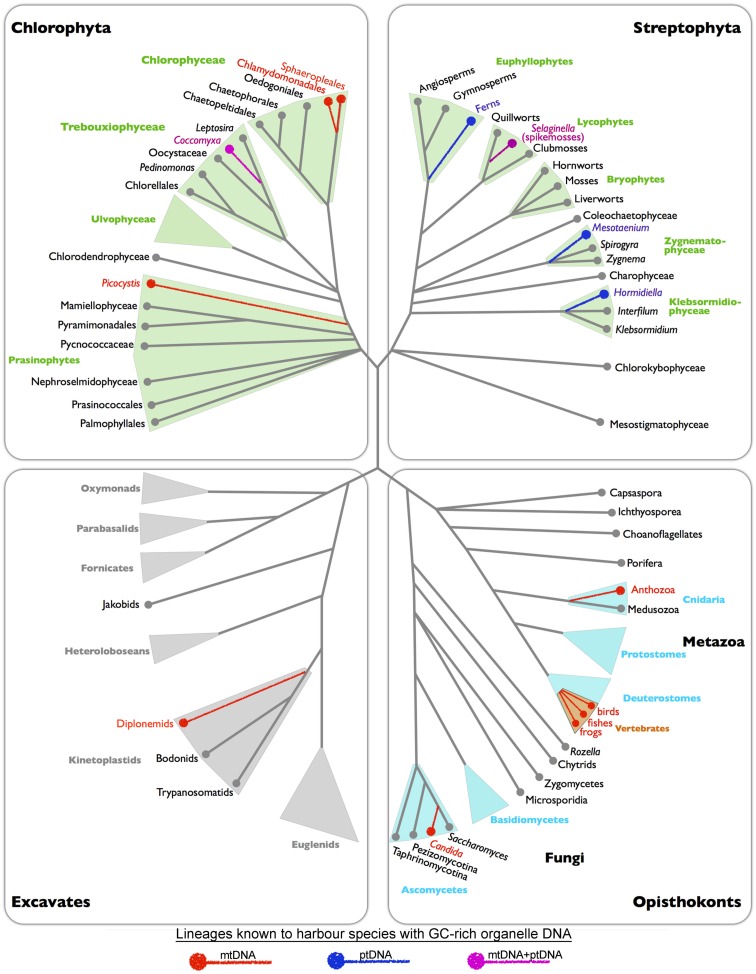
There are more than 40 complete organelle genome sequences in GenBank with GC contents exceeding 50% (Figure 1). These genomes come from various fish, green algae, and land plants as well as a fungus (Figure 1; Table 1). Moreover, the nucleotide composition of organelle genes, like those encoding the mitochondrial protein cytochrome c oxidase subunit I (cox1) and the large subunit of the plastid protein Rubisco (rbcL), have proven to be good predictors of overall organelle DNA nucleotide content (Min and Hickey, 2007; Clare et al., 2008; Smith, 2009). Analyses of cox1 and rbcL have revealed other lineages with GC-rich organelle genomes (Kerr et al., 2007; Borza et al., 2009; Figure 2; Table 1). The taxonomic groups containing (or predicted to contain) species with GC-rich organelle DNA are listed below and highlighted in Figures 1–3 and Table 1.
Table 1.
Examples of GC-rich organelle genomes and the species that harbor them.
| Taxonomy | GCTOT | GC1 | GC2 | GC3 | GCNC | GC-bias mt + pt2 | Other taxa3 | Genomic architecture | Organismal features | GenBank accession | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MtDNAs | |||||||||||
| Myobatrachus gouldii1 | Frog (turtle frog) | ∼55 | 59.3 | 44.4 | 69.5 | N/A | – | Yes | N/A | Small burrowing species found in sandy soil throughout Western Australia. | HQ584074-5 AY948768 |
| Candida subhashii strain CBS 10753 | Fungus (yeast) | 52.7 | 53.2 | 38.7 | 66.4 | 54.4 | – | No | 30 kb compact intron-less linear genome with inverted-repeat telomeres (Fricova et al., 2010). | Human pathogen. First isolated from a case of fungal peritonitis. | NC_014337 |
| Chirocentrus dorab | Fish (wolf herring) | 53.2 | 58.7 | 47.7 | 53.7 | 39.3 | – | Yes | 16 kb compact intron-less circular-mapping genome (Ishiguro et al., 2005). | Marine; brackish. Distribution Indo-Pacific. Observed in warm coastal waters. | NC_006913 AP006229 |
| Coccomyxa subellipsoidea C-169 | Green alga (trebouxiophyte) | 53.2 | 51.4 | 40.8 | 59.8 | 55.7 | Yes | Yes | 65 kb, circular-mapping genome with moderate amount of non-coding DNA. Similar repeat elements in mtDNA and ptDNA (Smith et al., 2011). | Free-living, unicellular species, isolated in Marble Point Antarctica. | HQ874522 |
| Diplonema papillatum ATCC 501621 | Protist (euglenozoan) | ∼55 | 60.9 | 53.0 | 53.2 | ∼54 | – | No | Multipartite genome comprised of circular-mapping chromosomes. Highly fragmented coding regions. U-insertion RNA editing (Vlcek et al., 2011). | Free-living, unicellular marine flagellate, isolated from the surface of eelgrass in New Hampshire. | HQ288819-33 EU123536-7 |
| Lobochlamys culleus SAG19.721 | Green alga (chlorophycean) | ∼60 | 53.4 | 40.5 | 93.0 | ∼59 | No | Yes | Multipartite genome comprised of linear chromosomes with overlapping homologies. Repeat dense (Borza et al., 2009). | Free-living, unicellular freshwater biflagellate. Isolated from pond in Florida, USA. | AF529310-6 FJ393025-57 |
| Mesozoanthus fossii1 | Coral (zoanthid) | ∼55 | 50.1 | 40.9 | 64.2 | N/A | – | Yes | N/A | Observed in fjords from Northern to Central Patagonia. | EF672653-5 EF687821-3 |
| Isoetes lacustris1 | Land plant (lycophyte) | ∼50 | 59.8 | 63.8 | 43.3 | ∼58 | No | Yes | ∼60 kb genome, comprising a complex network of recombinogenic mtDNA molecules. High levels of C-to-U RNA editing. Intron rich, but relatively compact (Grewe et al., 2009)4. | Boreal quillwort observed in Europe and North America. Grows on the bottom of ponds. | AM261455-6 Y17812-4 X92736 |
| Picocystis salinarum CCMP 18971 | Green alga (prasinophyte) | ∼60 | 55.4 | 41.1 | 91.7 | N/A | No | N/A | N/A | Unicellular picoplankton, isolated from a saline pond in San Francisco Bay. | AB491634 |
