Abstract
Crimean-Congo hemorrhagic fever (CCHF) is an important zoonotic viral disease that is asymptomatic in infected livestock, but poses a serious threat to humans. The high fatality rate may be due to phylogenetic variations in the virus, transmission routes, and a lack of an efficient surveillance system for the disease. The geographical features of the eastern and southeastern borders of Turkey may facilitate transmission of viruses between countries of the region. Therefore in this study we focused on the genetic relationship between Turkish and Iranian CCHF viruses based on their S-segment sequences. The research was performed on a total of 104 blood samples from small ruminants reared in southwest Iran. The results of phylogenetic analysis showed that Iranian CCHF virus isolates were closely related to human-originating Turkish Group II viruses from a European lineage reported previously.
Key Words: Crimean-Congo hemorrhagic fever, Iran, Phylogenetic analysis, S-segment, Turkey
Introduction
Crimean-Congo hemorrhagic fever (CCHF) is a zoonotic viral disease that poses a serious threat to humans, in contrast to its asymptomatic course in infected livestock. Human infections begin with nonspecific febrile symptoms, but progress to a serious hemorrhagic syndrome with a high case fatality rate of 2–50%. Although the causative virus is often transmitted by ticks, animal-to-human and human-to-human transmission also occurs. The disease is one of the most widely distributed viral hemorrhagic fevers occurring in Africa, the Middle East, Asia, and some parts of Europe (Chinikar et al. 2010a).
It is well documented that infections can travel long distances with infected individuals or via virus-carrying infected vectors. Infections may cross borders by regular or uncontrolled movement of infected persons, resulting in the emergence or recurrence of new strains or variants of viruses. The geographical features of the eastern borders of Turkey may facilitate uncontrolled and/or illegal animal movements between countries of the region (Fig. 1). It is well known that Turkey is a target in the region for the trade of uncontrolled small ruminants by its neighbors. For this reason, numerous important animal diseases are reported there each year. Economic analyses recently showed that approximately 50% of the meat consumed in Turkey is from illegally-imported animals (Anonymous, 2010). Like many other countries, Turkey has been suffering from these infectious diseases, particularly in animals. Foot-and-mouth disease (FMD) and peste des petits ruminants (PPR) are two of the most important viral diseases, and they cause several million dollars of economic losses each year in Turkey. In a previous study, FMD virus serotypes detected in Turkey were shown to originate from Afghanistan and get into Turkey through Iran (Klein et al. 2006), and probably spread to Bulgaria by the animal trade (Murray et al. 2011). Another serious instance of uncontrolled animal movement causing livestock problems is the detection of Asian lineage (IV) PPR virus in northeastern Africa, which threatens continental Europe (Kwiatek et al. 2011).
FIG. 1.
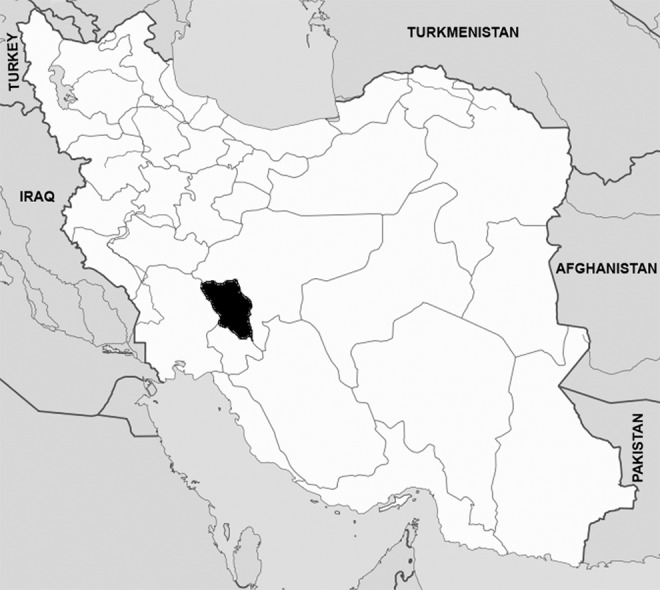
Map of Iran showing the location of Chaharmahal-va-Bakhtiari province (the black area in southwestern Iran), where the small ruminant samples were collected.
