Abstract
Crimean-Congo hemorrhagic fever (CCHF) is a zoonotic viral disease that is asymptomatic in infected livestock, but causes a serious threat to humans with a mortality rate up to 50%. Although the CCHF virus (CCHFV) is often transmitted by ticks, livestock-to-human and human-to-human transmission also occurs. In the current study, we focused on CCHF in the province of Isfahan, located in the center of Iran and deemed to be the second most infected province. Human and livestock sera and resident ticks in the livestock are collected from different regions of the province and analyzed with specific IgG ELISA and RT-PCR tests. Overall, 12% and 12.7% of studied human and livestock populations were IgG positive, respectively. The genome of CCHFV was detected in 9% of ticks resident in livestock involved in this survey. The CCHFV isolates from infected ticks were genetically examined. Nucleotide sequence of the S-segment revealed that the different isolates were closely related to each other, with nucleotide sequence identities higher than 98%. Phylogenetic analysis demonstrated that a variant isolate clustered with the Iraq strain. This high proportion of IgG-positive sera and nearly high proportion of infected ticks increases the risk of CCHF outbreaks in the province and probably posits a great danger to other provinces.
Key Words: CCHF in Isfahan, Molecular epidemiology, Serological epidemiology
Introduction
Crimean-Congo hemorrhagic fever (CCHF) is a tick-borne viral zoonosis with up to a 50% mortality rate in humans that occurs widely in Africa, Asia, and Eastern Europe (Hoogstraal 1979, Watts et al. 1988, Camicas et al. 1994). The disease is caused by CCHF virus (CCHFV), a segmented, negative-stranded RNA virus belonging to the family Bunyaviridae, genus Nairovirus (Donets et al. 1977, Martin et al. 1985, Drosten et al. 2002). The virus is transmitted to humans through the bite of Ixodid ticks (mainly of the genus Hyalomma) by contact with blood or tissues of infected or nosocomial livestock (Begum et al. 1970, Suleiman et al. 1980, van Eeden et al. 1985, Logan et al. 1989, Gonzalez et al. 1992, Chinikar et al. 2008). The main course of CCHF has been noted by authors as progressing through four distinct phases including incubation, prehemorrhagic, hemorrhagic, and convalescence (Swanepoel et al. 1987, 1989, Whitehouse et al. 2004, Ergonul 2006, Papa et al. 2006). Although CCHFV often causes fatal hemorrhagic manifestations in humans, there is no evidence of the disease in livestock.
With a high proportion of human confirmed CCHF cases, Isfahan, the central province of Iran, ranked the second most infected province (Chinikar et al. 2008). Similar to all other vector-borne diseases, the presence and persistence of zoonotic foci of CCHF infection depend on biological and ecological relationships between the CCHFV, ticks, and vertebrates, particularly cattle and sheep (Randolph and Rogers 2007). However, we hypothesize that the seroepidemiological survey in this endemic area will possibly reveal a prevalence of anti-CCHFV antibodies in people with high-risk professions and livestock. Therefore, we choose to perform a cross-sectional study in the high-risk places of the Isfahan province (e.g., slaughtering houses) as well as the human population in close contact with infected livestock (e.g., livestock breeders, abattoir workers) or with infected humans (e.g., health care workers). Since the genetic diversity of CCHFV in this geographic region is still unclear, we attempt to better understand the genetic relationships among the different CCHFV isolates, and to precisely determine the genotypes of circulating strains. In the current study, ticks are also collected from different high-risk regions of the province, and the detected CCHFV genome is genotyped on partial sequencing of the S-segment; these important data will subsequently help to implement more suitable procedures for the control and surveillance of CCHF in this province and adjacent provinces.
Materials and Methods
A cross-sectional study on human, livestock, and tick population was conducted in the spring and summer of 2008 in 8 townships of the Isfahan province, including Isfahan, Golpayegan, Kashan, Najaf Abad, Mobarakeh, Barkhovar and Meymeh, Khomeini Shahr, and Shahreza. Before sampling, a pretested self-administered questionnaire was used for data collection. The participants completed the questionnaire anonymously, and confidentiality was maintained. The questionnaire asked for information on age, sex, residential area, profession, clinical and epidemiological queries regarding the CCHF disease, some precautious cases in working on livestock, and history of tick contact. Similarly, a questionnaire was performed for livestock samples, consisting of information on species, sex, and age and tick infestation. Also, the collected ticks of the livestock were taken as a code, thus showing their correspondence to that host livestock.