| Polytomella capuana SAG 63-5 | Green alga (chlorophycean) | 57.2 | 52.2 | 41.3 | 76.0 | 61.0 | N/A | No | 13 kb highly reduced, linear genome with inverted-repeat telomeres (Smith and Lee, 2008). | Free-living, non-photosynthetic unicellular freshwater flagellate, isolated from ditch in Italy. | NC_010357 |
| Selaginella moellendorffii | Land plant (lycophyte) | 68.1 | 64.2 | 60.2 | 61.5 | 68.9 | Yes | Yes | Genome comprised of a complex network of recombinogenic mtDNA molecules. Repeat and intron dense. Unprecedented levels of C-to-U RNA editing (Hecht et al., 2011). | Seedless vascular plant. Model species, often used for cultivation. | GQ246802-8 JF338143-7 |
| Remiz pendulinus1 | Bird (penduline tit) | ∼55 | 58.6 | 42.2 | 65.4 | N/A | – | Yes | N/A | Tiny passerine observed in various regions throughout Eurasia. | GU572078-9 AY228081 |
| PtDNAs | |||||||||||
| Cheiropleuria integrifolia1 | Land plant (fern) | ∼50 | 59.6 | 45.7 | 45.9 | ∼50 | N/A | Yes | N/A | Terrestrial fern of moderate size. Collected in Japan, Kagoshima prefecture (Ebihara et al., 2010). | AB042569 EU328229 |
| Coccomyxa subellipsoidea C-169 | Green alga (trebouxiophyte) | 50.7 | 56.1 | 43.6 | 50.1 | 51.0 | Yes | Yes | 176 kb intron-poor circular-mapping genome. Similar repeat elements in mtDNA and ptDNA (Smith et al., 2011). | Free-living, unicellular species, isolated in Marble Point Antarctica. | NC_015084 |
| Coniogramme gracilis Ogata1 | Land plant (fern) | ∼50 | 59.3 | 43.8 | 48.7 | N/A | N/A | Yes | N/A | Narrow-leaf bamboo fern. Collected in Japan, Kagoshima prefecture (Ebihara et al., 2010). | AB574810 |
| Hormidiella attenuata strain M22141 | Green alga (klebsormidiophyte) | ∼51 | 58.4 | 42.5 | 52.9 | N/A | N/A | Yes | N/A | Freshwater species, forming multicellular, non-branching filaments. | HQ613235 |
| Mesotaenium braunii1 | Green alga (zygnemophyte) | ∼51 | 58.5 | 43.1 | 51.5 | N/A | N/A | No | N/A | Free-living, unicellular freshwater alga. Isolated from Eifel, Germany. | FM992358 FM992569 |
| Selaginella uncinata | Land plant (lycophyte) | 54.8 | 58.8 | 54.7 | 49.3 | 54.9 | Yes | Yes | 144 kb intron-poor circular-mapping genome. Reduced tRNA-coding content. High levels of C-to-U RNA editing (Tsuji et al., 2007). | Seedless vascular plant. Model species, often used for cultivation. | AB197035 |
Percentage of guanine and cytosine of entire genome (GCTOT), first-, second-, and third-position codon sites (GC1, GC2, GC3, respectively), non-coding regions (GCNC). Data not available (N/A).
1Statistics based on partial genome sequence data (e.g., rbcL or cox1).
2Reports if a GC bias is present in both the mitochondrial and plastid genomes (only applicable to plastid-bearing species).
3Reports if GC-rich organelle DNA has been observed in other members of the given lineage.
4Genomic architecture based on data for Isoetes engelmannii.
Figure 2.
Nucleotide composition continuum of the available cox1 and rbcL sequences from eukaryotic organelle genomes. The cox1 gene, which is located in the mitochondrial genome of all studied eukaryotes, encodes the protein cytochrome c oxidase subunit I. The rbcL gene, which is found in the ptDNA of most plastid-bearing eukaryotes, encodes the large subunit of Rubsico. The nucleotide content of cox1 and rbcL are good predictors of the overall mtDNA and ptDNA nucleotide composition, respectively (Min and Hickey, 2007; Smith, 2009). Complete and partial cox1 and rbcL sequences (minimum length = 400 nt) were downloaded from GenBank on January 1, 2012. Given the huge number of bilaterian cox1 sequences (>300,000), the chart only shows those for species from the Actinopterygii, Archosauria, and Amphibia – the bilaterians known to have cox1 sequences that can exceed 50% GC.