Crimean-Congo hemorrhagic fever (CCHF) has existed for many years in northwest Iran, in Ardebil, and in eastern and western Azerbaijan provinces under the local name “Kara-Mikh typhoid fever.” Numerous investigations beginning in the 1970s have reported the presence of CCHF in Iran (Hoogstraal, 1979; Sureau et al. 1980; Chinikar et al. 2005, 2008).
The epidemiology and phylogeny of the virus have been investigated in multiple reports, but these have generally concentrated on the phylogenetic relationships among sequences from a single segment, or have studied all three segments but with a limited number of sequences, or have concentrated on a specific region. Most recently, Han and Rayner (2011) collected all publicly-available full-length S segments for CCHF virus and projected phylogenetic trees based on the nucleotide alignments. Their trees predicted several major clades that were consistent with previous findings, and which supported geographical subdivision. Each of the trees exhibit clear geographic subdivisions, identifying seven clades that were named Asia 1, Asia 2, Europe 1, Europe 2, Africa 1, Africa 2, and Africa 3, and which are consistent with results from previous studies (Han and Rayner 2011). In another report, the global occurrence of CCHFV genotypes was described based on migration events between neighboring geographic areas, and this hypothesis was proved by analysis of tree topology of the maximum-likelihood phylogenies (Mild et al. 2010). These reports suggested that the most likely panmixia occurred in genotype Asia 1 for viruses reported from Iran, Pakistan, Oman, and Iraq. The clade Asia 2 includes viruses reported from China, Tajikistan, and Uzbekistan (Mild et al. 2010; Han and Rayner 2011). The Asian genotype was not reported from Turkey, and the European genotype was not reported from Iran in these articles.
In this article, we highlight the possible migration of CCHF virus between Iran and Turkey, where there have been recent increases in disease occurrence. Here we describe the phylogenetic relationships between CCHF viruses detected recently in Iranian sheep and goats, based on partial nucleotide sequences of the S-segments.
Materials and Methods
A total of 104 blood samples were collected from small ruminants reared in Chaharmal-Va-Bakhtiari province in southwest Iran (Fig. 1). Eleven of the samples were taken from goats and the rest were from sheep. The ages of the sampled animals varied between 5 months and 6 years in goats, and 3 months and 6 years in sheep. The majority of the animals were female. Viral RNA was isolated from serum and/or plasma samples using a viral RNA mini kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions, and RNA samples were immediately transferred the laboratories of the Virology Department of the Faculty of Veterinary Medicine, Ankara University, which performed all molecular virological studies. Synthesis of individual cDNAs, partial amplification of S-segments, and subsequent sequence analysis in positive samples, were carried out as described previously (Ozkaya et al. 2010). The S-segment sequences obtained from Iranian CCHF viruses were submitted to GenBank, and were used for phylogenetic evaluation in comparison to regional and global CCHF virus S-segment sequences that were also acquired from GenBank. The evolutionary history of the Turkish and Iran isolates, based on regional sequence comparisons, was inferred using the neighbor-joining (NJ) method (Saitou and Nei 1987).The evolutionary distances were computed using the maximum composite likelihood method (Tamura et al. 2004). Phylogenetic analyses were conducted using MEGA4 (Tamura et al. 2007).