Blood samples (10 mL each) were collected from the enrolled human and livestock subjects and immediately centrifuged at the local laboratory in a veterinary organization for serum taking. The sera were kept in −20°C and transmitted on ice to Arboviruses and Viral Hemorrhagic Fevers laboratory (National Ref. Lab) in Pasteur Institute of Iran. All laboratory tests including molecular and serological analyses were performed in biosafety level 2 under negative pressure, were previously validated (unpublished data), and are annually re-validated. The RT-PCR master mix solution was provided in a filtered clean chamber with positive pressure to avoid a false-positive result.
Serological assay
For IgG detection, ELISA plates were coated with mouse hyperimmune ascetic fluid diluted in 1×PBS and incubated overnight at 4°C. The native (produced in biosafety level 4 containment) or recombinant antigen (a recombinant nucleoprotein expressed in mammalian cells via the recombinant Semliki Forest alphavirus replicon), diluted in PBSTM (PBS containing 0.05% Tween 20 and 3% skim milk), was added to the plates, and the plates were incubated for 3 h at 37°C and extensively washed. Serum samples diluted in PBSTM were added, and the plates were incubated for 1 h at 37°C. After washing, the peroxidase-labeled anti-human or animal immunoglobulin diluted in PBSTM was added to each well, and the plates were incubated for 1 h at 37°C. The plates were then washed thrice with phosphate-buffered saline containing 0.5% Tween (PBST), which had also been used for washing the plates after each of the incubation periods. Finally, hydrogen peroxide and 3,3′,5,5′-tetramethylbenzedine were added, and the plates were incubated for 15 min at room temperature. The enzymatic reaction was stopped by the addition of sulfuric acid (4N). The plates were read by an ELISA reader at 450 nm. Positive and negative control sera were included in each test (Garcia et al. 2006, Chinikar et al. 2008). The sample with an OD higher than 0.15 was considered positive, and this is deducted of the OD of negative controls.
Molecular assay
Forty-four ticks were individually washed twice by PBS 1×(PBS, pH 7.4) and crushed with mortar and pestle in 200–300 μL of PBS1×. Then, Viral RNA was extracted from each tick by using the QIAamp Viral RNA Kit according to the manufacturer's instructions (QIAgen GmbH, Hilden, Germany). The extracted viral RNA was subsequently analyzed by Gel-Based and qualitative real-time RT-PCR with the One-Step RT-PCR Kit (QIAgen GmbH, Hilden, Germany) and the use of specific primers: F2 5′TGGACACCTTCACAAACTC3′ and R3 5′GACAATTCCCTACACC3′. These primers were selected to amplify a 536-bp fragment of a highly conserved region inside the S-segment of the CCHFV genome. The PCR was done in a total volume of 50 μl for 30 min at 50°C, 15 min at 95°C, and 40 cycles including 30 s at 95°C, 30 s at 50°C, 45 s at 72°C, and, finally, 10 min at 72°C as a final extension. The CCHFV genome extracted from an already confirmed RT-PCR-positive serum was used as the positive control, and no template control (NTC) and no template (NC) were used as the negative controls in RT-PCR tests (Chinikar et al. 2004, 2008). Those RT-PCR tests with a 536bp band that do not have the band in both NTC and NC are considered positive and in case of absence, they are evaluated as negative. Otherwise, tests would be failed because of the existence of the band in negative controls.
Sequencing and genetic analysis
PCR products were purified by using quick-spin PCR purification kit (Intron) and directly underwent sequencing by specific primers. For phylogenetic analysis, a 536 bp fragment of the S-segment was used. The sequences of CCHFVs were analyzed by a neighbor-joining method with Kimura two parameter distances by using Mega 4 software. Bootstrap confidence limits were based on 500 replicates. Evolutionary divergence, distance matrix, and subsequently sets of phylogenetic trees were calculated by the software (Tamura et al. 2007).