Figure 3.
Tree showing the eukaryotic lineages that have species with GC-rich organelle genomes. Colored branches represent lineages that are known to contain (based on complete organelle genome sequence data) or predicted to contain (based on cox1/rbcL sequences) species with GC-rich organelle genomes (red = mtDNA, blue = ptDNA, pink = mtDNA and ptDNA). Branching order based on published phylogenetic analyses.
Green algae
Some of the highest organelle genome GC contents come from green algae. The chlorophycean Polytomella capuana, a non-photosynthetic unicell closely related to the model organism Chlamydomonas reinhardtii, has an mtDNA GC content of 57% (Smith and Lee, 2008). All other investigated Polytomella species, however, have AT-rich mtDNAs (Smith et al., 2010). Partial mitochondrial genome sequences suggest that the freshwater biflagellates Oogamochlamys gigantea, Lobochlamys segnis, and Lobochlamys culleus, which are also close relatives of C. reinhardtii, have mtDNA GC compositions of approximately 50, 55, and 60%, respectively (Borza et al., 2009). The polar trebouxiophyte Coccomyxa subellipsoidea C-169 has a GC-bias in both its mitochondrial and plastid compartments (53 and 51% GC, respectively), and organelle gene sequencing indicate that Coccomyxa chodatii and Coccomyxa rayssiae have GC-rich organelle DNAs as well (Smith et al., 2011). The picoplankton Picocystis salinarum, a deep-branching prasinophyte, appears to have an mtDNA GC content exceeding 60%. And rbcL sequences imply that there are GC-enriched plastid genomes in select members of the charophyte genera Mesotaenium and Hormidiella (Gontcharov and Melkonian, 2010; Rindi et al., 2011).
Land plants
The highest recorded GC content in an mtDNA (68%) and a ptDNA (55%) belong to the seedless vascular plants Selaginella moellendorffii and Selaginella uncinata, respectively (Tsuji et al., 2007; Hecht et al., 2011). Like C. subellipsoidea, Selaginella species have a GC-bias in both their mitochondrial and plastid compartments (Smith, 2009). Plastid gene sequences of more than 100 Selaginella species from diverse regions revealed only one species without GC-biased ptDNA: the Chinese specimen Selaginella sinensis (Smith, 2009). Other lycophytes, including several Isoetes species, also have relatively high mtDNA GC contents (Malek and Knoop, 1998; Grewe et al., 2009). Analyses of rbcL genes suggests that some ferns from the genera Cheiropleuria, Coniogramme, Cystopteris, Dryopteris, and Monachosorum have a GC-bias in their ptDNA (Ebihara et al., 2010; de Groot et al., 2011).
Animals and fungi
There are at least 25 species of fish with overall mtDNA GC contents >50%, such as the wolf herring Chirocentrus dorab (53%), the Pacific porthole fish Diplophos taenia (52%), and the beaked salmon Gonorynchus greyi (52%; Miya and Nishida, 2000; Saitoh et al., 2003; Ishiguro et al., 2005). Moreover, cox1 nucleotide content analyses suggest that potentially hundreds, if not thousands, of other teleosts, from many different orders, have GC-biased mtDNA. Single-gene nucleotide content analyses have revealed various birds, frogs, and corals with GC-rich mitochondrial genomes (Kerr et al., 2007; Crawford et al., 2010). The European penduline tit Remiz pendulinus, the turtle frog Myobatrachus gouldii, and the zoanthid coral Mesozoanthus fossii all appear to have particularly high mtDNA GC contents. Among fungi, the pathogenic yeast Candida subhashii is the only species known to have GC-rich mtDNA (53%; Fricova et al., 2010).
Diplonemids
One of the earliest discoveries of GC-rich mtDNA came from the cox1 sequence of the euglenozoan Diplonema papillatum, a unicellular phagotrophic marine flagellate (Maslov et al., 1999). Further sequencing of mtDNA from this species has confirmed that its mitochondrial genome is enriched in G and C (∼55%; Vlcek et al., 2011). Other investigated members of the genus have AT-rich mtDNAs (Kiethega et al., 2011).
Organelle Genome Architecture and GC Content
The available GC-rich organelle genomes (Figures 1 and 2) vary greatly in size, gene content, and coding density (Table 1). For instance, the mtDNA of P. capuana is small and compact (13 kb, >80% coding, and no introns; Smith and Lee, 2008) whereas that of S. moellendorffii is large and distended (250 kb, >80% non-coding, and 37 introns; Hecht et al., 2011). There are, however, several reoccurring architectural themes among GC-biased organelle genomes (Table 1).
In the mtDNAs of the chlorophyceans L. culleus and P. capuana and the yeast C. subhashii, a high GC content is partnered with a linear genome conformation and, for the latter two species, distinct telomeric structures (Smith and Lee, 2008; Borza et al., 2009; Fricova et al., 2010). GC-rich mtDNAs are sometimes fragmented into multiple chromosomes, as seen in Oogamochlamys algae and the euglenozoan D. papillatum (Borza et al., 2009; Vlcek et al., 2011); these same taxa, along with P. capuana and S. moellendorffii, also contain fragmented and/or trans-spliced mtDNA genes (Kiethega et al., 2011).