Results
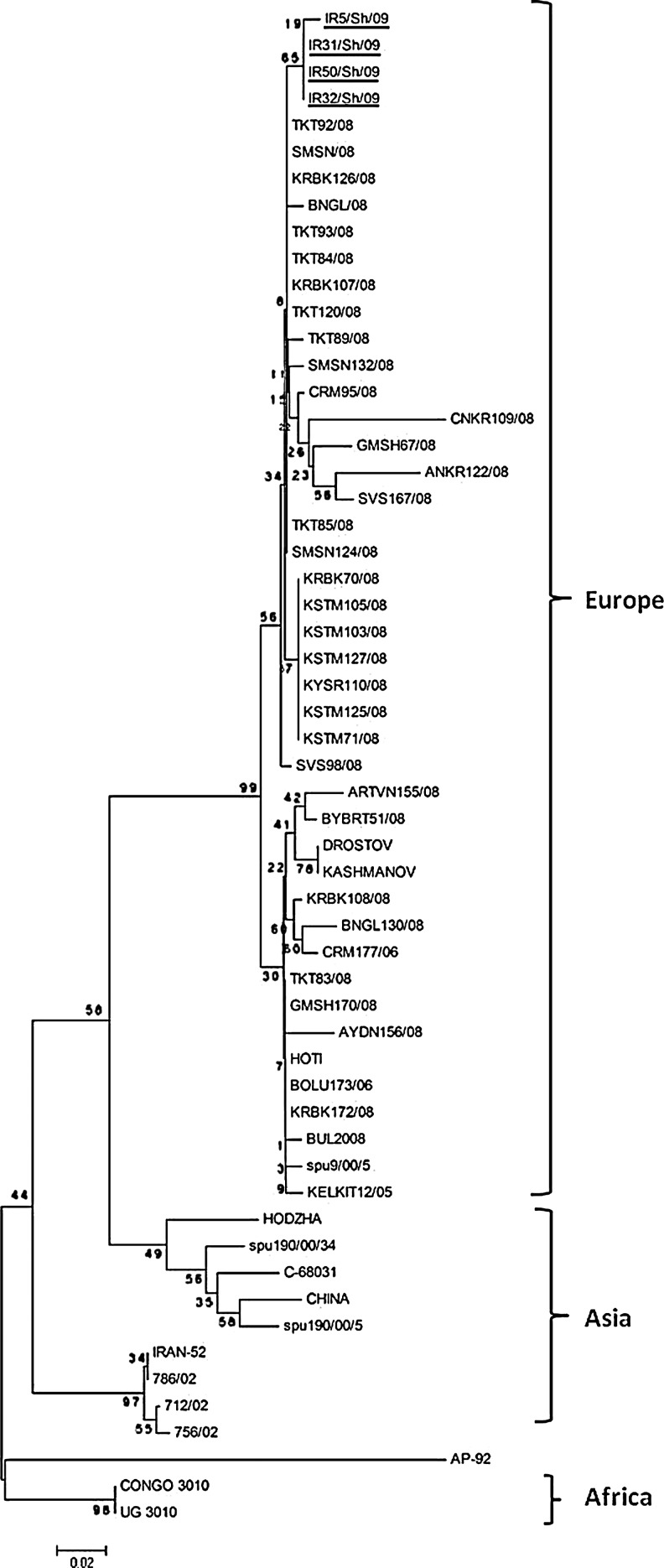
Four sheep were found to be positive for CCHF virus RNA, while all of the goats were negative. All viremic sheep were female. To investigate the genetic relationships of the CCHF virus strains circulating between Iran and Turkey, amplicons (536 nt long) obtained from four PCR positive samples were successfully sequenced and their accession numbers were assigned as HM853679, HM853680, HM853681, and HM853682 for CCHF virus Iranian isolates IR5/Sh/09, IR31/Sh/09, IR32/Sh/09, and IR50/Sh/09, respectively. The nucleotide identity of Iranian CCHF virus S-segment sequences was found to be between 98.4% and 100%. The NJ analysis of S-segment sequences of Iranian CCHF virus isolates showed that viruses from Iranian animals were closely related to previously reported Turkish Group II viruses (local topotype), belonging to a European lineage.
