Vertebrate hematopoiesis is a richly complex developmental system in which many transcription factors have essential and nonredundant roles (1). Some factors act as prominent controllers of differentiation in specific cell lineages, others are needed for stem cell generation, and others are needed for both. In this system, precise levels of one transcription factor relative to another in the same cell can control the direction of cell lineage choices, proliferation vs. apoptosis, lineage-specific malignant transformation, and the timing and sites of stem cell generation (2–9). Much has been learned in the past decade about the way the prominent transcription factors associated with particular hematopoietic cell types act on their target genes to execute lineage-specific differentiation programs. But lineage choice itself, the way progeny of the same pluripotent precursor adopt diverse fates, is less well understood. Its mechanism ultimately depends on the regulation of the key transcription factor genes themselves. There is very little information about the cis- and trans-acting elements that control expression of most of these genes. Furthermore, the genes encoding the relevant transcription factors are often large, and the sequences needed for correct expression in transgenic mice can be dispersed over several hundred kilobases (10), making the full regulatory system hard to define. It would be extremely valuable to devise a shortcut to help map the regulatory regions for such genes. In a paper in this issue of PNAS (11), the gene encoding an essential hematopoietic transcription factor, SCL, is used to illustrate a strategy that may provide such a shortcut.
If the pufferfish genomic DNA could be assayed for enhancer function by using a visible reporter, it would be possible to take advantage of the transparency of the living zebrafish embryo.
To map regulatory elements, two things are needed: first, the assurance that the essential regulatory sequences are actually present in the DNA to be tested, and second, an assay system that allows their functional activity to be read out. Even the first of these conditions is hard to satisfy for large genes with complex patterns of expression, because enhancer modules (12) can be dispersed among introns and sometimes distant flanking regions. The second condition can also be difficult to meet for regulatory elements of mammalian genes that act in multiple embryonic and adult developmental contexts, because cell lines cannot duplicate the developmental shifts in expression, and mammalian transgenesis is costly and slow. Barton et al. (11) suggest that, by focusing on teleost fish as sources of both genes and assay systems, both sets of conditions can be met.
The first element of this strategy is to clone the gene of interest from the pufferfish (Fugu rubripes), which has a genome about eight times smaller than that of a mammal. Nevertheless, coding sequences and intron/exon structures appear to be conserved: thus, essential regulatory elements may also be squeezed much closer to the genes they regulate than in mammalian genomes (13–17). At least some of the Fugu regulatory sequences remain similar enough to those in mammals to be recognizable by sequence and/or by function in transgenic rats or mice (16, 18). Of course, only regulatory elements that serve similar functions in fish and mammals are likely to be conserved, but studies of blood development in frogs and fish generally suggest that key hematopoietic transcription factor genes are used in encouragingly similar ways (19–22). Recent studies in cartilaginous fish and lampreys further suggest that some detailed aspects of transcription factor use in blood development may be shared among all jawed vertebrates (23, 24).
Whereas transgenic rodents can often read out the regulatory information in pufferfish DNA, they are too expensive, variable, and slow to map the borders of such elements purely on the basis of function. Pufferfish themselves are not yet adapted to gene transfer, or even to experimental embryology. Therefore, Barton et al. have taken advantage of the strong experimental embryology of another teleost, the zebrafish (Danio rerio). For example, the zebrafish GATA-1 5′-flanking region is able to drive apparently correct expression of a green fluorescent protein (GFP) transgene in transgenic zebrafish erythrocytes (25). By injecting pufferfish genomic cosmids into zebrafish zygotes and testing their expression in the resulting embryos by in situ hybridization, a rapid determination can be made of the location of sequences controlling expression in each of several embryonic domains at once (11). Thus, noncoding DNA regions can be scanned relatively easily for the sequences that are necessary or sufficient to drive a wide range of tissue-specific expression patterns. At a minimum, this approach should define most cis-regulatory elements of these pufferfish genes that are mutually compatible in the two kinds of teleost fish.
The gene used to illustrate this strategy is the SCL (Tal1) gene, which encodes a hematopoietic transcription factor with diverse developmental roles and complex regulation. SCL controls generation of hematopoietic stem cells of adult and fetal types, and both primitive hemangioblast and endothelial cell differentiation (26). SCL-deficient mice have defects in endothelial morphogenesis as well as a complete block in both primitive and definitive hematopoiesis. In later stages of mammalian hematopoietic differentiation, SCL overexpression drives erythromyeloid precursors toward erythroid and megakaryocytic fates at the expense of myeloid fates. In addition, SCL has several major expression sites in the central nervous system where its roles are less known.