For some species, a high organelle GC content is associated with a small number of tRNA-coding regions: P. capuana and S. moellendorffii have the most reduced mitochondrial tRNA-coding suites observed from the Archaeplastida: 1 and no tRNAs, respectively. A low tRNA content is also found in the Selaginella plastid genome (Tsuji et al., 2007; Smith, 2009) and the mtDNAs of D. papillatum and zoanthid corals (Sinniger et al., 2007; Vlcek et al., 2011).
In certain cases, organelle genome GC richness is allied with high levels of post-transcriptional editing, particularly cytosine-to-uracil changes. Hundreds of C-to-U editing sites have been identified in the GC-biased mitochondrial and plastid genomes of Selaginella species (Tsuji et al., 2007; Smith, 2009; Hecht et al., 2011). And for land plants as a whole there is a positive relationship between organelle GC content and the abundance of C-to-U editing sites (Jobson and Qiu, 2008). In the GC-rich mtDNA of D. papillatum, some mitochondrial transcripts experience U-insertion-type RNA editing (Kiethega et al., 2011). Given that organelle RNA editing tends to be a uracil-enriching process, it may turn out that some GC-rich mtDNAs and ptDNAs, once all of their edited sites are uncovered, have AT-rich transcriptomes.
What’s Causing Organelle Genome GC Enrichment?
Examining the distribution of GC among different regions within a genome, different genomes within a cell, and different species within a group can give insights into the forces that govern GC composition. The available GC-rich organelle DNAs come from an assortment of taxa belonging to disparate lineages (Figure 3). In some cases, the GC-bias is found in both the mitochondrial and plastid compartments of a species and in multiple species within a group, as observed for the spikemoss Selaginella and the trebouxiophyte Coccomyxa (Figure 3). In other examples, the GC-bias is restricted to either the mtDNA or ptDNA and/or is present in only a single species within the group, as seen for the green algae Polytomella and Picocystis (Figure 3). This variation in the presence and absence of GC-rich organelle DNA indicates that the processes biasing mitochondrial and plastid genomes in GC likely differ between lineages.
For many GC-rich organelle genomes, particularly those of green algae and the coral Mesozoanthus fossii (Figures 1 and 2), the concentration of GC is highest at silent sites, such as non-coding and synonymous sites (Table 1). This implies that in some organelle systems there is a non-adaptive underpinning to the GC-bias (Kimura, 1983). Two non-adaptive processes that can influence nucleotide landscape are biased mutation pressure and biased gene conversion. In most species, mtDNA and ptDNA mutation pressure seems to be skewed toward A and T (discussed above). Gene conversion, however, favors G and C in most genomes in which it has been studied (Mancera et al., 2008; Duret and Galtier, 2009; Muyle et al., 2011), with the exception of the tobacco ptDNA where it is AT biased (Khakhlova and Bock, 2006). Genomic regions with high rates of recombination undergo more gene conversion events than those with low recombination rates. In this context, it is noteworthy that some GC-rich organelle genomes are highly recombinogenic (Dieckmann and Gandy, 1987; Smith and Lee, 2008; Borza et al., 2009; Hecht et al., 2011), which may be a sign of a GC-biased conversion process. Moreover, in a variety of organelle genomes, including AT-rich ones, repeat elements (sequences that presumably undergo high levels of recombination) often have inflated GC contents (de Zamaroczy and Bernardi, 1986; Nedelcu and Lee, 1998). DNA methylation can also influence GC content – by promoting cytosine deamination events – but GC-rich mtDNAs and ptDNAs, like those of S. moellendorffii, do not have lower levels of methylation than those that are AT-rich (Zemach et al., 2010).
In other organelle DNAs, like those from land plants, the GC content is highest at functionally constrained sites, such as first and second codon positions (Table 1), suggesting that the GC-bias is the product of natural selection. Complicating this interpretation, however, is the fact that many of the cytosines residues at the non-silent sites from these taxa are post-transcriptionally edited to uracil (Jobson and Qiu, 2008; Smith, 2009; Hecht et al., 2011). Other adaptive hypotheses for a high GC composition include increased DNA thermo stability and UV tolerance. But these arguments seem implausible given that many GC-rich organelle DNAs come from species living in extremely cold habitats (e.g., C. subellipsoidea originates from Marble Point Antarctica) or environments with little UV light (e.g., the pathogenic yeast C. subhashii; Table 1).
For some species there is a correlation between lifestyle and organelle DNA GC content. Within the Coccomyxa genus, the three taxa known to have GC-rich organelle genomes are non-lichenized, free-living species, whereas all investigated symbiont Coccomyxa species have AT-rich organelle DNA (Smith et al., 2011). In the case of Candida, a parasitic lifestyle correlates with extreme organelle genome nucleotide compositions: the mtDNA of C. subhashii has one of the highest GC contents observed from the opisthokonts (Fricova et al., 2010) and that of its close relative Candida castellii is remarkably AT-rich (87%; Figure 1; Bouchier et al., 2009). The high mitochondrial GC contents of certain animals, such as frogs and fish (Figures 1 and 2), may be a reflection of them having low metabolic rates and consequently reduced mtDNA damage from oxygen free radicals (Martin, 1995).