Discussion
Our findings show that the livestock sequences we studied may be new variants, which are related to a European lineage of viruses, and did not show any similarities to the previously-sequenced human-originating Iranian samples of Asian lineage. This finding is in accord with those of a previously published study (Chinikar et al. 2008), indicating that a new variant may be circulating in Iran. To understand the migration patterns of CCHF virus between countries, phylogenetic and statistical studies have recently been published (Hewson et al. 2004; Deyde et al. 2006; Mild et al. 2010; Chinikar et al. 2010b). In one of those studies, Turkey was proposed to be a potential source for CCHF throughout Europe (genotype 4) in the west, and Iran in the east (Mild et al. 2010). The results of our study also showed that Iranian CCHF viruses of animal origin were found to be closely related to the Turkish CCHF viruses of human origin in Group II reported previously (Ozkaya et al. 2010). On the other hand, as illustrated in Figure 2, the underlined sequences obtained in this study were not located together with other human-originating Iranian CCHF viruses detected in 2002 and 2005, which have an Asian lineage. This finding might indicate that the virus could be imported into Turkey via trans-border animal movements, as there are many uncontrolled animal movements into Turkey via its eastern borders. East and southeast Iran, where Sistan and Baluchistan provinces are located, have many cases of suspected, confirmed, and reported cases of CCHF (Izadi et al. 2004). This region borders Afghanistan and Pakistan, where CCHF is endemic, and there are nomadic populations there who traditionally eat sheep and goat meat (Alavi-Naini et al. 2006). In addition, sheep and goats that are reared in Chaharmal-Va-Bakhtiari do not move into northwest regions, although many annually move south (to Fars and Khuzestan provinces) during the cold season.
FIG. 2.
Phylogenetic analysis of CCHF virus isolates (underlined) from the Chaharmahal-va-Bakhtiari province of southwest Iran. The GeneBank accession numbers for the nucleotide sequences of the CCHF viruses used in the phylogenetic analysis were given in a previous report (Ozkaya et al. 2010). Bootstrap values are given at the nodes of the internal branches. The bar indicates 0.02 nucleotide substitutions per site.
The results of our study also suggest that livestock traditionally reared between the Turkey, Iran, and Iraq borders, might serve as a biological mixer for near and far eastern CCHF virus strains, and these animals may facilitate the spread of viruses west or east. These imported, or migrating, strains might also have caused changes in the gene pools in Turkey, resulting in the occurrence of a local topotype (Ozkaya et al. 2010).
In conclusion, CCHF is endemic in Iran, and also in neighboring Turkey, which is closer to Europe. Thus precautionary measures and control programs for CCHF in both countries could be beneficial to the eastern part of continental Europe. These findings will help to determine the virulence factors and pathogenesis of local CCHF viruses at the molecular level. Consequently, understanding the genetic relationships among CCHF viral strains from different regions will help in the development of molecular diagnostic strategies for vaccine development and control of disease. Although our results agree with those of others (Mild et al. 2010), which indicate that CCHF virus is spreading between neighboring countries, analysis of these patterns and estimates of the rates of spread require more biological exploration.
Acknowledgment
This study was supported in part by grants from the Research Fund of Shahrekord University and the State Planning Organization (project no. 2011K120220), Prime Ministry of the Turkish Republic.
Author Disclosure Statement
No competing financial interests exist.