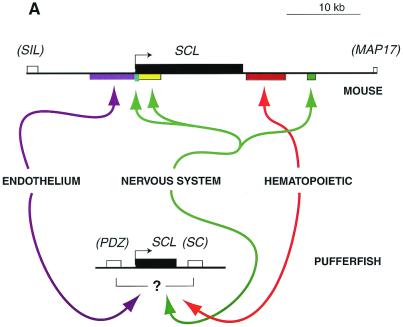
This is a particularly good gene to use in demonstrating the pufferfish/zebrafish strategy for several reasons. First, all its sites of expression appear to be conserved between mammals and teleost fish (11, 27). Second, the regulation of the mammalian SCL gene is probably better understood than that of most other transcription factor genes. Previous studies by the same group have defined multiple discrete enhancer regions, both upstream and downstream of the murine gene, that drive expression in endothelial, central nervous system, and hematopoietic cell types (27–31). Collectively, these enhancers are spread over 30 kb of mouse DNA. Using the zebrafish assay for pufferfish regulatory sequences needed in these tissues, Barton et al. confirm that the full set of positive elements is contained within ≈10 kb in the Fugu genome (11). Although the mapping of specific functions to particular subregions has not been done yet, the results show that this kind of information should be easy to obtain using the same strategy.
This approach has a number of possible extensions. Repeating the expression analysis in various mutant zebrafish embryos could potentially identify genes that are needed to act “upstream” of particular regulatory elements. The responses could further help to reveal functional boundaries between enhancer modules that depend on different upstream regulators. Also, if the pufferfish genomic DNA were assayed for enhancer function by using a GFP reporter (25), it would be possible to take advantage of the transparency of the living zebrafish embryos and to track the regulation of expression patterns over time. With the kind of rapid, multidomain expression analysis that could be possible in this system, the pufferfish enhancer sequences might be used as a guide to seek the mammalian counterparts of fish regulatory elements in the first place. The initial scan for pufferfish regulatory elements should identify specific, limited DNA sequences that can be tested for functional activity in transgenic mammals, then used to seek corresponding mammalian sequences. Sequence comparisons could locate conserved regions around the corresponding mammalian genes as likely starting points for functional analysis (17, 18, 31). Recognition of such features in long-range sequence comparisons is inefficient now, but likely to improve substantially with ongoing development of genomic informatics tools.
Is this, then, the new way to map complex regulatory regions for less well-studied mammalian genes? The answer will depend on case-by-case testing of a few critical assumptions. First is that the expression sites that are of interest in mammals (e.g., hematopoietic cells) will be broadly conserved—and recognizable enough—to be scored in fish embryos. This appears to be true for early-embryonic expression sites of the SCL gene, but it cannot be generalized. For example, hematopoietic cell types that can be distinguished only in mammals by cell surface markers will not be distinguished at all in the less-studied fish system. Furthermore, redeployment of transcriptional regulators to novel tissues is a common mechanism of evolutionary change (32, 33).
Another assumption is that the individual protein–DNA interactions that promote expression in conserved sites will themselves remain conserved across the large phylogenetic distance between fish and mammal. If the gene is regulated by multiple tissue-specific enhancer modules (12), it is also assumed that the mapping of particular functions to particular modules will be retained. But this need not be the case. Ironically, whereas expression of related genes is often used as evidence of homology of body parts across wide phylogenetic distances, only occasional studies have indicated whether genes that are expressed in corresponding body parts of different animals are actually using the same regulatory sequences and transcription factors. Precisely because of the combinatoriality of transcriptional regulation, there is considerable room for evolutionary drift in the importance of particular sites or whole modules: for example, the Ig heavy chain enhancer of channel catfish has a completely different structure from those of mammals, with apparently enhanced roles for octamer factors and reduced roles for Ets family factors (34). Thus, whereas it is likely that some regulatory relationships will be conserved, there could be considerable alteration in the relative prominence of the roles given to particular cis/trans interactions.
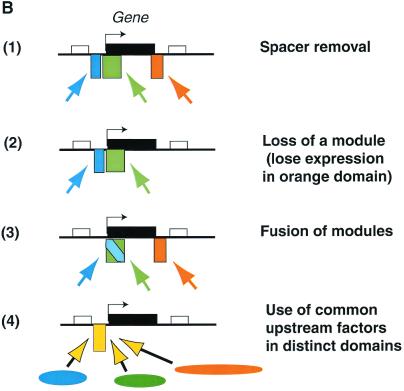
A final, still-untested assumption relates particularly to the small pufferfish genome. This is the assumption that the compression of the genome in this organism has been achieved without any radical simplification of gene regulatory mechanisms (Fig. 1B, 1), and that pufferfish genome organization is similar to the mammalian one, but minus the “junk.” Synteny between the pufferfish and mammalian genomes in certain regions supports this picture (14, 17). But a caveat is introduced by the new findings for SCL (11), where the pufferfish ortholog of the mammalian gene is flanked by completely different neighboring genes than those in either mammalian or avian genomes. Something has changed in this genomic context that could split off a regulatory element: is it due simply to amniote/teleost divergence, or to the unusual events that created the compressed genome of Fugu? Here, our limited information about pufferfish development and gene expression is a serious problem. The problem is exacerbated by the lack of comparative genome organization information for other teleosts with more typical genome sizes, including the zebrafish.