Results
On the basis of the epidemiological data obtained from the questionnaire, 100 individuals with high-risk professions who had been already exposed to the blood and the tissues of livestock and also had a history of knife-cut hand during their works were selected. Among them, 12 individuals tested IgG positive but none of them with fever history or the other clinical symptoms characteristic of CCHF disease. All seropositive cases originated from regions in the vicinity of the Isfahan province (Table 1). In addition to human samples, livestock sera were collected from different parts of high-risk regions in Isfahan including Fereydonshahr, Zarrinshahr, Semirom, and Varzaneh. In this seroepidemiological survey on livestock, 41 out of 322 livestock sera tested IgG positive, which comprised 36 sheep, 2 goats, and 3 cows.
Table 1.
IgG-Positive Human Serum Samples
| Location | Profession | Age | Human serum code | No. |
|---|---|---|---|---|
| Varzaneh | Farmer | 78 | 1VH1 | 1 |
| Fasaran | Slaughterer | 40 | HA7 | 2 |
| Fasaran | Slaughterer | 24 | HA26 | 3 |
| Fasaran | Slaughterer | 53 | HA29 | 4 |
| Fasaran | Slaughterer | 22 | HA40 | 5 |
| Fasaran | Technician | 28 | HA42 | 6 |
| Zarrinshahr | Slaughterer | 39 | HA65 | 7 |
| Zarrinshahr | Slaughterer | 34 | HA69 | 8 |
| Zarrinshahr | Farmer | 26 | HA76 | 9 |
| Zarrinshahr | Butcher | 40 | HA86 | 10 |
| Zarrinshahr | Butcher | 38 | HA92 | 11 |
| Semirom | Farmer | 60 | SH9 | 12 |
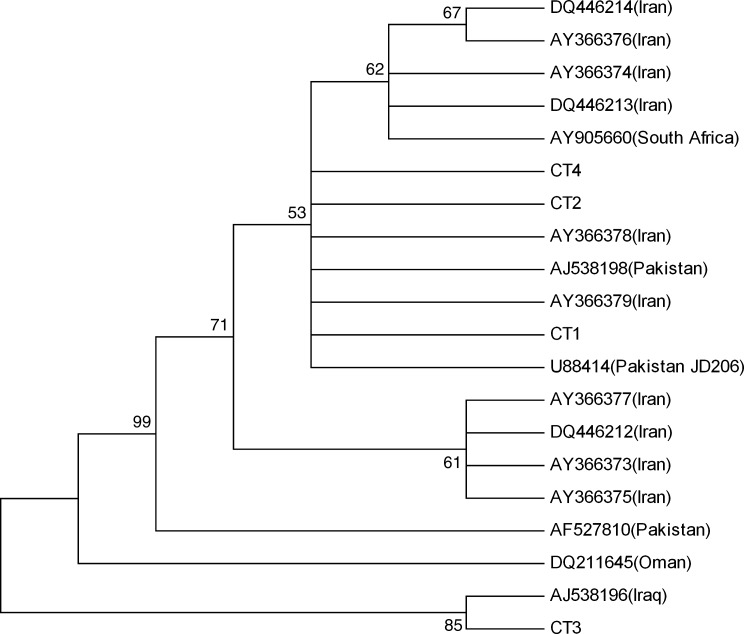
Soft and hard ticks were also collected in the survey and analyzed for the presence of the viral genome. Single PCR bands corresponding to the expected 536 bp product of the S-segment were detected in 4 of 44 analyzed hard ticks, but all the 15 soft ticks were found negative. The entire hard ticks positive for CCHFV genome were Rhipicephalus bursa, and they were collected only in the Fereydonshahr region (Table 2). The PCR products, CT1, CT2, CT3, and CT4, were sequenced and analyzed. Comparative analysis of the sequences from the collected hard ticks showed a close relationship among them except for one. Nucleotide sequence diversities ranged from 0.2 to 5.3%. Moreover, pairwise comparisons of S-segment nucleotide sequences detected in the hard ticks and representative sequences from different parts of the world demonstrated that CT1, CT2, and CT4 isolates were very closely related to S-segment sequences of Iran and Pakistan in GenBank. As illustrated in Figure 1, phylogenetic analysis using S-segment sequences of CCHFV implied that CT1, CT2, and CT4 isolates formed a distinct lineage together with Iran and Pakistan sequences, whereas CT3 isolate was clustered with Iraq strain (AJ538196).