References
- Alavi-Naini R. Moghtaderi A. Koohpayeh HR, et al. Crimean-Congo hemorrhagic fever in southeast of Iran. J Infect. 2006;52:378–382. doi: 10.1016/j.jinf.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Anonymous; Ankara Trade Chamber (ATO) Livestock report. 2010. http://www.atonet.org.tr/turkce/bulten/bulten.php3?sira=524. [Aug 14;2011 ]. http://www.atonet.org.tr/turkce/bulten/bulten.php3?sira=524
- Chinikar S. Ghiasi SM. Hewson R, et al. Crimean-Congo hemorrhagic fever in Iran and neighboring countries. J Clin Virol. 2010a;47:110–114. doi: 10.1016/j.jcv.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Chinikar S. Goya MM. Shirzadi MR, et al. Surveillance and laboratory detection system of Crimean-Congo haemorrhagic fever in Iran. Transbound Emerg Dis. 2008;55:200–204. doi: 10.1111/j.1865-1682.2008.01028.x. [DOI] [PubMed] [Google Scholar]
- Chinikar S. Mirahmadi R. Mazaheri V, et al. A serological survey in suspected human patients of Crimean-Congo hemorrhagic fever in Iran by determination of IGM specific ELISA method during 2000–2004. Arch Ir Med. 2005:52–55. [Google Scholar]
- Chinikar S. Mojtaba Ghiasi S. Moradi M, et al. Phylogenetic analysis in a recent controlled outbreak of Crimean-Congo haemorrhagic fever in the south of Iran, December (2008) Euro Surveill. 2010b;15:47. doi: 10.2807/ese.15.47.19720-en. [DOI] [PubMed] [Google Scholar]
- Chinikar S. Persson SM. Johansson M, et al. Genetic analysis of Crimean-Congo hemorrhagic fever virus in Iran. J Med Virol. 2004;73:404–411. doi: 10.1002/jmv.20106. [DOI] [PubMed] [Google Scholar]
- Deyde V. Khristova M. Rollin ML, et al. Crimean-Congo hemorrhagic fever virus genomics and global diversity. J Virol. 2006;80:8834–8842. doi: 10.1128/JVI.00752-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han N. Rayner S. Epidemiology and mutational analysis of global strains of Crimean-Congo haemorrhagic fever virus. Virologica Sinica. 2011;26:229–244. doi: 10.1007/s12250-011-3211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson R. Chamberlain J. Mioulet V, et al. Crimean-Congo hemorrhagic fever virus: sequence analysis of the small RNA segments from a collection of viruses world wide. Virus Res. 2004;102:185–189. doi: 10.1016/j.virusres.2003.12.035. [DOI] [PubMed] [Google Scholar]
- Hoogstraal H. The epidemiology of tick-borne Crimean-Congo hemorrhagic fever in Asia, Europe, and Africa. J Med Entomol. 1979;15:307–417. doi: 10.1093/jmedent/15.4.307. [DOI] [PubMed] [Google Scholar]
- Izadi S. Naieni K. Madjdzadeh SR, et al. Crimean-Congo hemorrhagic fever in Sistan and Baluchestan Province of Iran, a case-control study on epidemiological characteristics. J Inf Dis. 2004;8:299–306. doi: 10.1016/j.ijid.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Klein J. Parlak U. Ozyoruk F, et al. The molecular epidemiology of foot-and-mouth disease virus serotypes A and O from 1998 to 2004 in Turkey. BMC Vet Res. 2006;2:35–48. doi: 10.1186/1746-6148-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatek O. Ali YH. Saeed IK, et al. Asian lineage of peste des petits ruminants virus, Africa. EID. 2011;17:1223–1231. doi: 10.3201/eid1707.101216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mild M. Simon M. Albert J, et al. Towards an understanding of the migration of Crimean-Congo hemorrhagic fever virus. J Gen Virol. 2010;91:199–207. doi: 10.1099/vir.0.014878-0. [DOI] [PubMed] [Google Scholar]
- Murray T. Carbon M. Hartley M, et al. Update on foot and mouth disease in wild boar in Bulgaria. Preliminary Outbreak Assessment. Jan, 2011. http://archive.defra.gov.uk/foodfarm/farmanimal/diseases/monitoring/documents/fmd-bulgaria110120.pdf. [Nov 4;2011 ]. http://archive.defra.gov.uk/foodfarm/farmanimal/diseases/monitoring/documents/fmd-bulgaria110120.pdf
- Ozkaya E. Dincer E. Carhan A, et al. Molecular epidemiology of Crimean-Congo hemorrhagic fever virus in Turkey: Occurrence of local topotype. Virus Res. 2010;149:64–70. doi: 10.1016/j.virusres.2009.12.014. [DOI] [PubMed] [Google Scholar]
- Saitou N. Nei N. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sureau P. Klein JM. Casals J, et al. Isolation of Thogoto, Wad Medani, Wanowrie, and Crimean-Congo hemorrhagic fever viruses from ticks of domestic animals in Iran. Ann Virol (Inst Pasteur) 1980;131:185–200. [Google Scholar]
- Tamura K. Nei M. Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Nat Acad Sci USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. Dudley J. Nei M, et al. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]