Figure 1.
Genome compression in Fugu. (A) Comparison between the murine and pufferfish SCL genes. (B) Three hypothetical ways that compression of the genome could affect regulatory sequence organization: (1) shrinking spacer regions between tissue-specific modules (no simplification) vs. simplification by (2) deletion of selected modules, (3) fusion of modules, or (4) broadening the activity of an upstream regulator for use in different tissues. In both A and B, rectangles above the lines represent genes (exons and introns together), with the gene of interest shown in black. Colored rectangles below the lines show the locations of enhancer modules that drive expression in different tissues. Colored arrows indicate the tissue-specific transcription factors that engage these respective enhancer modules. In B, three hypothetical tissues, and the transcription factors and modules active in each tissue respectively, are coded as orange, blue, and green.
Simplification could involve changes in either cis-acting or trans-acting mechanisms. As diagrammed in Fig. 1B, 2 and 3, cis-acting elements could be changed by deletion of whole modules, leading to loss of a whole expression domain, or by compressing two distinct modules into one by intercalating the binding sites for two different sets of tissue-specific transcription factors. In the latter case, some globally active protein–DNA interactions within the module could collaborate with either set of tissue-specific regulators, so that less cis-regulatory sequence is needed overall. Simplification could also involve broadening the expression pattern of an upstream regulator that is more narrowly tissue-specific in higher vertebrates: i.e., in the pufferfish the same regulator acting on the same cis-element could drive expression in multiple tissue sites, and additional enhancer modules would become superfluous (Fig. 1B, 4). The corresponding protein–DNA interaction in the mammalian regulatory system would control expression in only a subset of the territories where it acts in the fish.
When these assumptions are tested, the answers could limit the straightforward applicability of Fugu gene regulatory element mapping to the mammalian context. But from another vantage point, such answers would go to the heart of the evolutionary changes that have occurred because the last common ancestor of mammals and these peculiar teleost fish. Any change in the expression domain of a gene as important as SCL should be a cause of evolutionary change in the features controlled by that gene. Any change in the expression pattern of regulators controlling SCL should have coordinate effects on other genes in the network of which SCL is a part. Thus, if pursued appropriately, even the “problems” that could arise in the application of this ambitious, cross-phylogenetic analysis of gene regulation should be scientifically illuminating. This approach offers a path to many evolutionary and molecular insights.
Footnotes
See companion article on page 6747.
References
- 1.Shivdasani R A, Orkin S H. Blood. 1996;87:4025–4039. [PubMed] [Google Scholar]
- 2.Cai Z L, de Bruijn M, Ma X Q, Dortland B, Luteijn T, Downing J R, Dzierzak E. Immunity. 2000;13:423–431. doi: 10.1016/s1074-7613(00)00042-x. [DOI] [PubMed] [Google Scholar]
- 3.Barton K, Nucifora G. BioEssays. 2000;22:214–218. doi: 10.1002/(SICI)1521-1878(200003)22:3<214::AID-BIES2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 4.Nerlov C, Graf T. Genes Dev. 1998;12:2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang P, Behre G, Pan J, Iwama A, Wara-aswapati N, Radomska H S, Auron P E, Tenen D G, Sun Z. Proc Natl Acad Sci USA. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rekhtman N, Radparvar F, Evans T, Skoultchi A. Genes Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sieweke M H, Graf T. Curr Opin Genet Dev. 1998;8:545–551. doi: 10.1016/s0959-437x(98)80009-9. [DOI] [PubMed] [Google Scholar]
- 8.Enver T, Greaves M. Cell. 1998;94:9–12. doi: 10.1016/s0092-8674(00)81215-5. [DOI] [PubMed] [Google Scholar]
- 9.Cross M A, Enver T. Curr Opin Genet Dev. 1997;7:609–613. doi: 10.1016/s0959-437x(97)80007-x. [DOI] [PubMed] [Google Scholar]