Table 2.
Crimean-Congo Hemorrhagic Fever Virus Genome-Positive Ticks
| Location | Host | Sex | Tick Species | Sequence code |
|---|---|---|---|---|
| Fereydonshahr | Sheep | Female | Rhipicephalus bursa | CT1 |
| Fereydonshahr | Sheep | Male | R. bursa | CT2 |
| Fereydonshahr | Goat | Female | R. bursa | CT3 |
| Fereydonshahr | Goat | Female | R. bursa | CT4 |
FIG. 1.
Phylogenetic tree (neighbor-joining) calculated for Crimean-Congo hemorrhagic fever virus S-segment sequences. New sequences in this paper (CT1, CT2, CT3, and CT4) and previously described sequences from Iran (AY 366373-8, DQ446212-14), Oman (DQ211645), Pakistan (AJ538198, U88414, and AF527810), South Africa (AY905660), and Iraq (AJ538196) are shown.
Discussion
Serological and molecular epidemiology survey will certainly help in determining CCHFV in the Isfahan country. For instance, in endemic areas, assessment of livestock antibodies against CCHFV appears to be one of the best indicators of CCHF risk to humans. During the last decade, an increasing number of human CCHFV infections have been reported from various regions of Iran (Chinikar et al. 2005). Most of the cases have been reported to occur in the south-Eastern province of Sistan-va-Baluchistan, in areas close to the border of Afghanistan and Pakistan. Second most cases were reported from the province of Isfahan, located in the central part of Iran (Chinikar et al. 2004). The reason for the increasing number of human CCHFV infections in Isfahan during the last decade remains obscure, especially as infections were rarely reported in Iran before 1999. The occurrence of human CCHFV infections in Isfahan is newly recognized, whereas virus circulation in ticks and in livestock may have been present and unrecognized for many years. An early seroepidemiological survey in northern/central parts of the country was carried out in 1974, thus showing the presence of virus in local livestock (Saidi et al. 1975). With regard to a study conducted in 2003–2004 in the Sistan-va-Baluchistan province, among 285 human volunteers, 6.3% appeared seropositive for CCHF infection. A serosurvey among livestock in the Isfahan province between 2004 and 2005 showed seropositivity in almost 56% (unpublished data). During the years 2003 to 2005, of the 448 livestock sera collected from the Khorasan province, northeast part of Iran, 77.5% of 298 sheep samples and 46% of 150 goat samples were positive, thus implying that this region is hyper enzootic for CCHF.
As represented in Table 3, 12% of collected human sera and 12.7% of livestock sera from different parts of the Isfahan province were tested seropositive, and the CCHFV genome was detected in 9.1% of collected ticks. The percentage of the seropositive vertebrate hosts, human and livestock, and CCHF infectivity of the ticks indicated that CCHFV is effectively established in this region. As summarized in the Table 3, by comparing the sampled sera from slaughtering houses in Fasaran, Zarrinshar and Khomeinishahr, the percentage of IgG-positive human cases was higher than that of IgG-positive livestock cases. It is still unclear in evidence as to when and how seropositive humans have been infected without any course of disease and clinical symptoms; so, further study needs to be done to show protection in the high-risk professionals in the country. Of note, most slaughtered livestock had been brought from the Sistan-va-Baluchistan province, the most infected province of CCHF in Iran.
Table 3.