Although data are limited, organelle DNA GC enrichment does not appear to be associated with nuclear DNA GC enrichment: C. subellipsoidea and S. moellendorffii have had their nuclear genomes completely sequenced (Banks et al., 2011; Blanc et al., 2012), revealing overall GC contents of ∼50%, which is unremarkable relative to the nuclear genomes of other green plants. However, the availability of these nuclear sequences will allow researchers to explore the full complement of nuclear-encoded mitochondrial- and plastid-targeted proteins, which should give insight into the biochemical and metabolic processes occurring within these organelles. Already, it has been revealed that the C. subellipsoidea nuclear genome lacks the plastid-targeted gene for the photosystem 1 (PSI) reaction center subunit N (psaN), which codes for a protein involved in the docking of plastocyanin. Interestingly, psaN-lacking strains of Arabidopsis, although maintaining a functional PSI complex, have reduced rates of electron transfer from plastocyanin to PSI (Haldrup et al., 1999). It is hypothesized that for C. subellipsoidea the unique loss of psaN may lead to reduced ROS formation (Blanc et al., 2012), which could help explain the high GC content of its organelle DNAs.
Concluding Remarks
Organelle genomes are models for studying the evolution of genome size and structure (Nosek and Tomáska, 2003; Lynch et al., 2006). Now, with the discovery of GC-rich mtDNAs and ptDNAs, they have established themselves as excellent systems for exploring the origins of nucleotide landscape. The presence of GC-biased organelle DNA in key research lineages, like Selaginella, Candida, and Chlamydomonadalean algae, and the availability of complete organelle and nuclear genome sequences from these groups provide promising avenues for future studies on nucleotide composition. I predict that in the years to come GC-rich organelle DNAs will help further our understanding of nucleotide composition and its relationship with other aspects of genome architecture.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
David Roy Smith is supported by postdoctoral fellowships from the Natural Sciences and Engineering Research Council of Canada and the Izaak Walton Killam Trusts.
References
- Ames B. N., Shigenaga M. K., Hagen T. M. (1995). Mitochondrial decay in aging. Biochim. Biophys. Acta 1271, 165–170 10.1016/0925-4439(95)00024-X [DOI] [PubMed] [Google Scholar]
- Archibald J. M. (2009). The puzzle of plastid evolution. Curr. Biol. 19, R81–R88 10.1016/j.cub.2008.11.067 [DOI] [PubMed] [Google Scholar]
- Asada K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141, 391–396 10.1104/pp.106.082040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks J. A., Nishiyama T., Hasebe M., Bowman J. L., Gribskov M., dePamphilis C., Albert V. A., Aono N., Aoyama T., Ambrose B. A., Ashton N. W., Axtell M. J., Barker E., Barker M. S., Bennetzen J. L., Bonawitz N. D., Chapple C., Cheng C., Correa L. G., Dacre M., DeBarry J., Dreyer I., Elias M., Engstrom E. M., Estelle M., Feng L., Finet C., Floyd S. K., Frommer W. B., Fujita T., Gramzow L., Gutensohn M., Harholt J., Hattori M., Heyl A., Hirai T., Hiwatashi Y., Ishikawa M., Iwata M., Karol K. G., Koehler B., Kolukisaoglu U., Kubo M., Kurata T., Lalonde S., Li K., Li Y., Litt A., Lyons E., Manning G., Maruyama T., Michael T. P., Mikami K., Miyazaki S., Morinaga S., Murata T., Mueller-Roeber B., Nelson D. R., Obara M., Oguri Y., Olmstead R. G., Onodera N., Petersen B. L., Pils B., Prigge M., Rensing S. A., Riaño-Pachón D. M., Roberts A. W., Sato Y., Scheller H. V., Schulz B., Schulz C., Shakirov E. V., Shibagaki N., Shinohara N., Shippen D. E., Sørensen I., Sotooka R., Sugimoto N., Sugita M., Sumikawa N., Tanurdzic M., Theissen G., Ulvskov P., Wakazuki S., Weng J. K., Willats W. W., Wipf D., Wolf P. G., Yang L., Zimmer A. D., Zhu Q., Mitros T., Hellsten U., Loqué D., Otillar R., Salamov A., Schmutz J., Shapiro H., Lindquist E., Lucas S., Rokhsar D., Grigoriev I. V. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963 10.1126/science.1203810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendich A. J. (2010). Mitochondrial DNA, chloroplast DNA and the origins of development in eukaryotic organisms. Biol. Direct 5, 42. 10.1186/1745-6150-5-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky C. W. (2001). The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu. Rev. Genet. 35, 125–148 10.1146/annurev.genet.35.102401.090231 [DOI] [PubMed] [Google Scholar]
- Blanc G., Agarkova I., Grimwood J., Kuo A., Brueggeman A., Dunigan D. D., Gurnon J., Ladunga I., Lindquist E., Lucas S., Pangilinan J., Pröschold T., Salamov A., Schmutz J., Weeks D., Yamada T., Lomsadze A., Borodovsky M., Claverie J. M., Grigoriev I. V., Van Etten J. L. (2012). The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 13, R39. 10.1186/gb-2012-13-5-r39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borza T., Redmond E. K., Laflamme M., Lee R. W. (2009). Mitochondrial DNA in the Oogamochlamys clade (Chlorophyceae): high GC content and unique genome architecture for green algae. J. Phycol. 45, 1323–1334 10.1111/j.1529-8817.2009.00753.x [DOI] [PubMed] [Google Scholar]