- 10.Lakshmanan G, Lieuw K H, Lim K C, Gu Y, Grosveld F, Engel J D, Karis A. Mol Cell Biol. 1999;19:1558–1568. doi: 10.1128/mcb.19.2.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barton L M, Göttgens B, Gering M, Gilbert J G R, Grafham D, Rogers J, Bentley D, Patient R, Green A R. Proc Natl Acad Sci USA. 2001;98:6747–6752. doi: 10.1073/pnas.101532998. . (First Published May 29, 2001; 10.1073/pnas.101532998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnone M I, Davidson E H. Development. 1997;124:1851–1864. doi: 10.1242/dev.124.10.1851. [DOI] [PubMed] [Google Scholar]
- 13.Aparicio S, Morrison A, Gould A, Gilthorpe J, Chaudhuri C, Rigby P, Krumlauf R, Brenner S. Proc Natl Acad Sci USA. 1995;92:1684–1688. doi: 10.1073/pnas.92.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elgar G, Sandford R, Aparicio S, Macrae A, Venkatesh B, Brenner S. Trends Genet. 1996;12:145–150. doi: 10.1016/0168-9525(96)10018-4. [DOI] [PubMed] [Google Scholar]
- 15.Baxendale S, Abdulla S, Elgar G, Buck D, Berks M, Micklem G, Durbin R, Bates G, Brenner S, Beck S. Nat Genet. 1995;10:67–76. doi: 10.1038/ng0595-67. [DOI] [PubMed] [Google Scholar]
- 16.Venkatesh B, Si-Hoe S L, Murphy D, Brenner S. Proc Natl Acad Sci USA. 1997;94:12462–12466. doi: 10.1073/pnas.94.23.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gellner K, Brenner S. Genome Res. 1999;9:251–258. [PMC free article] [PubMed] [Google Scholar]
- 18.Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Dev Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- 19.Tracey W D, Jr, Pepling M E, Horb M E, Thomsen G H, Gergen J P. Development. 1998;125:1371–1380. doi: 10.1242/dev.125.8.1371. [DOI] [PubMed] [Google Scholar]
- 20.Huber T L, Zon L I. Semin Immunol. 1998;10:103–109. doi: 10.1006/smim.1998.0111. [DOI] [PubMed] [Google Scholar]
- 21.Gering M, Rodaway A R F, Göttgens B, Patient R K, Green A R. EMBO J. 1998;17:4029–4045. doi: 10.1093/emboj/17.14.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen J D, Zapata A G. Immunol Rev. 1998;166:199–220. doi: 10.1111/j.1600-065x.1998.tb01264.x. [DOI] [PubMed] [Google Scholar]
- 23.Anderson M K, Sun X, Miracle A L, Litman G W, Rothenberg E V. Proc Natl Acad Sci USA. 2001;98:553–558. doi: 10.1073/pnas.021478998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shintani S, Terzic J, Saraga-Babic M, O'hUigin C, Tichy H, Klein J. Proc Natl Acad Sci USA. 2000;97:7417–7422. doi: 10.1073/pnas.110505597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long Q, Meng A, Wang H, Jessen J R, Farrell M J, Lin S. Development. 1997;124:4105–4111. doi: 10.1242/dev.124.20.4105. [DOI] [PubMed] [Google Scholar]
- 26.Begley C G, Green A R. Blood. 1999;93:2760–2770. [PubMed] [Google Scholar]
- 27.Sinclair A M, Göttgens B, Barton L M, Stanley M L, Pardanaud L, Klaine M, Gering M, Bahn S, Sanchez M-J, Bench A J, Fordham J L, Bockamp E-O, Green A R. Dev Biol. 1999;209:128–142. doi: 10.1006/dbio.1999.9236. [DOI] [PubMed] [Google Scholar]
- 28.Bockamp E-O, McLaughlin F, Göttgens B, Murrell A M, Elefanty A G, Green A R. J Biol Chem. 1997;272:8781–8790. doi: 10.1074/jbc.272.13.8781. [DOI] [PubMed] [Google Scholar]
- 29.Göttgens B, McLaughlin F, Bockamp E-O, Fordham J L, Begley C G, Kosmopoulos K, Elefanty A G, Green A R. Oncogene. 1997;15:2419–2418. doi: 10.1038/sj.onc.1201426. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez M-J, Göttgens B, Sinclair A M, Stanley M, Begley C G, Hunter S, Green A R. Development. 1999;126:3891–3904. doi: 10.1242/dev.126.17.3891. [DOI] [PubMed] [Google Scholar]
- 31.Göttgens B, Barton L M, Gilbert J G R, Bench A J, Sanchez M-J, Bahn S, Mistry S, Grafham D, McMurray A, Vaudin M, Amaya E, Bentley D R, Green A R. Nat Biotechnol. 2000;18:181–186. doi: 10.1038/72635. [DOI] [PubMed] [Google Scholar]
- 32.Wray G A, Abouheif E. Curr Opin Genet Dev. 1998;8:675–680. doi: 10.1016/s0959-437x(98)80036-1. [DOI] [PubMed] [Google Scholar]
- 33.Davidson E H. Genomic Regulatory Systems. San Diego: Academic; 2001. , ch. 4, 5. [Google Scholar]
- 34.Magor B G, Ross D A, Middleton D L, Warr G W. Immunogenetics. 1997;46:192–198. doi: 10.1007/s002510050261. [DOI] [PubMed] [Google Scholar]