Comparison Results of Molecular and Serological Survey on the Collected Samples
| Location | Human IgG + (%) | Livestock IgG+ (%) | Tick RT-PCR+ (n) | High-risk profession |
|---|---|---|---|---|
| Varzaneh | 8.3 | 29.3 | — | Husbandries |
| Fasaran | 41.7 | — | — | Slaughter house |
| Zarrinshahr | 41.7 | 7.3 | — | Slaughter house |
| Semirom | 8.3 | 19.5 | — | Husbandries |
| Fereydonshahr | — | 36.6 | 4 | Husbandries |
| Khomeinishahr | — | 7.3 | — | Slaughter house |
The hard ticks collected In the study were Rhipicephalus bursa and Hyalomma detritum, whereas the soft ticks were Ornithodoros lahorensis and Ornithodoros canistrini. Although Hyalomma ticks are considered the most important vector and reservoir in the epidemiology of CCHFV, the virus has also been reported from other genera of ticks. In 1979, CCHF was first isolated in Iran from Ornithodoros lahorensis (Sureau et al. 1980). The year 2004 showed CCHFV infection in 22.8% of soft ticks in the Chaharmahal Bakhtiaryi province, south east of the Hamadan province (Shirani et al. 2004). In another study in the Ardabil province, North West of Hamadan, 33.3% of the ticks including Hyalomma sp., Rhipicephalus sp., and Ornithodoros sp. were infected by CCHFV (Telmadarraiy et al. 2010). A survey showed CCHFV infection in 25.5% of ticks including Hyalomma, Dmarginatus, and Ornithodoros species (Tahmasebi et al. 2010). In another study in Bahar (Central part of Hamadan Province), 11.3% of ticks including Hyalomma marginatum, Hyalomma dromedarii, Hyalomma anatolicum, R. bursa, Rhipicephalus sanguineus, and Hyalomma pucntata were CCHFV positive (Moradi et al. 2008).
In this molecular survey, all CCHFV-positive ticks were collected from the Fereydonshahr region, whereas those collected from the other regions tested negative. Moreover, a high percentage of seropositive livestock from this region indicated CCHFV circulation in livestock populations. Also, it is essential to note that there is a report of a death case of CCHF from Fereydonshahr in 2007, verifying already established circulation of the CCHFV in the region (unpublished data from National Reference Laboratory of Arboviruses in Pasteur Institute of Iran).
Pairwise S-segment sequence comparisons with a representative set of CCHFV sequences from Iran and Asia1 countries clade demonstrated that the isolates CT1, CT2, and CT4 had been closely related to Pakistani and some South African strains, whereas the isolate CT3 clustered with Iraqi strain (accession number AJ538196) (Tahmasebi et al. 2010). Phylogenetic analysis could further strengthen this close relationship, thus placing the isolates CT1, CT2, and CT4 and previously characterized Iranian sequences and Pakistani sequences within the same phylogenetic clade (Fig. 1). The data currently available on the geographical distribution, genetic diversity, and prevalence of CCHFV in the Isfahan province are, however, very limited. Before our study, CCHFV genome S-segment sequences obtained from Iranian patients has been genetically characterized and very close to Matin strain (Pakistan strain), whereas only one Iranian strain (ArTeh 193-3) obtained from ticks in 1978 by Sureau et al. was very similar to the senegal strain (Chinikar et al. 2004). Further, the available serum and the extracted genome of ticks will be addressed for analysis of whole CCHFV genome sequencings and probably virus isolation in biosafety level 4 containments. The results indicated that at least three genetic lineages of CCHFV are circulating in Iran. Also, the increase in human infections during the last decade, especially in Sistan-va-Baluchistan and Isfahan as well as the other provinces of Iran, might be due to the new introduction of CCHFV into Iran through its eastern and western borders, possibly through infected ticks and/or livestock from Afghanistan, Pakistan, and Iraq.
Acknowledgments
This study is granted by project number 381 of Pasteur Institute of Iran. The authors appreciate Dr. P. Adibi and the staff of Infectious Diseases and Tropical Medicine Research Center of Isfahan University of Medical Sciences, and Dr. S.A. Husseini and the other staff of Isfahan Veterinary Organization for their supportive assistance. They are also grateful to the members of Parasitology department and Arboviruses and Viral Hemorrhagic Fevers Laboratory (Nat. Ref. Lab) of Pasteur Institute of Iran for their technical support.
Disclosure Statement
The authors declare that they have no conflicts of interest.