- Bouchier C., Ma L., Créno S., Dujon B., Fairhead C. (2009). Complete mitochondrial genome sequences of three Nakaseomyces species reveal invasion by palindromic GC clusters and considerable size expansion. FEMS Yeast Res. 9, 1283–1292 10.1111/j.1567-1364.2009.00551.x [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D. (2010). Elements of Evolutionary Genetics. Greenwood Village: Roberts and Company Publishers [Google Scholar]
- Clare E. L., Kerr K. C. R., Königslöw T. E., Wilson J. J., Hebert P. D. N. (2008). Diagnosing mitochondrial DNA diversity: applications of a sentinel gene approach. J. Mol. Evol. 66, 362–367 10.1007/s00239-008-9088-2 [DOI] [PubMed] [Google Scholar]
- Covello P. S., Gray M. W. (1989). RNA editing in plant mitochondria. Nature 341, 662–666 10.1038/341662a0 [DOI] [PubMed] [Google Scholar]
- Crawford A. J., Lips K. R., Bermingham E. (2010). Epidemic disease decimates amphibian abundance, species diversity, and evolutionary history in the highlands of central Panama. Proc. Natl. Acad. Sci. U.S.A. 107, 13777–13782 10.1073/pnas.0914115107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot G. A., During H. J., Maas J. W., Schneider H., Vogel J. C., Erkens R. H. J. (2011). Use of rbcL and trnL-F as a two-locus DNA barcode for identification of NW-European ferns: an ecological perspective. PLoS ONE 6, e16371. 10.1371/journal.pone.0016371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. (1986). The GC clusters of the mitochondrial genome of yeast and their evolutionary origin. Gene 41, 1–22 10.1016/0378-1119(86)90262-3 [DOI] [PubMed] [Google Scholar]
- Dieckmann C. L., Gandy B. (1987). Preferential recombination between GC clusters in yeast mitochondrial DNA. EMBO J. 6, 4197–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin G., Daoud H., Xia J. (2008). Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 49, 827–831 10.1016/j.ympev.2008.09.009 [DOI] [PubMed] [Google Scholar]
- Duret L., Galtier N. (2009). Biased gene conversion and the evolution of mammalian genomic landscapes. Annu. Rev. Genomics Hum. Genet. 10, 285–311 10.1146/annurev-genom-082908-150001 [DOI] [PubMed] [Google Scholar]
- Ebihara A., Nitta J. H., Ito M. (2010). Molecular species identification with rich floristic sampling: DNA barcoding the pteridophyte flora of Japan. PLoS ONE 5, e15136. 10.1371/journal.pone.0015136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith J. J., Pollock D. D. (2003). Likelihood analysis of asymmetrical mutation bias gradients in vertebrate mitochondrial genomes. Genetics 165, 735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. G., Hickey D. A. (1999). Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J. Mol. Evol. 48, 284–290 10.1007/PL00006471 [DOI] [PubMed] [Google Scholar]
- Frank A. C., Lobry J. R. (1999). Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene 238, 65–77 10.1016/S0378-1119(99)00297-8 [DOI] [PubMed] [Google Scholar]
- Fricova D., Valach M., Farkas Z., Pfeiffer I., Kucsera J., Tomaska L., Nosek J. (2010). The mitochondrial genome of the pathogenic yeast Candida subhashii: GC-rich linear DNA with a protein covalently attached to the 5′ termini. Microbiology 156, 2153–2163 10.1099/mic.0.038646-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson A. (2005). A Comprehensive analysis of mammalian mitochondrial genome base composition and improved phylogenetic methods. Mol. Biol. Evol. 22, 251–264 10.1093/molbev/msi012 [DOI] [PubMed] [Google Scholar]
- Gontcharov A. A., Melkonian M. (2010). Molecular phylogeny and revision of the genus Netrium (Zygnematophyceae, Streptophyta): Nucleotaenium gen. nov. J. Phycol. 46, 346–362 10.1111/j.1529-8817.2010.00814.x [DOI] [Google Scholar]
- Gray M. W., Burger G., Lang B. F. (1999). Mitochondrial evolution. Science 283, 1476–1481 10.1126/science.283.5407.1476 [DOI] [PubMed] [Google Scholar]
- Gray M. W., Lang B. F., Burger G. (2004). Mitochondria of protists. Annu. Rev. Genet. 38, 477–524 10.1146/annurev.genet.37.110801.142526 [DOI] [PubMed] [Google Scholar]
- Green B. R. (2011). Chloroplast genomes of photosynthetic eukaryotes. Plant J. 66, 34–44 10.1111/j.1365-313X.2011.04541.x [DOI] [PubMed] [Google Scholar]
- Grewe F., Viehoever P., Weisshaar B., Knoop V. (2009). A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 37, 5093–5104 10.1093/nar/gkp532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldrup A., Naver H., Scheller H. V. (1999). The interaction between plastocyanin and photosystem I is inefficient in transgenic Arabidopsis plants lacking the PSI-N subunit of photosystem. Plant J. 17, 689–698 10.1046/j.1365-313X.1999.00419.x [DOI] [PubMed] [Google Scholar]
- Handa H. (2008). Linear plasmids in plant mitochondria: peaceful coexistences or malicious invasions. Mitochondrion 8, 15–25 10.1016/j.mito.2007.10.002 [DOI] [PubMed] [Google Scholar]