References
- Begum F. Wisseman CL., Jr. Casals J. Tick-borne viruses of West Pakistan-II- Hazara virus, a new agent isolated from Ixodes redikorzevi ticks from the Kaghan Valley, W. Pakistan. Am J Epidemiol. 1970;92:192–194. doi: 10.1093/oxfordjournals.aje.a121197. [DOI] [PubMed] [Google Scholar]
- Camicas JL. Cornet JP. Gonzalez JP. Wilson ML, et al. Crimean-Congo hemorrhagic fever in Senegal. Latest data on the ecology of the CCHF virus. Bull Soc Pathol Exot. 1994;87:11–16. [PubMed] [Google Scholar]
- Chinikar S. Goya MM. Shirzadi MR. Ghiasi SM, et al. Surveillance & Laboratory Detection System of Crimean–Congo Hemorrhagic Fever (CCHF) in Iran. Transbound Emerg Dis. 2008;55:200–204. doi: 10.1111/j.1865-1682.2008.01028.x. [DOI] [PubMed] [Google Scholar]
- Chinikar S. Mirahmadi R. Mazaheri V. Nabeth P, et al. A serological survey in suspected human patients of Crimean-Congo hemorrhagic fever in Iran by determination of IgM specific ELISA method during 2000–2004. Arch Ir Med. 2005;8:52–55. [Google Scholar]
- Chinikar S. Persson SM. Johansson M. Bladh L, et al. Genetic analysis of Crimean- Congo Hemorrhagic Fever virus in Iran. J Med Virol. 2004;73:404–411. doi: 10.1002/jmv.20106. [DOI] [PubMed] [Google Scholar]
- Donets MA. Chumakov MP. Korolev MB. Rubin SG. Physicochemical characteristics, morphology and morphogenesis of virions of the causative agent of Crimean hemorrhagic fever. Interviology. 1977;8:294–308. doi: 10.1159/000148904. [DOI] [PubMed] [Google Scholar]
- Drosten C. Minnak D. Emmerich P. Schmitz H. Reinicke T. Crimean-Congo hemorrhagic fever in Kosovo. J Clin Microbiol. 2002;40:1122–1123. doi: 10.1128/JCM.40.3.1122-1123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergonul O. Crimean-Congo hemorrhagic fever. Lancet Infect Dis. 2006;6:203–214. doi: 10.1016/S1473-3099(06)70435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia S. Chinikar S. Coudrier D. Billecocq A, et al. Evaluation of a Crimean-Congo hemorrhagic fever virus recombinant antigen expressed by Semliki Forest Suicide virus for IgM and IgG antibody detection in human and animal sera collected in Iran. J Clin Virol. 2006;35:154–159. doi: 10.1016/j.jcv.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Gonzalez JP. Camicas JL. Cornet JP. Faye O. Wilson ML. Sexual and transovarian transmission of Crimean-Congo hemorrhagic fever virus in Hyalomma truncatum ticks. Res Virol. 1992;143:23–28. doi: 10.1016/s0923-2516(06)80073-7. [DOI] [PubMed] [Google Scholar]
- Hoogstraal H. The epidemiology of tick-born Crimean-Congo hemorrhagic fever in Asia, Europe, and Africa. J Med Entomol. 1979;15:307–417. doi: 10.1093/jmedent/15.4.307. [DOI] [PubMed] [Google Scholar]
- Logan TM. Linthicum KJ. Bailey CL. Watts DM. Moulton JR. Experimental transmission of Crimean-Congo hemorrhagic fever virus by Hyalomma truncatum Koch. Am J Trop Med Hyg. 1989;40:207–212. doi: 10.4269/ajtmh.1989.40.207. [DOI] [PubMed] [Google Scholar]
- Martin ML. Lindsey-Regnery H. Sasso DR. McCormick JB. Palmer E. Distinction between Bunyaviridae genera by surface structure and comparison with Hantaan Virus using negative stain electron microscopy. Arch Virol. 1985;86:17–28. doi: 10.1007/BF01314110. [DOI] [PubMed] [Google Scholar]