- Hecht J., Grewe F., Knoop V. (2011). Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol. Evol. 3, 344–358 10.1093/gbe/evr027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro N. B., Miya M., Inoue J. G., Nishida M. (2005). Sundasalanx (Sundasalangidae) is a progenetic clupeiform, not a closely-related group of salangids (Osmeriformes): mitogenomic evidence. J. Fish Biol. 67, 561–569 10.1111/j.0022-1112.2005.00746.x [DOI] [Google Scholar]
- Jobson R. W., Qiu Y. L. (2008). Did RNA editing in plant organellar genomes originate under natural selection or through genetic drift? Biol. Direct 3, 43. 10.1186/1745-6150-3-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T. H., Bhushan V. (1986). Silent nucleotide substitutions and G + C content of some mitochondrial and bacterial genes. J. Mol. Evol. 24, 39–44 10.1007/BF02099949 [DOI] [PubMed] [Google Scholar]
- Jukes T. H., Osawa S. (1993). Evolutionary changes in the genetic code. Comp. Biochem. Physiol. B 106, 489–494 10.1016/0305-0491(93)90122-L [DOI] [PubMed] [Google Scholar]
- Keeling P. J. (2010). The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 729–748 10.1098/rstb.2009.0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr K. C. R., Stoeckle M. Y., Dove C. J., Weigt L. A., Francis C. M., Hebert P. D. N. (2007). Comprehensive DNA barcode coverage of North American birds. Mol. Ecol. Notes 7, 535–543 10.1111/j.1471-8286.2007.01670.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhlova O., Bock R. (2006). Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 46, 85–94 10.1111/j.1365-313X.2006.02673.x [DOI] [PubMed] [Google Scholar]
- Kiethega G. N., Turcotte M., Burger G. (2011). Evolutionarily conserved cox1 trans-splicing without cis-motifs. Mol. Biol. Evol. 28, 2425–2428 10.1093/molbev/msr075 [DOI] [PubMed] [Google Scholar]
- Kimura M. (1983). The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press [Google Scholar]
- Kleine T., Maier U. G., Leister D. (2009). DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu. Rev. Plant Biol. 60, 115–138 10.1146/annurev.arplant.043008.092119 [DOI] [PubMed] [Google Scholar]
- Kusumi J., Tachida H. (2005). Compositional properties of green-plant plastid genomes. J. Mol. Evol. 60, 417–425 10.1007/s00239-004-0086-8 [DOI] [PubMed] [Google Scholar]
- Lang B. F., Gray M. W., Burger G. (1999). Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 33, 351–397 10.1146/annurev.genet.33.1.351 [DOI] [PubMed] [Google Scholar]
- Liu P., Demple B. (2010). DNA repair in mitochondria: much more than we thought? Environ. Mol. Mutagen. 51, 417–426 [DOI] [PubMed] [Google Scholar]
- Lynch M. (2007). The Origins of Genome Architecture. Massachusetts: Sinauer Associates, Inc [Google Scholar]
- Lynch M., Koskella B., Schaack S. (2006). Mutation pressure and the evolution of organelle genomic architecture. Science 311, 1727–1730 10.1126/science.1118884 [DOI] [PubMed] [Google Scholar]
- Malek O., Knoop V. (1998). Trans-splicing group II introns in plant mitochondria: the complete set of cis-arranged homologs in ferns, fern allies, and a hornwort. RNA 4, 1599–1609 10.1017/S1355838298981262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M. (2008). High-resolution mapping of meiotic crossovers and noncrossovers in yeast. Nature 454, 479–485 10.1038/nature07135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A. P. (1995). Metabolic rate and directional nucleotide substitution in animal mitochondrial DNA. Mol. Biol. Evol. 12, 1124–1131 [DOI] [PubMed] [Google Scholar]
- Maslov D. A., Yasuhira S., Simpson L. (1999). Phylogenetic affinities of Diplonema within the Euglenozoa as inferred from the SSU rRNA gene and partial COI protein sequences. Protist 150, 33–42 10.1016/S1434-4610(99)70007-6 [DOI] [PubMed] [Google Scholar]
- McCutcheon J. P., Moran N. A. (2010). Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol. Evol. 2, 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min X. J., Hickey D. A. (2007). DNA barcodes provide a quick preview of mitochondrial genome composition. PLoS ONE 2, e325. 10.1371/journal.pone.0000325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya M., Nishida M. (2000). Use of mitogenomic information in teleostean molecular phylogenetics: a tree-based exploration under the maximum-parsimony optimality criterion. Mol. Phylogenet. Evol. 17, 437–455 10.1006/mpev.2000.0839 [DOI] [PubMed] [Google Scholar]
- Moore C. E., Archibald J. M. (2009). Nucleomorph genomes. Annu. Rev. Genet. 43, 251–264 10.1146/annurev-genet-102108-134809 [DOI] [PubMed] [Google Scholar]
- Morton B. R. (1998). Selection on the codon bias of chloroplast and cyanelle genes in different plant and algal lineages. J. Mol. Evol. 46, 449–459 10.1007/PL00006325 [DOI] [PubMed] [Google Scholar]
- Muller H. J. (1964). The relation of recombination to mutational advance. Mutat. Res. 106, 2–9 [DOI] [PubMed] [Google Scholar]
- Murphy M. P. (2009). How mitochondria produce reactive oxygen species. Biochem. J. 417, 1–13 10.1042/BJ20081386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyle A., Serres-Giardi L., Ressayre A., Escobar J., Glémin S. (2011). GC-biased gene conversion and selection affect GC content in the Oryza genus (rice). Mol. Biol. Evol. 28, 2695–2706 10.1093/molbev/msr104 [DOI] [PubMed] [Google Scholar]