- Moradi AR. Chinikar S. Oshaghi MA. Vatandoost H, et al. Molecular detection of Crimean–Congo Hemorrhagic Fever (CCHF) virus in Ticks (Ixodidae, Argasidae) of Hamedan Province, Iran. Biochem Cell Arch. 2008;8:119–123. [Google Scholar]
- Papa A. Bino S. Velo E. Harxhi A, et al. Cytokine levels in Crimean-Congo hemorrhagic fever. J Clin Virol. 2006;36:272–276. doi: 10.1016/j.jcv.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Randolph SE. Rogers DJ. Ecology of tick-borne disease and the role of climate. In: Ergonul O, editor; Whitehouse CA, editor. Crimean Congo Hemorrhagic Fever—A Global Perspective. The Netherlands: Springer; 2007. pp. 167–186. [Google Scholar]
- Saidi S. Casals J. Faghih MA. Crimean hemorrhagic fever-Congo (CHF-C) virus antibodies in man and domestic and small mammals in Iran. Am J Trop Med Hyg. 1975;24:353–357. doi: 10.4269/ajtmh.1975.24.353. [DOI] [PubMed] [Google Scholar]
- Shirani M. Asmar M. Chinikar S. Mirahmadi R, et al. Detection of CCHF virus in soft ticks (Argasidae) by RT-PCR method. J Infect Dis Trop Med. 2004;9:11–15. [Google Scholar]
- Suleiman MN. Muscat-Baron JM. Harries JA. Satti AG, et al. Congo-Crimean hemorrhagic fever in Dubai. An outbreak at the Rashid hospital. Lancet. 1980;2:939–941. [PubMed] [Google Scholar]
- Sureau P. Klein JM. Casals J. Digoutte JOP, et al. Isolation of Thogoto, Wad medani, Wanowrie, and Crimean-Congo hemorrhagic fever viruses from ticks of domestic animals in Iran. Ann Virol (Inst Pasteur) 1980;131E:185–200. [Google Scholar]
- Swanepoel R. Gill DE. Shepherd AJ. Leman PA, et al. The clinical pathology of Crimean-Congo hemorrhagic fever. Rev Infect Dis. 1989;11(Suppl 4):S794–S800. doi: 10.1093/clinids/11.supplement_4.s794. [DOI] [PubMed] [Google Scholar]
- Swanepoel R. Shepherd AJ. Leman PA. Shepherd SP, et al. Epidemiologic and clinical features of Crimean-Congo hemorrhagic fever in southern Africa. Am J Trop Med Hyg. 1987;36:120–132. doi: 10.4269/ajtmh.1987.36.120. [DOI] [PubMed] [Google Scholar]
- Swanepoel R. Nairovirus infections. In: Porterfield JS, editor. Exotic Viral Infections. London: Chapman and Hall; 1995. pp. 285–293. [Google Scholar]
- Tahmasebi F. Ghiasi SM. Mostafavi E. Moradi M, et al. Molecular epidemiology of Crimean-Congo hemorrhagic fever virus genome isolated from ticks of Hamadan province of Iran. J Vector Borne Dis. 2010;47:211–216. [PubMed] [Google Scholar]
- Tamura K. Dudley J. Nei M. Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Telmadarraiy Z. Ghiasi SM. Moradi M. Vatandoost H, et al. A survey of Crimean-Congo haemorrhagic fever in livestock and ticks in Ardabil Province, Iran during 2004–2005. Scand J Infect Dis. 2010;42:137–141. doi: 10.3109/00365540903362501. [DOI] [PubMed] [Google Scholar]
- van Eeden PJ. van Eeden SF. Joubert JR. King JB, et al. A nosocomial outbreak of Crimean-Congo haemorrhagic fever at Tygerberg Hospital. Part II. Management of patients. S Afr Med J. 1985;68:718–721. [PubMed] [Google Scholar]
- Watts DM. Ksiazek TG. Linthicum KJ. Hoogstraal H. Crimean-Congo hemorrhagic fever. In: Monath TP, editor. The Arboviruses: Epidemiology and Ecology. Vol. 2. Boca Raton, FL: CRC Press; 1988. pp. 177–260. [Google Scholar]
- Whitehouse CA. Crimean-Congo hemorrhagic fever. Antiviral Res. 2004;64:145–160. doi: 10.1016/j.antiviral.2004.08.001. [DOI] [PubMed] [Google Scholar]