- Nedelcu A. M., Lee R. W. (1998). Short repetitive sequences in green algal mitochondrial genomes: potential roles in mitochondrial genome evolution. Mol. Biol. Evol. 15, 690–701 10.1093/oxfordjournals.molbev.a025972 [DOI] [PubMed] [Google Scholar]
- Nosek J., Tomáska L. (2003). Mitochondrial genome diversity: evolution of the molecular architecture and replication strategy. Curr. Genet. 44, 73–84 10.1007/s00294-003-0426-z [DOI] [PubMed] [Google Scholar]
- Nowack E. C. M., Melkonian M., Glöckner G. (2008). Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol. 18, 410–418 10.1016/j.cub.2008.02.051 [DOI] [PubMed] [Google Scholar]
- Pallen M. J., Wren B. W. (2007). Bacterial pathogenomics. Nature 449, 835–842 10.1038/nature06248 [DOI] [PubMed] [Google Scholar]
- Palmer J. D. (1985). Comparative organization of chloroplast genomes. Annu. Rev. Genet. 19, 325–354 10.1146/annurev.ge.19.120185.001545 [DOI] [PubMed] [Google Scholar]
- Palmer J. D. (2003). The symbiotic birth and spread of plastids: how many times and whodunit? J. Phycol. 39, 1–9 10.1111/j.0022-3646.2003.03906001_2.x [DOI] [Google Scholar]
- Piganeau G., Gardner M., Eyre-Walker A. (2004). A broad survey of recombination in animal mitochondria. Mol. Biol. Evol. 21, 2319–2325 10.1093/molbev/msh244 [DOI] [PubMed] [Google Scholar]
- Rand D. M. (2001). The units of selection on mitochondrial DNA. Annu. Rev. Ecol. Syst. 32, 415–448 10.1146/annurev.ecolsys.32.081501.114109 [DOI] [Google Scholar]
- Rindi F., Mikhailyuk T. I., Sluiman H. J., Friedl T., López-Bautista J. M. (2011). Phylogenetic relationships in Interfilum and Klebsormidium (Klebsormidiophyceae, Streptophyta). Mol. Phylogenet. Evol. 58, 218–231 10.1016/j.ympev.2010.11.030 [DOI] [PubMed] [Google Scholar]
- Rocha E. P., Danchin A. (2002). Base composition bias might result from competition for metabolic resources. Trends Genet. 34, 377–378 [DOI] [PubMed] [Google Scholar]
- Saitoh K., Miya M., Inoue J. G., Ishiguro N. B., Nishida M. (2003). Mitochondrial genomics of ostariophysan fishes: perspectives on phylogeny and biogeography. J. Mol. Evol. 56, 464–472 10.1007/s00239-002-2417-y [DOI] [PubMed] [Google Scholar]
- Shokolenko I., Venediktova N., Bochkareva A., Wilson G. L., Alexeyev M. F. (2009). Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 37, 2539–2548 10.1093/nar/gkp100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L., Thiemann O. H. (1995). Sense from nonsense: RNA editing in mitochondria of kinetoplastid protozoa and slime molds. Cell 81, 837–840 10.1016/0092-8674(95)90003-9 [DOI] [PubMed] [Google Scholar]
- Sinniger F., Chevaldonné P., Pawlowski J. (2007). Mitochondrial genome of Savalia savaglia (Cnidaria, Hexacorallia) and early metazoan phylogeny. J. Mol. Evol. 64, 196–203 10.1007/s00239-006-0015-0 [DOI] [PubMed] [Google Scholar]
- Smith D. R. (2009). Unparalleled GC content in the plastid DNA of Selaginella. Plant Mol. Biol. 71, 627–639 10.1007/s11103-009-9545-3 [DOI] [PubMed] [Google Scholar]
- Smith D. R., Burki F., Yamada T., Grimwood J., Grigoriev I. V., Van Etten J. L., Keeling P. J. (2011). The GC-rich mitochondrial and plastid genomes of the green alga Coccomyxa give insight into the evolution of organelle DNA nucleotide landscape. PLoS ONE 6, e23624. 10.1371/journal.pone.0023624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. R., Hua J., Lee R. W. (2010). Evolution of linear mitochondrial DNA in three known lineages of Polytomella. Curr. Genet. 56, 427–438 10.1007/s00294-010-0311-5 [DOI] [PubMed] [Google Scholar]
- Smith D. R., Lee R. W. (2008). Mitochondrial genome of the colorless green alga Polytomella capuana: a linear molecule with an unprecedented GC content. Mol. Biol. Evol. 25, 487–496 10.1093/molbev/msm245 [DOI] [PubMed] [Google Scholar]
- Tsuji S., Ueda K., Nishiyama T., Hasebe M., Yoshikawa S., Konagaya A., Nishiuchi T., Yamaguchi K. (2007). The chloroplast genome from a lycophyte (microphyllophyte), Selaginella uncinata, has a unique inversion, transpositions and many gene losses. J. Plant Res. 120, 281–290 10.1007/s10265-006-0055-y [DOI] [PubMed] [Google Scholar]
- Vlcek C., Marande W., Teijeiro S., Lukes J., Burger G. (2011). Systematically fragmented genes in a multipartite mitochondrial genome. Nucleic Acids Res. 39, 979–988 10.1093/nar/gkq883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. H. (1991). Mammalian DNA replication: mutation biases and the mutation rate. J. Theor. Biol. 149, 441–451 10.1016/S0022-5193(05)80092-X [DOI] [PubMed] [Google Scholar]
- Wu M., Zhang L., Li G., Jiang D., Ghabrial S. A. (2010). Genome characterization of a debilitation-associated mitovirus infecting the phytopathogenic fungus Botrytis cinerea. Virology 406, 117–126 10.1016/j.virol.2010.07.010 [DOI] [PubMed] [Google Scholar]
- Zemach A., McDaniel I. E., Silva P., Zilberman D. (2010). Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916–919 10.1126/science.1186366 [DOI] [PubMed] [Google Scholar]